Abstract
The c-Myc oncoprotein functions as a transcription factor that can transform normal cells into tumor cells, as well as playing a direct role in normal cell proliferation. The c-Myc protein transactivates cellular promoters by recruiting nuclear cofactors to chromosomal sites through an N-terminal transactivation domain. We have previously reported the identification and functional characterization of four different c-Myc cofactors: TRRAP, hGCN5, TIP49, and TIP48. Here we present the identification and characterization of the actin-related protein BAF53 as a c-Myc-interacting nuclear cofactor that forms distinct nuclear complexes. In addition to the human SWI/SNF-related BAF complex, BAF53 forms a complex with TIP49 and TIP48 and a separate biochemically distinct complex containing TRRAP and a histone acetyltransferase which does not contain TIP60. Using deletion mutants of BAF53, we show that BAF53 is critical for c-Myc oncogenic activity. Our results indicate that BAF53 plays a functional role in c-Myc-interacting nuclear complexes.
Cell cycle progression is normally a carefully regulated succession of growth and genome replication steps that is monitored by different checkpoint pathways. Cancer arises through the accumulation of mutations that disrupt the normal balance of growth-promoting or -suppressive signals and the different checkpoints. Transcription factors ultimately mediate most of these signals, either as the direct sites of mutation in tumor cells or as downstream effectors of signaling or checkpoint pathways. The Myc transcription factor family is among the most frequently disrupted networks in human and animal cancers (15). Myc plays a direct role in G1-to-S progression by regulating sets of target genes required for growth and DNA replication, although the precise set of genes involved in various steps remains poorly defined. Myc can also induce programmed cell death (apoptosis), which may oppose the outgrowth of cancer cells that suffer mutations in this network (27).
The biological activities of the c-Myc protein have been dissected primarily through mutational analysis. Disruption of the C-terminal DNA binding domain destroys both oncogenic and apoptotic activities, establishing a requirement for the recognition of chromosomal sites (2, 32). Beyond DNA binding, the function of transcription factors is to recruit other proteins to specific sites to modulate transcription and/or alter chromatin structure. Consistent with this role, the N-terminal portion of c-Myc interacts with a number of nuclear factors (28). Important insight into the functional significance of individual cofactor interactions is provided by mutational analysis of the c-Myc N-terminal domain. A number of studies have identified an evolutionarily conserved sequence called Myc homology box II (MBII) as essential for oncogenic and apoptotic activities, as well as for blocking of differentiation and stimulation of cell proliferation (7, 9, 19, 25, 32). Studies centered on understanding the function of MBII led to the purification of TRRAP, a 430-kDa nuclear protein with homology to the ATM/phosphatidylinositol 3-kinase family (21). Inhibition of TRRAP synthesis or function blocks Myc-mediated oncogenesis, establishing an essential role for TRRAP in c-Myc activity (21). The identification of TRRAP provided a key mechanistic link to c-Myc function when it was found that TRRAP was a component of the SAGA chromatin-modifying complex in both yeast and mammalian cells (12, 22, 29, 33). The yeast ortholog of TRRAP (Tra1p) is also part of a separate chromatin-modifying complex called NuA4 (1). These large complexes regulate gene expression through enzymatic subunits with histone acetylation activity, either GCN5/PCAF in SAGA or Esa1p in NuA4. The acetylation of histones apparently stimulates transcription by reducing the affinity of nucleosomes for DNA and facilitating the access of other transcription factors or the movement of the transcription complex along chromatin. It was subsequently shown that c-Myc recruits the hGCN5 histone acetyltransferase (HAT) and that this enzyme is critical for Myc oncogenic activity (22). More recent data demonstrate the recruitment of HAT activity to Myc target genes in vivo (4, 8, 35). Since the Myc-related Mad/Mxi proteins can suppress Myc activity through the recruitment of histone deacetylases, chromatin remodeling through the modification of histone tails is clearly linked to oncogenic activity (18).
Although the recruitment of HAT activity offers one mechanism for Myc function, this is not sufficient to account for all of its oncogenic activity. Affinity purification of other nuclear factors that bind tightly to the c-Myc N terminus identified the TIP49 and TIP48 proteins as critical cofactors (34). Like TRRAP, TIP49 and TIP48 are highly conserved in evolution and essential for viability in yeast, but the latter proteins contain ATPase/helicase motifs rather than histone-modifying activity. Mutation of the ATPase motif in TIP49 creates a dominant inhibitor of c-Myc oncogenic activity, establishing a critical role for this enzyme in Myc function (34). The precise role of these ATPase/helicase family proteins in cell physiology requires further study, but they have also been reported to bind to other transcription factors (3) and have recently been shown to be present in a chromatin-remodeling complex in yeast (30) as well as in a complex with TIP60, a HAT (16). Here we report the identification and characterization of two new Myc cofactors, BAF53 and β-actin, that complex with c-Myc in vivo and are critical for Myc-mediated oncogenesis.
MATERIALS AND METHODS
Biochemical purification of c-Myc-associated nuclear proteins.
Large-scale affinity chromatography was performed as previously described (34). Briefly, FLAG-GAL4 and FLAG-GAL4/c-Myc were mixed with HeLa nuclear extract and supplemented with anti-FLAG antibody. For microsequencing, eluted proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide), visualized by Coomassie staining, excised, and submitted to the Harvard University Microchemistry Facility. Peptide sequences QGGPTYYIDTNALR and VDFPTAIGMVVER identified BAF53; peptide sequence AGFAGDDAPR identified actin, and monoclonal β-actin antibodies identified the polypeptide as β-actin. Full-length human cDNA for BAF53 was obtained by ordering an expressed sequence tag from the American Type Culture Collection (GenBank no. AA360918).
Transfection and immunoprecipitation.
293 cells were cultured in Dulbecco's modified Eagle's media supplemented with 10% fetal calf serum (GIBCO-BRL). Cells were transfected with 2 to 4 μg of each indicated expression vector by using the calcium phosphate method and lysed with F buffer (31). For immunoprecipitations, lysates were incubated with anti-FLAG or anti-HA antibodies in conjunction with protein G beads. Precipitates were then analyzed by Western blotting with anti-FLAG, anti-HA, anti-BAF53 (generated against the internal peptide GKQGGPTYYIDTNALRVPRE), anti-β-actin, anti-TIP49 (34), or anti-TIP48 (34). Protein expression was determined by analyzing the lysates by Western blotting with the appropriate antibody. For the in vivo interaction experiment, 293 cells were lysed in F buffer and subjected to immunoprecipitation using anti-Myc-conjugated beads (C33; Santa Cruz Biotechnology) or anti-FLAG-conjugated beads (Sigma). Antibody detection was performed using enhanced chemiluminescence (Amersham).
Chromatography and HAT assay.
Nuclei were isolated by the Dignam procedure (6) and extracted with soft lysis buffer (0.1% NP-40, 10 mM NaCl, 20 mM Tris [pH7.0], 0.05% 2-mercaptoethanol, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 40 mM NaF, 0.5 mM Na3VO4), which did not release TRRAP or TRRAP/BAF HAT activity from nuclei. Then the nuclear pellet was extracted with F buffer and loaded onto Q-Sepharose that was equilibrated with F buffer (pH 7.5). After unbound proteins were collected as flowthrough, bound proteins were sequentially eluted with 0.15, 0.4, and 1.0 M NaCl step gradients in F buffer. The 0.15 M NaCl fraction from Q-Sepharose was dialyzed with P11 binding buffer (20 mM HEPES [pH 7.8], 0.25 mM EDTA, 20% glycerol, 0.1% Tween 20) and loaded onto a P11 phosphocellulose column. Bound proteins from the P11 column were sequentially eluted with 0.2, 0.4, and 0.75 M KCl step gradients in P11 binding buffer. For the HA-BAF53-associated HAT assay, aliquots of each fraction were dialyzed with soft lysis buffer, 20 μl of anti-HA antibody-agarose conjugated beads was added, and bound proteins were eluted by adding HA peptide (0.2 mg/ml). The HAT assay and relevant Ponceau S staining procedure were described previously (24).
Transformation experiments.
Rat embryo fibroblast transformation assays were performed as previously described (21). Transfections included a cytomegalovirus (CMV) promoter-driven FLAG epitope-tagged c-myc expression vector (6 μg) and H-rasG12V (6 μg) supplemented with HA-BAF53 vectors (3 μg) or empty vector control. The HA-BAF53 constructs included wild-type, Δ39-67, Δ171-179, Δ233-255, and Δ319-324 constructs. Transfections were performed in triplicate using the calcium phosphate method. For the colony growth assay, early-passage primary rat embryo fibroblast cells were transfected with the same CMV-driven cDNA expression vectors used in the rat embryo fibroblast transformation assay. Expression constructs (6 μg) were transfected into the cells along with RSV-neo (1μg) by the calcium phosphate method. Transfected cells were selected in 400 μg of G418 (GIBCO-BRL) per ml for 14 days, at which time the number of colonies per plate was determined.
RESULTS
Purification of Myc N-terminus-interacting factors.
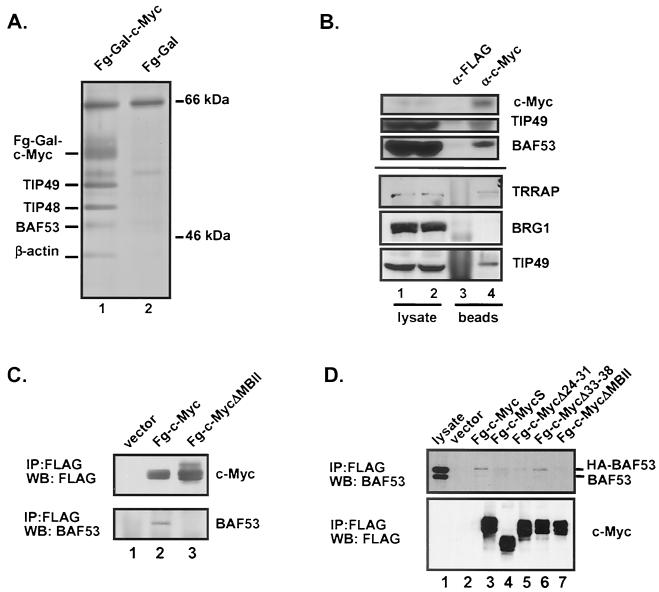
We have previously described the affinity purification of nuclear proteins that form a stable complex with the c-Myc transactivation domain (amino acids 1 to 262 fused to a FLAG epitope-tagged GAL4 DNA-binding domain) (21, 34). The complex was assembled in vitro, and proteins associated with the c-Myc N terminus were eluted using the FLAG peptide. In addition to the previously characterized proteins, TRRAP, TIP49, and TIP48 (21, 34), two other polypeptides with apparent molecular masses of 45 and 41 kDa were purified using the FLAG-GAL4-c-Myc fusion protein (Fig. 1A, lane 1). The only other specific proteins in the immunoprecipitates migrated as a doublet with an apparent molecular mass of approximately 60 kDa, which corresponds to the FLAG-GAL4-c-Myc fusion protein used as the affinity substrate. The binding specificity was assessed by using a control affinity matrix, FLAG-GAL4, which failed to recruit polypeptides of similar size.
FIG. 1.
Affinity purification of nuclear proteins interacting with the Myc amino terminus. (A) Affinity purification performed as previously described (34). Lanes 1, elution from the FLAG-GAL4/Myc (Fg-Gal-c-Myc) reaction; 2, elution from the FLAG-GAL4 reaction. The molecular mass (indicated in kilodaltons) was determined by coelectrophoresis of a protein molecular mass marker (Amersham). (B) 293 cells were lysed and subjected to immunoprecipitation using either anti-FLAG/agarose-conjugated beads or anti-Myc (C33)/agarose-conjugated beads. Precipitated proteins were resolved by SDS-PAGE and Western blotted for c-Myc, TIP49, and BAF53 or for TRRAP, Brg1, and TIP49 as indicated. Lanes: 1 and 2, lysates alone; 3 and 4, lysates subject to immunoprecipitation using the antibodies indicated. (C) 293 cells were transiently transfected with CMV-driven expression vectors for FLAG-tagged full-length c-Myc or an N-terminal mutant. Lysates were prepared, and c-Myc proteins were immunoprecipitated (IP) with anti-FLAG antibody. Precipitated proteins were resolved by SDS-PAGE and Western blotted (WB) for either c-Myc (with anti-FLAG antibody) or BAF53. Lanes: 1, immunoprecipitate from cells transfected with a vector expressing the FLAG epitope alone; 2, immunoprecipitate from cells expressing FLAG-c-Myc; 3, immunoprecipitate from cells expressing FLAG-c-Myc(Δ118-152). (D) 293 cells stably expressing HA-BAF53 were transiently transfected with CMV-driven expression vectors for FLAG-c-Myc or the N-terminal mutants indicated, and BAF53 coprecipitation was assayed by Western blotting. The MycS protein corresponds to the Δ1-110 mutant.
Identification of p45 as BAF53 and p41 as β-actin.
The immunoprecipitation experiment was scaled up to obtain sufficient quantities of the 45- and 41-kDa polypeptides for sequencing. Two independent peptides unambiguously identified the 45-kDa polypeptide as a 429-amino-acid actin-related protein named BAF53. BAF53 was originally identified as a component of the mammalian SWI/SNF-related chromatin remodeling complex known as the BAF complex (for “Brg1-associated factors”) (36). BAF53 shows extensive homology to actin and other actin-related proteins. Despite the general homology to actin, the ATP-binding pocket in actin is poorly conserved in BAF53, suggesting that BAF53 may not have ATP-binding activity (reference 36 and data not shown). To confirm that the 45-kDa protein was BAF53, we performed a Western blot analysis using two different anti-BAF53 antibodies which both detected the same 45-kDa band isolated through c-Myc interaction.
Sequences derived from the 41-kDa polypeptide proved to correspond to actin. However, insufficient sequence was obtained to distinguish between β-actin and α-actin. Western blotting with monoclonal antibodies to both proteins indicated that the 41-kDa protein is β-actin (data not shown). The isolation of β-actin as a component of a nuclear protein complex is consistent with the identification of β-actin and actin-related proteins in the BAF complex and in chromatin-remodeling complexes from yeast (11, 26, 36).
c-Myc interacts with BAF53 in vivo.
The affinity purification demonstrated that BAF53 formed a stable complex with the N terminus of c-Myc in vitro. To determine whether this interaction existed in vivo, we precipitated c-Myc from lysates prepared from 293 cells and probed the precipitates for BAF53 (Fig. 1B). As a control, we also probed the same blot for the previously characterized cofactor TIP49. This experiment provided clear evidence for the specific coprecipitation of c-Myc and BAF53, demonstrating that these proteins interact in vivo when expressed at normal physiological levels. Based on the prior link between BAF53 and the BAF remodeling complex, we probed the same immunoprecipitates for the Brg1 protein (Fig. 1B). No specific precipitation of c-Myc and Brg1 was observed, supporting the observation that no polypeptides comparable in size to Brg1 were isolated by affinity purification with the c-Myc N terminus. Western blots of in vitro affinity-purified Myc complexes also failed to detect Brg1 or Brm (data not shown). Therefore we conclude that c-Myc binds to the actin family proteins BAF53 and β-actin independently of any association with the BAF complex itself.
Deletion mutants of c-Myc were used to map the domain on the N terminus of Myc required for interaction with BAF53. Expression vectors for FLAG epitope-tagged wild-type or mutant Myc proteins were transiently transfected into 293 cells, the cells were lysed under native conditions, and the Myc proteins were immunoprecipitated with anti-FLAG antibodies. The precipitated material was then resolved by SDS-PAGE and Western blotted using anti-BAF53 antibodies to determine the extent of coprecipitation. The same membrane was subsequently probed with anti-FLAG antibodies to show equivalent protein expression between the different constructs. The full-length c-Myc protein coprecipitated with endogenous BAF53 (Fig. 1C, lane 2). In contrast, a deletion mutant with a mutation in the MBII domain (MycΔ118-152; Fig. 1C, lane 3) was defective for binding to BAF53. On the other hand, both MycS and a deletion mutant with a mutation in the c-Myc N terminus (Δ24-31) lack TRRAP-binding activity and the ability to transform primary cells in cooperation with an H-rasG12V oncogene (21). The binding of BAF53 was similar to but distinguishable from that of TRRAP because a FLAG-tagged MycS protein and c-Myc(Δ24-31) were defective for efficient binding to TRRAP but still retained weak BAF53-binding activity (Fig. 1D, lanes 4 and 5). Another mutant with a deletion in the c-Myc N terminus (Δ33-38) retained the same BAF53-binding activity (lane 6) as that of full-length c-Myc protein. TIP49 and TIP48 bind to MycS and N-terminal mutants in similar assays (data not shown).
A TIP49-TIP48-BAF53 complex is distinct from TRRAP complexes.
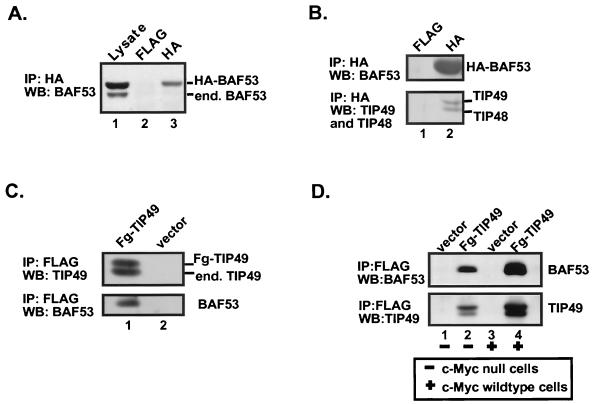
We were interested in determining if the different cofactors that bound to the c-Myc transactivation domain existed as independent subunits or as distinct complexes. To answer this question, we established a cell line with stable expression of HA-tagged BAF53 at a level approximately equal to that of the endogenous BAF53 protein (Fig. 2A, lane 1). HA-tagged BAF53 was immunoprecipitated from lysates, and the precipitates were probed by Western blotting for different proteins of interest. Interestingly, the HA-BAF53 protein did not coprecipitate with the endogenous BAF53 protein, which can be resolved from the epitope-tagged exogenous protein by relative migration on the gel (lanes 1 and 3). This observation suggests that BAF53 exists in nuclear complexes that are unlikely to contain polymeric actin-related proteins.
FIG. 2.
TIP49, TIP48, and BAF53 form a distinct complex. (A) 293 cells stably expressing HA-BAF53 were lysed and subjected to immunoprecipitation (IP) using either anti-FLAG antibodies or anti-HA antibodies and protein G beads. Precipitated proteins were resolved by SDS-PAGE and Western blotted (WB) for BAF53. Lanes: 1, lysates subject to immunoprecipitation using the antibodies indicated; 2 and 3, immunoprecipitation using anti-FLAG and anti-HA antibodies, respectively. (B) Same experimental procedure as for panel A, except that the Western blot was probed for BAF53, TIP49, and TIP48. Lanes 1 and 2, immunoprecipitation using the antibodies indicated. (C) HeLa cells stably expressing FLAG-TIP49 (Fg-TIP49) or FLAG epitope alone were lysed and subjected to immunoprecipitation using anti-FLAG antibodies. Precipitated proteins were resolved by SDS-PAGE and Western blotted for TIP49 and BAF53, as indicated. Lanes 1, immunoprecipitate from cells stably expressing FLAG-TIP49; 2, immunoprecipitate from cells stably expressing FLAG epitope alone. (D) Parental c-myc diploid (+) or c-myc null (−) cells, both stably expressing either FLAG-TIP49 or the FLAG epitope alone, were lysed and subjected to immunoprecipitation using anti-FLAG antibodies. Precipitated proteins were resolved by SDS-PAGE and Western blotted for BAF53 and TIP49, as indicated. Lanes: 1 and 3, immunoprecipitate from lysates prepared from null cells and diploid cells, respectively, expressing the FLAG epitope alone; 2 and 4, immunoprecipitate from lysates prepared from c-myc null cells and diploid cells, respectively, expressing FLAG-TIP49.
We next considered if BAF53 might exist in a stable complex with the TIP49 and TIP48 ATPase cofactors. Interactions between nuclear cofactors could be mediated by direct protein contacts or by simultaneous recruitment to the same transactivation domain. Immunoprecipitation with anti-HA from the HA-tagged BAF53 cell line demonstrated that BAF53 exists in a complex with both TIP49 and TIP48. (Fig. 2B, lane 2). To perform the reciprocal immunoprecipitation and explore the interaction between BAF53 and TIP49 in an additional cell type, we constructed a HeLa cell line with stable expression of FLAG-tagged TIP49. Immunoprecipitation with anti-FLAG antibodies confirmed the association between TIP49 and endogenous BAF53 shown above (Fig. 2C, lane 1, bottom panel), as well as the association of FLAG-TIP49 with the endogenous TIP49 (top panel) and TIP48 (data not shown) proteins described previously (34). However, these experiments do not rule out the possibility that the BAF53-TIP49 protein interaction was bridged by endogenous c-Myc. To exclude the latter possibility, we took advantage of companion Rat1-derived cell lines, in one of which the endogenous c-myc genes have been knocked out by targeted recombination (20). The c-myc-null cells also lack any detectable expression of other myc family members. The c-myc-expressing and c-myc-null cell lines were each engineered to stably express FLAG-TIP49. Lysates were prepared, and the FLAG-TIP49 protein was immunoprecipitated using anti-FLAG antibody. Coprecipitation of endogenous BAF53 and FLAG-TIP49 was observed in both cell lines (Fig. 2D, lanes 2 and 4), irrespective of the presence or absence of endogenous c-Myc expression (compare lanes 2 and 4). These experiments establish that the BAF53-TIP49 interaction is not mediated by c-Myc, but it remains possible that other cellular transactivation domains could bridge the two proteins.
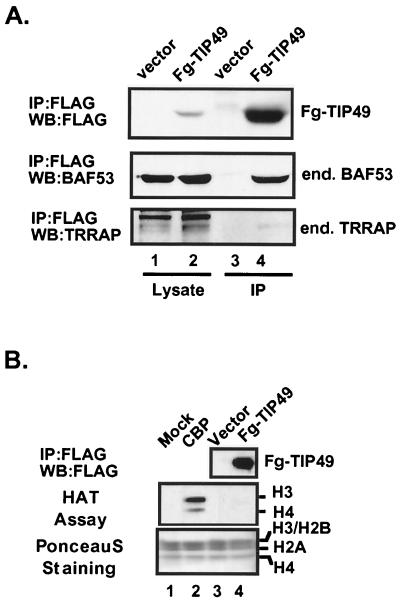
We wanted to determine if the nuclear cofactor complexes that contain BAF53 and TIP49 were distinct from SAGA or other TRRAP complexes. Previous studies showed that FLAG-TIP49 does not bind to endogenous TRRAP in 293 cells (34). To extend this, we retested to see if there was any coprecipitation of TRRAP with the TIP49-BAF53 complex. Transient expression of FLAG-TIP49 and immunoprecipitation with anti-FLAG antibodies precipitated endogenous BAF53 but no TRRAP (Fig. 3A, lane 4). Endogenous proteins were clearly visible in the cellular lysates prior to immunoprecipitation (lanes 1 and 2). Because TRRAP exists in multisubunit HAT complexes in yeast and humans, we also examined if TIP49 is associated with any HAT activity in vivo. FLAG-tagged TIP49 protein was transiently expressed in 293 cells, immunoprecipitated with anti-FLAG antibodies, eluted from the beads with an excess of FLAG peptide, and subjected to the HAT using core histones as substrates (Fig. 3B). We found that immunoprecipitates of FLAG-TIP49 protein from the 293 cells did not bring down any significant HAT activity (Fig. 3B, lane 4), suggesting that the TIP49-TIP48-BAF53 complex is biochemically distinct from the TRRAP HAT complex. Immunoprecipitates from the HeLa and 293 cells that stably express FLAG-tagged TIP49 and TIP48 proteins also did not contain TRRAP or other SAGA components (data not shown). Conversely, immunoprecipitation of ectopically expressed FLAG-tagged TRRAP cDNA does not precipitate TIP49 and TIP48. Finally, cotransfection of FLAG-TIP49 with wild-type c-Myc does not stimulate the binding of TIP49 or BAF53 to endogenous TRRAP, indicating that the c-Myc transactivation domain does not bridge the TIP49/BAF53 complex to TRRAP (data not shown). These data imply that c-Myc binds independently to distinct nuclear cofactor complexes and does not simultaneously recruit both complexes. However, it remains possible that these complexes are sequentially recruited by Myc to an overlapping set of chromosomal sites.
FIG. 3.
BAF53 associates with TIP49, but the complex lacks HAT activity. (A) 293 cells transiently expressing FLAG-tagged TIP49 (Fg-TIP49) or FLAG epitope alone were lysed, and anti-FLAG immunoprecipitates (IP) were resolved on SDS-PAGE and probed for BAF53 and TRRAP by Western blotting (WB). (B) 293 cells transiently expressing FLAG-tagged TIP49 or FLAG epitope alone were lysed and immunoprecipitated with anti-FLAG antibody-conjugated beads, and FLAG-tagged proteins were eluted by FLAG peptide. The eluates were subjected to a HAT assay and fluorography. As controls, buffer alone and recombinant CBP were included in lanes 1 and 2.
BAF53 forms a new HAT complex with TRRAP.
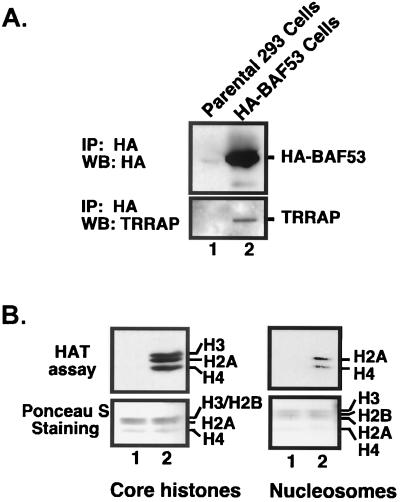
The actin-related protein Act3/Arp4 is a stable subunit of the NuA4 complex in yeast and is proposed to play a role in the integrity of the NuA4 complex (11). We are interested in determining if BAF53, the human protein closest to Act3/Arp4, is present in similar complexes in humans. HA-tagged BAF53 protein was immunoprecipitated from cells stably expressing HA-BAF53, eluted with an excess of HA peptide, and subjected to a HAT assay using core histones and mononucleosomes as substrates (Fig. 4). Interestingly, BAF53 immunoprecipitates contained TRRAP protein (Fig. 4A) and a unique HAT activity (Fig. 4B). The BAF53-associated HAT activity has an equal preferences for histones H3, H2A, and H4, but not for H2B, in core histones, and this specificity is similar to the substrate specificity of TIP60 but not GCN5 (16, 23). When the HAT was assayed with mononucleosomal substrates, H3 acetylation was completely suppressed and approximately equal intensities of H2A and H4 acetylation were obtained (Fig. 4B, right panel). These results, along with the immunoprecipitation data in Fig. 3B, indicate that BAF53 is a stable subunit of a new TRRAP-HAT complex that does not have TIP49-TIP48 or TIP60 (see below).
FIG. 4.
BAF53 forms a new HAT complex with TRRAP. (A) HA-tagged proteins were immunoprecipitated (IP) with anti-HA antibodies from 293 cells stably expressing HA-BAF53 and the parental 293 cells, respectively. Bound HA-BAF53 was released by adding an excess amount of HA peptide and subjected to Western blotting (WB) with probes for HA tag and TRRAP. (B) Proteins bound on anti-HA beads were tested for HAT activity using core histones (left panel) and mononucleosomes (right panel) as substrates.
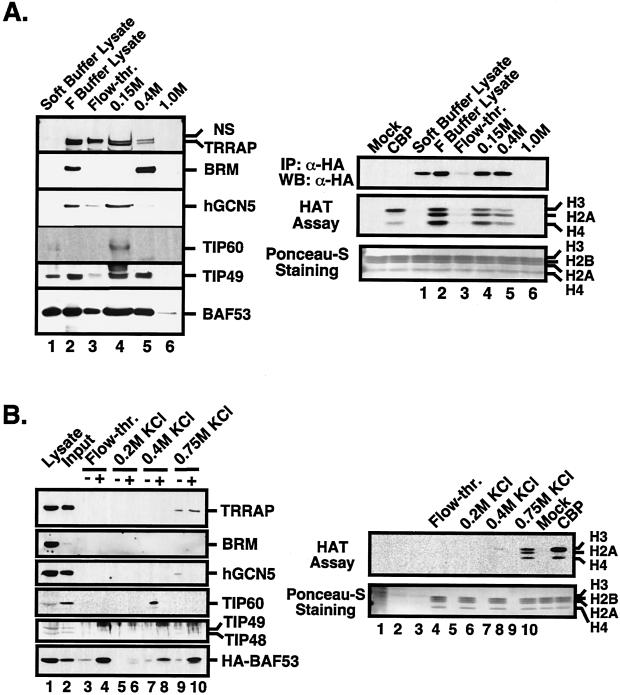
To resolve the different HAT complexes more thoroughly, we used conventional chromatography to monitor the fractionation pattern of BAF53-containing complexes. Nuclear lysates from 293 cells stably expressing HA-tagged BAF53 protein were prepared by sequential lysis, loaded onto a Q-Sepharose column, and eluted with a 0.15, 0.4, and 1.0 M NaCl step gradient. Aliquots of each fraction were analyzed by Western blot and probed for key proteins (Fig. 5A, left panel). To analyze the BAF53-associated HAT activity, HA-tagged BAF53 protein was immunoprecipitated with anti-HA antibodies from each fraction and subjected to a HAT assay (right panel). All of the proteins of interest to this study, TRRAP, hGCN5, TIP60, TIP49, and BAF53, coeluted at 0.15 M NaCl, but BRM did not. TIP49 and BAF53 are equally distributed at 0.15 and 0.4 M NaCl as well as released by soft lysis buffer from the nucleus, suggesting that complexes containing these proteins are heterogeneous with respect to their subunit composition and cellular localization. Consistent with the previous finding that BAF53 forms a HAT complex with TRRAP, the relative HAT activity associated with BAF53 matched the amount of TRRAP protein in each fraction (Fig. 5A, lanes 3 and 4).
FIG. 5.
Biochemical separation of a BAF53-TRRAP HAT complex. (A) Q-Sepharose column chromatography was conducted as described in Materials and Methods, and aliquots of each fraction were analyzed by SDS-PAGE and probed for antigens by Western blotting as indicated (left panel). TRRAP gave a doublet antigenic signal with crude antiserum, where the upper band is nonspecific (NS). HA-BAF53 was immunoprecipitated with anti-HA antibodies, released by HA peptide, and subjected to a HAT assay using core histones as substrate (right panel). The amount of HA-BAF53 in each immunoprecipitate (IP) was analyzed by Western blotting (WB) (right, top panel). As a control, mock and recombinant CBP were included in the assay reactions. (B) The 0.15 M NaCl fraction of Q-Sepharose in panel A was used as input for a P11 phosphocellulose column. F buffer lysate was included in lane 1 of the Western blot analysis to identify the respective antigenic signals. Each fraction from KCl step gradients was dialyzed with soft lysis buffer and immunoprecipitated with anti-HA antibody-conjugated beads. HA-BAF53 protein was released by HA peptide, and eluates were analyzed by SDS-PAGE and probed for antigens by Western blotting as indicated (left panel). An aliquot of each fraction before immunoprecipitation was paired with the corresponding immunoprecipitate (indicated as − and +, respectively). The same eluates from anti-HA immunoprecipitates were subjected to a HAT assay using core histones as substrates (right panel).
The 0.15 M NaCl fraction from Q-Sepharose provided a source of protein devoid of the BRM ATPase, so we further fractionated this sample using a P11 phosphocellulose column. The 0.15 M NaCl fraction of Q-Sepharose was dialyzed, loaded onto the P11 column, and eluted with a 0.2, 0.4, and 0.75 M KCl step gradient (Fig. 5B). Each fraction was analyzed by Western blotting for relevant proteins and for BAF53-associated HAT activity. For comparison, an aliquot of each fraction before anti-HA immunoprecipitation (− lanes) was paired with the corresponding HA-tagged BAF53 immunoprecipitates (+ lanes) in the Western blot analysis (Fig. 5B, left panel). The majority of the bound HA-tagged BAF53 eluted in the 0.75 M KCl fraction, where it coimmunoprecipitated with TRRAP but not with hGCN5 (Fig. 5B, lane 10). TIP60 eluted in the 0.4 M KCl fraction, which lacks TRRAP, TIP49-TIP48, and BAF53-associated HAT activity (lanes 7 and 8). Since BRM was immunoprecipitated with HA-BAF53 in the 0.4 M NaCl Q-Sepharose fraction, this separates the human SWI-SNF-related complex from the TIP49-TIP48-BAF53 complex and the TRRAP-BAF53 HAT complex. Although we could not separate the TIP49-TIP48-BAF53 complex from the TRRAP-BAF53 HAT complex by conventional chromatography, based on reciprocal immunoprecipitation and the HAT assay it is evident that these are biochemically distinct complexes. Therefore, our results indicate that BAF53 is a stable subunit of three distinct nuclear complexes: the human SWI-SNF complex, the TIP49-TIP48-BAF53 complex, and the TRRAP-BAF53 HAT complex.
Targeted BAF53 deletion mutants inhibit transformation by c-Myc.
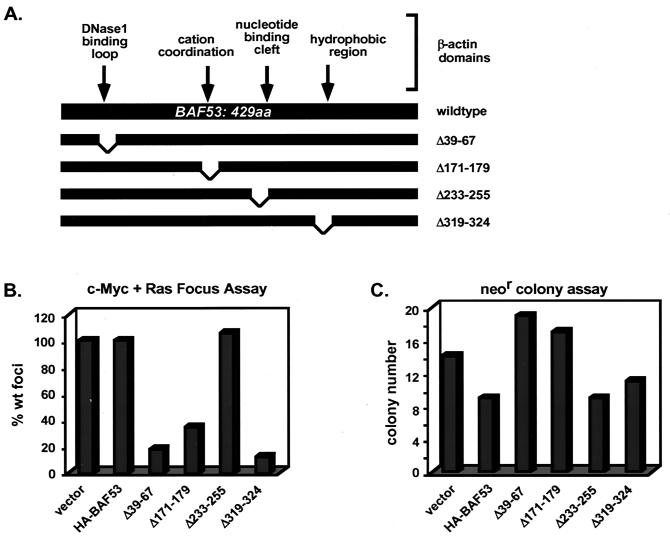
Since BAF53 interacts with cofactors that are essential for oncogenesis (34), it was of great interest to determine whether BAF53 might also play a role in Myc-mediated transformation. The extensive homology between BAF53 and β-actin provided a means of predicting critical regions in BAF53. The regions of BAF53 that place it in the actin-related gene family can be superimposed onto the β-actin crystal structure to create a hypothetical model. Virtually all of the major differences between BAF53 and β-actin can be modeled as BAF53-specific loops on the surface of the protein. We predict that these loops are important protein-protein interaction domains that could be mutated without disrupting the underlying actin folds. Four deletion mutants with deletions in BAF53 were constructed and tested in Myc-mediated transformation assays (Fig. 6A). The first mutant (Δ39-67) has a deletion of the BAF53-specific insertion found in the equivalent position of the DNase 1-binding domain of β-actin. The BAF53(Δ171-179) mutant has a deletion of the residues that would be equivalent to those coordinating Ca2+ or Mg2+ ions in β-actin. The BAF53(Δ233-255) mutant has a deletion of an insertion found in the equivalent of the nucleotide-binding cleft of β-actin. The BAF53(Δ319-324) mutant has a deletion of an insertion in a hydrophobic region of β-actin that may mediate protein-protein interactions. All of these BAF53 deletion mutants are stably expressed and coprecipitate with c-Myc in transient assays (data not shown).
FIG. 6.
BAF53 deletion mutants block the transformation of rat embryo fibroblasts by c-myc and H-ras. (A) Schematic diagram of BAF53 and β-actin. The amino acid (aa) sequence of BAF53 was aligned with β-actin to overlay known domains of β-actin onto BAF53. BAF53 deletions were created by site-directed mutagenesis. (B) Primary rat embryo fibroblasts were transfected with expression vectors for c-Myc, H-RasG12V, HA-BAF53, and deletion mutants of this construct in the different combinations indicated. wt, wild type. Three plates were assayed for each bar in the graph. (C) Duplicate plates of rat embryo fibroblasts were cotransfected with pRSV-neo and CMV promoter-driven HA-BAF53 or deletion mutants of this construct. Transfected cells were selected for 14 days, at which time the number of colonies per plate was determined. Two plates were assayed for each bar in the graph.
The oncogenic activity of c-Myc can be assayed by cotransfection of c-myc and the H-rasG12V oncogene into early-passage rat embryo fibroblasts, which leads to the formation of foci that are strictly dependent on Myc protein function. The number of oncogenic foci was scored after cotransfection of c-myc and H-rasG12V with the BAF53 wild-type and deletion mutant expression vectors (Fig. 6B). Cotransfection of a BAF53 wild-type vector with c-myc and H-rasG12V had no effect on focus formation. In contrast, cotransfection with three of the BAF53 deletion mutants (Δ39-67, Δ171-179, and Δ319-324) resulted in substantial inhibition of Myc-mediated transformation to 18, 35, and 12% of wild-type levels, respectively (Fig. 6B). However, BAF53 (Δ233-255) had no effect on transformation, indicating that the projected BAF53-specific insertions in the nucleotide-binding cleft region of β-actin are not critical for Myc oncogenesis. A trivial explanation for the inhibitory effect of the deletion mutants could be a general growth inhibition or toxicity. Both BAF53 wild-type and deletion mutant proteins were tested for general growth inhibition by assessing the efficiency of G418-resistant colony formation in the primary rat embryo fibroblasts used in the transformation experiment. When the proteins were cotransfected with pRSV-neo, nearly equal numbers of G418-resistant colonies were observed with all expression vectors compared to the empty vector control (Fig. 6C). This result indicates that there was no inherent general growth inhibition or toxicity involved with the BAF53 deletion mutants, although a minor suppression of colony formation was observed with wild-type BAF53 and BAF53(Δ233-255) that had no effect on oncogenesis. We conclude that the wild-type BAF53 protein functions in vivo as a cofactor for c-Myc in oncogenic transformation.
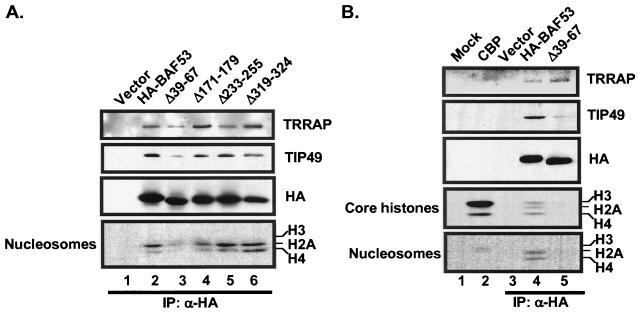
A BAF53 mutant is defective for HAT recruitment.
To explore the molecular basis for the dominant inhibitory activity of the BAF53 mutants, we tested each mutant for binding to TRRAP and recruitment of HAT activity. All four deletion mutants continued to bind to TRRAP and TIP49 (Fig. 7A). There was no distinction in TRRAP binding between mutants that were dominant inhibitors [BAF53(Δ171-179) and BAF53(Δ319-324)] and the one mutant that was not inhibitory (BAFΔ233-255). We presume that the dominant inhibitory activity of these mutants is due to the direct involvement of BAF53 in functional activity of the HAT complex or in some other aspect of complex assembly. One mutant, BAF53(Δ39-67), consistently failed to recruit HAT activity and had reduced binding to TIP49, even though as binding to TRRAP was undiminished compared to that of wild-type wtBAF53 (Fig. 7). This observation provides a molecular basis for the dominant inhibition of Myc-dependent oncogenic activity by this mutant, since Myc is known to recruit an H4-specific HAT activity to cellular target genes (4, 8; M. A. Nikiforov, S. Chandriani, J. Park, I. Kotenko, D. Matheos, A. Johnsson, S. B. McMahon, and M. D. Cole, submitted for publication). Further studies are required to unravel the dominant inhibitory activity of the other mutants.
FIG. 7.
A dominant inhibitory BAF53 mutant is defective for HAT activity but not TRRAP binding. (A) HA-tagged wild-type BAF53, HA-tagged BAF53 deletion mutants (Δ39-67, Δ171-179,Δ233-255, and Δ319-324), and empty vector were transiently expressed in HEK293 cells and immunoprecipitated (IP) with anti-HA antibodies. Samples were eluted with HA peptide and either electrophoresed for Western blotting (top three panels, with the antibody indicated at the right) or assayed for HAT activity with core histones or nucleosomes as indicated. All proteins were expressed at similar levels (HA panel). (B) HA-tagged wild-type BAF53, HA-tagged BAF53(Δ39-67), and empty vector were transiently expressed in HEK293 cells, immunoprecipitated with anti-HA antibodies, and eluted as for panel A. Mock and recombinant CBP were included as controls. HA-tagged wild-type BAF53 (lane 4) immunoprecipitated with both TRRAP, TIP49, and a HAT activity that specifically acetylates H2A and H4 in nucleosomes. The HA-tagged BAF53(Δ39-67) mutant (lane 5) immunoprecipitated with TRRAP but showed reduced binding to TIP49 and no detectable HAT activity. The wild-type and mutant proteins were expressed at similar levels (HA panel).
DISCUSSION
Functional data indicate that BAF53 is an integral component of a nuclear complex required for c-Myc-mediated oncogenic activity. BAF53 was originally identified as a component of the mammalian BAF chromatin-remodeling complex, which contains the Swi2/Snf2-related Brg1 or Brm ATPases (36). β-Actin was also identified as a subunit of the BAF complex, and together with BAF53 these actin family proteins are necessary for the full ATPase activity of Brg1 and targeting of the BAF complex to chromatin (36). BAF53 and β-actin were also found in a complex with TIP60, although no functional role was established (16). The studies presented here establish a new role for BAF53 as a component of nuclear cofactor complexes recruited by c-Myc. A fraction of BAF53 is found in the BAF complex, but separate fractions are found in complexes with c-Myc, TRRAP, and TIP49-TIP48. Since the BAF53-TIP49-TIP48 complex exists in the absence of c-Myc, this complex presumably functions as a cofactor for other transcription factors. The in vitro binding of β-actin to c-Myc and BAF53 indicates that it also serves as a c-Myc cofactor, although the abundance of β-actin in other structures makes it difficult to establish a direct functional role.
BAF53 belongs to a family of actin-related proteins (ARPs) with extensive identity (30 to 40%) to β-actin. Many ARPs have recently been found predominantly in the nucleus as components of chromatin-remodeling complexes. In yeast, Act3/Arp4p, Arp7p, and Arp9p are essential for viability, and their only known function is in chromatin modification or remodeling and the regulation of transcription (5, 11, 17, 26, 30). The function of nuclear ARPs in transcriptional regulation remains enigmatic. An important feature of β-actin is an inherent ATPase activity, which is likely to induce dynamic structural changes in the protein. However, attempts to detect ATP binding or ATPase activity by BAF53 and other nuclear ARPs have been unsuccessful to date. A study of potential ATP binding by Arp7p and Arp9p involved the introduction of specific mutations targeted to the residues corresponding to those that may be involved in ATP binding in actin, but these mutations had no effect on Arp7p and Arp9p with respect to in vivo function (5).
The Saccharomyces cerevisiae protein with the greatest similarity to BAF53 is Act3p/Arp4p. Like BAF53, Act3p has insertions into the actin homology domains that are likely to form loops on the surface of protein (13), but the sequences of the loops are not homologous between the yeast and human proteins. ACT3 is an essential gene identified in a screen for transcriptional defects in yeast that are similar to position effect variegation in metazoans (17). More recently, Act3p was found with Act1p (cellular actin) as a component of two different chromatin-modifying or -remodeling complexes. Act3p and Act1p are part of the NuA4 complex, which also contains the yeast homolog of the essential Myc/E2F1 cofactor TRRAP (Tra1p) and Esa1p, an H4-specific HAT (11). Act3p and Act1p are also found in a chromatin-remodeling complex containing the Swi2/Snf2-related Ino80p ATPase and the yeast homologs of ATPase proteins TIP49 (Rvb1p) and TIP48 (Rvb2p) (30). The overlap in shared components between these yeast complexes and the c-Myc cofactors described in this study is striking, although the precise complexes are apparently not identical. Despite the differences in the enzymatic activities of the two yeast complexes, one common element is the utilization of nucleosomes for a substrate. A role for Act3p in binding to core histones, specifically involving residues within one of the proposed loops, has been proposed (14). No comparable domain has been shown to exist in Arp7p or Arp9p, which also function in nuclear cofactor complexes (5, 26). The most compelling evidence of a direct role for Act3p in a chromatin-modifying complex comes from an analysis of temperature-sensitive mutations in Act3p, which disrupt the integrity of the NuA4 complex at the nonpermissive temperature (11). Thus, ARPs may be involved in both the structure and function of chromatin-modifying complexes, supporting the importance of BAF53 as a cofactor for the c-Myc transcription factor.
Biochemical fractionation identified at least three distinct fractions that contain BAF53, one containing Brm, one containing TIP49-TIP48, and one containing TRRAP and a presently unidentified HAT. Although TRRAP, BAF53, and TIP60 were previously described in a HAT complex (16), our data show no TIP60 in the complex. Furthermore, the yeast homologs of TIP48(Rvb2p) and TIP49(Rvb1p) were not found in the yeast NuA4 complex (11). BAF53 was also found more recently in a complex with TRRAP and the E1A-associated p400 protein; however, this complex contained no detectable HAT activity (10). The finding that the c-Myc transactivation domain recruits TRRAP, hGCN5, TIP49-TIP48, BAF53 (our studies), and p400 (10) makes it difficult to determine exactly which complexes are predominantly bound to c-Myc in vivo. A further complication may arise from the different extraction and immunoprecipitation conditions used in different laboratories, which may dissociate less stable components. We are presently purifying the BAF53-associated HAT detected in our studies, and functional studies implicate a specific BAF53 domain (amino acids 39 to 67 [Fig. 7]) in the recruitment of HAT activity into a TRRAP complex. However, existing data support the recruitment of several separate complexes by c-Myc. The TRRAP-hGCN5 complex was described previously (22), but this complex does not contain the other Myc-associated cofactors described here. TIP49 and TIP48 also associate with c-Myc in vivo (34), and these proteins are also present in association with BAF53 (Fig. 2). However, TIP49-TIP48 does not associate with HAT activity in our hands (Fig. 3B), even though c-Myc immunoprecipitates contain both H3- and H4-specific HAT activities (Nikiforov et al., submitted). Myc proteins promote the acetylation of both H3 and H4 at chromosomal sites (4, 8; Nikiforov et al., submitted). Further studies should provide more detailed functional insight into the specific protein-protein interactions mediated by BAF53 in chromatin-modifying complex that participate in Myc oncogenic activity.
Acknowledgments
We thank Gerald Crabtree for antibodies to BAF53.
This work was supported by grants from the National Cancer Institute and the New Jersey Commission on Cancer Research to M.D.C. M.A.W. was supported by a predoctoral fellowship from the New Jersey Commission on Cancer Research.
J.P. and M.A.W. contributed equally to this work.
REFERENCES
- 1.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Cote. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18:5108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amati, B., M. W. Brooks, N. Levy, T. D. Littlewood, G. I. Evans, and H. Land. 1993. Oncogenic activity of the c-Myc protein requires dimerization with max. Cell 72:233-245. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, A., O. Huber, and R. Kemler. 1998. Pontin52, an interaction partner of β-catenin, binds to the TATA box binding protein. Proc. Natl. Acad. Sci. USA 95:14787-14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchard, C., O. Dittrich, A. Kiemaier, K. Dohmann, A. Menkel, M. Eilers, and B. Luscher. 2001. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 15:2042-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairns, B. R., H. Erdjument-Bromage, P. Tempst, F. Winston, and R. D. Kornberg. 1998. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol. Cell 2:639-651. [DOI] [PubMed] [Google Scholar]
- 6.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evan, G. I., A. H. Wyllie, C. S. Gilbert, T. D. Littlewood, H. Land, M. Brooks, C. M. Waters, L. Z. Penn, and D. C. Hancock. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69:119-128. [DOI] [PubMed] [Google Scholar]
- 8.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freytag, S. O., C. V. Dang, and W. M. F. Lee. 1990. Definition of the activities and properties of c-myc required to inhibit cell differentiation. Cell Growth Differ. 1:339-343. [PubMed] [Google Scholar]
- 10.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 11.Galarneau, L., A. Nourani, A. Boudreault, Y. Zhang, L. Heliot, S. Allard, J. Savard, W. S. Lane, D. J. Stillman, and J. Cote. 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5:927-937. [DOI] [PubMed] [Google Scholar]
- 12.Grant, P. A., D. Schieltz, M. G. Pray-Grant, J. R. R. Yates, and J. L. Workman. 1998. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2:863-867. [DOI] [PubMed] [Google Scholar]
- 13.Harata, M., A. Karwan, and U. Wintersberger. 1994. An essential gene of Saccharomyces cerevisiae coding for an actin-related protein. Proc. Natl. Acad. Sci. USA 91:8258-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harata, M., Y. Oma, S. Mizuno, Y. W. Jiang, D. J. Stillman, and U. Wintersberger. 1999. The nuclear actin-related protein of Saccharaomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol. Cell. Biol. 10:2595-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henriksson, M., and B. Luscher. 1996. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv. Cancer Res. 68:109-182. [DOI] [PubMed] [Google Scholar]
- 16.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, Y. W., and D. J. Stillman. 1996. Epigenetic effects on yeast transcription caused by mutations in an actin-related protein present in the nucleus. Genes Dev. 10:604-619. [DOI] [PubMed] [Google Scholar]
- 18.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 19.Li, L. H., C. Nerlov, G. Prendergast, D. MacGregor, and E. B. Ziff. 1994. c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 13:4070-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mateyak, M. K., A. J. Obaya, S. Adachi, and J. M. Sedivy. 1997. Phenotypes of Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 8:1039-1048. [PubMed] [Google Scholar]
- 21.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 22.McMahon, S. B., M. A. Wood, and M. D. Cole. 2000. The c-Myc cofactor TRRAP recruits the histone acetylase hGCN5. Mol. Cell. Biol. 20:556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, J. Quin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 24.Park, J., S. Kunjibettu, S. B. McMahon, and M. D. Cole. 2001. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 15:1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penn, L. J. Z., M. W. Brooks, E. M. Laufer, and H. Land. 1990. Negative autoregulation of c-myc transcription. EMBO J. 9:1113-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson, C. L., Y. Zhao, and B. T. Chait. 1998. Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J. Biol. Chem. 273:23641-23644. [DOI] [PubMed] [Google Scholar]
- 27.Prendergast, G. 1999. Mechanisms of apoptosis by c-Myc. Oncogene 18:2967-87. [DOI] [PubMed]
- 28.Sakamuro, D., and G. Prendergast. 1999. New Myc-interacting proteins: a second Myc network emerges. Oncogene 18:2942-2954. [DOI] [PubMed] [Google Scholar]
- 29.Saleh, A., D. Schieltz, N. Ting, S. B. McMahon, D. W. Litchfield, J. R. Yates III, S. P. Lees-Miller, M. D. Cole, and C. J. Brandl. 1998. Tra1p is a component of the yeast ADA/SPT transcriptional regulatory complexes. J. Biol. Chem. 273:26559-26570. [DOI] [PubMed] [Google Scholar]
- 30.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodeling complex involved in transcription and DNA processing. Nature 406:541-545. [DOI] [PubMed] [Google Scholar]
- 31.Sommer, A., K. Bousset, E. Kremmer, M. Austen, and B. Luscher. 1998. Identification and characterization of specific DNA-binding complexes containing members of the Myc/Max/Mad network of transcriptional regulators. J. Biol. Chem. 273:6632-6642. [DOI] [PubMed] [Google Scholar]
- 32.Stone, J., T. De Lange, G. Ramsay, E. Jakobovits, J. M. Bishop, H. Varmus, and W. Lee. 1987. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol. Cell. Biol. 7:1697-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vassilev, A., J. Yamauchi, T. Kotani, C. Prives, M. L. Avantaggiati, J. Qin, and Y. Nakatani. 1998. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol. Cell 2:869-875. [DOI] [PubMed] [Google Scholar]
- 34.Wood, M. A., S. B. McMahon, and M. D. Cole. 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5:321-330. [DOI] [PubMed] [Google Scholar]
- 35.Xu, D., N. Popov, M. Hou, Q. Wang, M. Bjorkholm, A. Gruber, A. R. Menkel, and M. Henriksson. 2001. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc. Natl. Acad. Sci. USA 98:3826-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao, K., W. Wang, O. J. Rando, Y. Xue, K. Swiderek, A. Kuo, and G. R. Crabtree. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95:625-636. [DOI] [PubMed] [Google Scholar]