Abstract
Glucocorticoid-induced leucine zipper (GILZ) is a leucine zipper protein, whose expression is augmented by dexamethasone (DEX) treatment and downregulated by T-cell receptor (TCR) triggering. Stable expression of GILZ in T cells mimics some of the effects of glucocorticoid hormones (GCH) in GCH-mediated immunosuppressive and anti-inflammatory activity. In fact, GILZ overexpression inhibits TCR-activated NF-κB nuclear translocation, interleukin-2 production, FasL upregulation, and the consequent activation-induced apoptosis. We have investigated the molecular mechanism underlying GILZ-mediated regulation of T-cell activation by analyzing the effects of GILZ on the activity of mitogen-activated protein kinase (MAPK) family members, including Raf, MAPK/extracellular signal-regulated kinase (ERK) 1/2 (MEK-1/2), ERK-1/2, and c-Jun NH2-terminal protein kinase (JNK). Our results indicate that GILZ inhibited Raf-1 phosphorylation, which resulted in the suppression of both MEK/ERK-1/2 phosphorylation and AP-1-dependent transcription. We demonstrate that GILZ interacts in vitro and in vivo with endogenous Raf-1 and that Raf-1 coimmunoprecipitated with GILZ in murine thymocytes treated with DEX. Mapping of the binding domains and experiments with GILZ mutants showed that GILZ binds the region of Raf interacting with Ras through the NH2-terminal region. These data suggest that GILZ contributes, through protein-to-protein interaction with Raf-1 and the consequent inhibition of Raf-MEK-ERK activation, to regulating the MAPK pathway and to providing a further mechanism underlying GCH immunosuppression.
Glucocorticoid hormones (GCH) regulate cell growth, survival of normal and neoplastic cells (26, 37, 48, 63), and inflammatory and immune processes. Immunosuppression is due, at least in part, to the ability of GCH to regulate T-cell activation, growth, and development. For these reasons, GCH are used as therapeutic agents in several acute and chronic inflammatory and autoimmune diseases, in organ transplantation, and in the treatment of leukemia and lymphoma (4, 10, 22, 37, 63).
Most of the effects of GCH are mediated by modulation of gene transcription, through interaction with the glucocorticoid receptor (GR) (5, 14, 51), which functions as a ligand-dependent transcription factor that regulates gene expression directly by binding to DNA or indirectly by protein-to-protein interaction with other transcription factors (1, 5, 14, 51). Thus, GCH inhibit T-cell activation and proliferation, cytokine production, and transactivation of several transcription factors such as, for example, NF-κB and AP-1 (50, 51, 55, 56).
GILZ (glucocorticoid-induced leucine zipper), one of the GCH-induced genes, codes for a leucine zipper protein and was first isolated as a dexamethasone (DEX)-responsive gene from a thymus subtraction library (11). Increased by DEX, GILZ expression inhibits anti-CD3-induced interleukin-2 (IL-2) production, FasL upregulation, and activation-induced cell death (AICD) (2, 11, 51, 52). GILZ overexpression also inhibits NF-κB nuclear translocation, which is induced by triggering the T-cell receptor (TCR)/CD3 complex (2). GILZ expression is downregulated by anti-CD3 stimulation, suggesting a role for GILZ in controlling T-cell activation and development (2, 51).
The inhibition of IL-2 production and FasL expression, induced by GILZ overexpression, appears to be an event contributing to the block of T-cell activation and the consequent AICD. In fact, IL-2 contributes to FasL upregulation and is involved in the propriocidal regulation of AICD (3, 31). Therefore, overexpression of GILZ mimics GCH inhibition of T-cell activation.
T-cell activation, IL-2 production, and FasL expression are regulated at the transcriptional level (25, 28). IL-2 promoter activation requires, in particular, the cooperative interaction of several transcription factors, such as, for example, NF-κB, AP-1, and NF-AT (16, 20, 41, 59). AP-1 proteins play a role in IL-2 regulation by binding to the AP-1 site in the IL-2 promoter and by helping to form transcriptionally active NF-AT (6, 29, 32, 43).
The AP-1 complex activity is controlled by regulation of Jun and Fos transcription and by posttranslation modification. Both phenomena are controlled by mitogen-activated protein kinase (MAPK) pathways (9, 15, 61). MAPKs are evolutionarily conserved through the plant and animal kingdoms and the wide MAPK family includes the p44/p42 extracellular signal-regulated kinases 1/2 (ERK-1/2), p38 MAPK, and c-Jun NH2-terminal protein kinase (JNK). These MAPKs are regulated by small G proteins of the Ras family; have significant roles in mediating signals triggered by cytokines, growth factors, and environmental stress; and are involved in controlling cell proliferation, cell differentiation, and death (18, 54).
The Ras proto-oncogene encodes a small GTP-binding protein, whose activation initiates a complex array of signal transduction events. Active Ras stimulates the classical MAPK pathway, which acts via the kinases Raf, MAPK/ERK-1/2 (MEK-1/2), and ERK-1/2. This results in phosphorylation and activation of ELK-1, which, in turn, induces transcription of c-Fos and JunB genes (36).
TCR triggering also stimulates the so-called alternative or stress-activated MAPK pathway, which, via p95Vav, Rac, MEKK-1, and SEK-1/2, activates JNK-1/2, which in turn regulates c-Jun phosphorylation and transcription (27).
Many studies have indicated that GCH interferes with different components of the MAPK cascade (8, 39, 53, 62), and GR and Raf have been found within the same protein complex (receptosome) in rat liver cells (62). In the present study, we address the question of whether GILZ interferes with MAPK cascade activation. The results indicate that GILZ binds and inhibits Raf-1 and, consequently, the downstream activation pathway.
MATERIALS AND METHODS
Cell line and animals.
A spontaneously dividing CD3+ CD4+ CD2+ CD44+ subline of the ovum-specific hybridoma T-cell line 3DO (60) was used for the experiments. Cells were maintained in logarithmic growth in RPMI 1640 supplemented with 10% fetal calf serum, 20 mM HEPES, and various antibiotics. COS-7 cells were maintained in culture in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum.
Thymus cells were obtained from 4- to 6-week-old C3H/HeN mice (Charles River, Calco, Milan, Italy).
Antibody cross-linking and cell treatment.
Hamster anti-mouse CD3ɛ monoclonal antibody (MAb; Pharmingen, San Diego, Calif.) at 1 μg/ml was allowed to adhere in flat-bottom, high-binding, 96-well plates (Costar, Cambridge, Mass.) at 4°C in 100 μl of phosphate-buffered saline (PBS). After 20 h, MAb-coated plates were washed, and transfected clones were plated at 105 cells/well and incubated at 37°C for different times as indicated.
Cytokine assays.
Supernatants from clones left untreated or treated with anti-CD3 MAb for 18 h were tested for IL-2 concentration by two-site enzyme-linked immunosorbent assay (ELISA) with MAb JES6-1A12 as the primary reagent and biotinylated monoclonal S4B6 as the secondary reagent. In gamma interferon (IFN-γ) measurement, two-site ELISA involved the use of XMG1.2 as the primary reagent and biotinylated monoclonal AN18.17.24 antibody as the secondary MAb. Antibodies were purchased from Pharmingen. Cytokine titers, expressed as picograms or nanograms per milliliter, were calculated with reference to standard curves constructed with known amounts of rIL-2 or rIFN-γ.
Transfection of cultured cells and clone preparation.
Transfected clones were prepared as previously described (11). Briefly, mouse GILZ cDNA-coding sequence (414 bp) was cloned into a pcDNA3 plasmid (Invitrogen, San Diego, Calif.) for expression in 3DO cells. Cells were transfected by electroporation (300 mA, 960 μF) with 15 μg of linearized pcDNA3 vector (control clones) or 15 μg of linearized pcDNA3 vector expressing GILZ cDNA. After 36 h, cells were cultured in medium containing G418 at 0.8 mg/g (active form)/ml (Gibco-BRL/Life Technologies, Paisley, Scotland), and a 100-μl/ml cell suspension was plated in 96-well plates (four for each transfection). After 15 to 20 days, no more than 15% of the wells presented live growing cells. These cells were considered clones and were analyzed by RNase protection assay (RPA) for the expression of exogenous GILZ.
COS-7 cells were cotransfected by using DEAE-dextran as previously described (39) with the expression vector pUSEamp containing human Raf-1 (2 μg; Upstate Biotechnology, Lake Placid, N.Y.) and a myc-tagged GILZ (myc-GILZ; 2 μg), obtained by inserting full-length GILZ cDNA as a BamHI-XbaI fragment in pcDNA3.1/Myc-His. The vector was purchased from Invitrogen.
In some experiments, transfected cells were serum starved and treated for 15 min with phorbol 12-myristate 13-acetate (PMA; 10 ng/ml; Sigma-Aldrich, Milan, Italy).
Indirect immunofluorescence and microscope analysis.
myc-GILZ/pUSEamp-Raf-1-cotransfected COS-7 cells were cytospun for 5 min at 400 rpm directly onto glass slides. Cells were permeabilized by incubation in methanol for 20 min at −20°C. After three washes in PBS and a blocking step in PBS containing 3% bovine serum albumin and 1% glycine (blocking buffer), the cells were incubated for 1 h at room temperature with monoclonal mouse anti-myc antibody (1:200; Invitrogen) and polyclonal rabbit anti-human Raf-1 antibody (1:50; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.). Cells were then incubated for 1 h with secondary antibodies (i.e., anti-mouse fluorescein-conjugated and anti-rabbit Texas red-conjugated antibodies) in blocking buffer containing 1 ng of DAPI (4′,6′-diamidino-2-phenylindole)/ml and then washed; the slides were then mounted with a coverglass. The slides were analyzed by a Zeiss Axioplan fluorescence microscope, and the images were acquired by using a Spot-2 cooled camera (Diagnostic Instruments).
Luciferase assay.
3DO cells were cotransfected with either pcDNA3-GILZ constructed as described above or pcDNA3 and a reporter vector containing tandem repeats of the murine AP-1 site in which the promoter drives the expression of the firefly luciferase (obtained from F. D'Adamio, Albert Einstein College of Medicine, New York, N.Y.). Some groups were cotransfected with activated Raf-1 cDNA: 5 μg in pUSEamp (Upstate Biotechnology) in which the activating mutation is a substitution of aspartic acid for tyrosine at residue 340. Transfection was performed by electroporation as described above. The transfection efficiency, as assessed by transfection of a green fluorescent protein vector and quantitated by fluorescence-activated cell sorting, ranged between 65 and 80%. Cells were purified by Ficoll to eliminate dead cells resulting from transfection and stimulated with anti-CD3 MAb (1 μg/ml) for 4 and 18 h.
Each transfection was performed in triplicate. Cell lysis and luciferase quantification were performed with commercial reagents (Luciferase Reporter Gene Assay; Roche Diagnostics, Monza, Italy). The values are expressed as the increase (n-fold) above the level of luciferase activity of untransfected cells or of cells transfected with the reporter plasmids without anti-CD3 MAb stimulation (values are comparable).
Nuclear extracts and electrophoresis mobility shift assay (EMSA).
Cells were stimulated for 1 h with plastic-bound anti-CD3 MAb, and nuclear extracts were prepared as previously described (2).
All DNA-binding reactions were conducted for 20 min at room temperature in a final volume of 20 μl. The reactions were started by adding 10 μg of nuclear protein extract to a reaction mix containing 2 μg of poly(dI-dC), 4 μl of 5× binding buffer (50 mM Tris, 250 mM NaCl, 5 mM EDTA, 25% glycerol, 5 mM dithiothreitol), and ca. 20,000 cpm of the respective [γ-32P]ATP-labeled double-stranded DNA oligonucleotide. Cold competitor oligonucleotides were added to the reaction mix before the radiolabeled probe. The sample was then loaded on 5% native polyacrylamide gel in Tris-borate-EDTA buffer. After electrophoresis for 2.5 h at room temperature and 10 V/cm, gels were dried and separated protein-DNA complexes were visualized by autoradiography by using Kodak XAR5 films. The following double-stranded DNA oligonucleotides (Promega, Milan, Italy) were used in EMSA analyses both as labeled or competitor cold probes: AP-1 (5′-CTAGTGATGAGTCAGCCGGAT-3′) and NF-AT (5′-AAGAGGAAAATTTGTTTCATACAG-3′). For antibody-induced supershift assays, 2 μl of antibody (0.2 μg/ml), monoclonal mouse anti-human c-Fos antibody (Santa Cruz), or polyclonal rabbit anti-avian c-Jun antibody (Upstate Biotechnology) was incubated with 10 μg of nuclear extract for 30 min at room temperature after the addition of radiolabeled probe.
Construction of GILZ mutants.
Deletion and substitution mutants of pCR3.1-GILZ were generated by PCR with pCR3.1-GILZ as a template and mutant synthetic oligonucleotides as primers.
The COOH-terminal proline-rich region (amino acids 98 to 137) was deleted in mutant 6; the NH2-terminal region (amino acids 9 to 73) was deleted in mutant 13. In mutant 2 an aspargine in position 87 was replaced by aspartic acid; in mutant 117 the leucines in positions 76, 83, and 90 were replaced by alanines.
In vitro protein-binding assays.
Extracts were made from 3DO cells or thymocytes left untreated or treated with DEX (10 μM), as previously described (2). Glutathione S-transferase (GST)-GILZ fusion protein was prepared as previously described (11); GST-Raf fusion protein corresponding to the human Ras-binding domain (RBD; residues 1 to 149) (13) of Raf-1 (GST-Raf-RBD) was from Upstate Biotechnology. Pulldown assays were performed by incubating the fusion proteins, loaded on glutathione-Sepharose beads, with cellular lysates in binding buffer (250 mM NaCl, 50 mM HEPES [pH 7.5], 0.5 mM EDTA, 0.1% [vol/vol] NP-40, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 100 μg of bovine serum albumin/ml) for 18 h at 4°C. The beads were washed extensively, resuspended in sample buffer, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting with polyclonal rabbit anti-mouse Raf-1 antibody (1 μg/ml; Upstate Biotechnology), polyclonal rabbit anti-mouse MEK-1/2 or polyclonal rabbit anti-mouse ERK-1/2 (1 μg/ml; Cell Signaling Technology, Beverly, Mass.), or polyclonal rabbit anti-mouse GILZ antibody (1:5,000), prepared as previously reported (11). Briefly, GST-GILZ containing the full GILZ amino acid sequence was used to immunize New Zealand White rabbits. The purified antiserum was tested by Western blotting with GST-GILZ fusion protein, cellular extracts from clones overexpressing GILZ, and DEX-treated thymocytes (11). Full-length and mutant (i.e., mutants 6, 13, 2, and 117) GILZ forms were translated in vitro with [35S]methionine by using the rabbit reticulocyte-coupled in vitro transcription translation system under the T7 promoter, according to the manufacturer's instructions (Promega, Madison, Wis.). In vitro-translated proteins were diluted with binding buffer (25 mM HEPES [pH 7.5], 10% glycerol, 50 mM NaCl, 0.05% NP-40, 1 mM dithiothreitol) and precleared with glutathione-Sepharose beads for 45 min at 4°C. GST or GST-Raf-RBD bound to glutathione-Sepharose beads was incubated with in vitro-translated proteins for 18 h at 4°C. The beads were subsequently washed five times with 0.5 ml of binding buffer; bound proteins were recovered by boiling in SDS sample buffer, analyzed by SDS-PAGE, and visualized by autoradiography.
For competition experiments, COS-7 cells were transfected with activated-Ras at 2 μg in pUSEamp (Upstate Biotechnology). The Ras-activating mutation is a substitution of leucine for glutamine at position 61. Cell extracts (100 μg) were incubated with 2.5 μg of GST-Raf-RBD for 1 h at 4°C. The GST-Raf-RBD-Ras complex was purified by adsorption to glutathione-Sepharose beads, washed, and resuspended in PBS. Serial dilutions of purified GILZ were added. The GST portion of GST-GILZ was removed by trombin cleavage. After 1 h at 4°C, the GST-Raf-RBD beads were washed and examined for associated proteins by Western blotting with anti-GILZ, anti-Ras (1 μg/ml; Upstate Biotechnology), or anti-GST (1 μg/ml; Santa Cruz) antibodies.
Kinase assays.
The kinase activity of Raf-1 immunoprecipitates was measured by using a Raf-1 immunoprecipitation kinase cascade assay kit according to the manufacturer's instructions (Upstate Biotechnology). Briefly, kinase reactions were carried out in 30 μl of assay dilution buffer, supplemented with 10 μl of magnesium-ATP cocktail (500 μM ATP and 75 mM magnesium chloride) and 10 μCi of [γ-32P]ATP with GST-MEK-1, GST-ERK-1, and myelin basic protein (MBP) as substrates. Reaction mixtures were incubated for 30 min at 30°C and resolved on SDS-12% acrylamide gels. MEK-1 and MBP phosphorylation was then quantitated by using a Fuji phosphorimager. As a positive control, Raf-1 active, recombinant enzyme (10 U; Upstate Biotechnology) expressed in Sf9 was used in the assay.
Immunoprecipitation and coimmunoprecipitation.
Whole-cell extracts were prepared. Immunoprecipitations were performed in radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 5 mM EDTA) supplemented with 1 mM phenylmethylsulfonyl fluoride. Antigen-antibody complexes were precipitated with protein A bound to Sepharose beads (Santa Cruz) prior to SDS-PAGE. For immunoprecipitation, anti-Raf antibody and control isotype antibody were used at concentrations of 4 μg/500 μg of protein; the Raf-bound proteins were detected by Western blotting with anti-GILZ antibody. COS-7 lysates (500 μg) were immunoprecipitated with anti-myc antibody (3 μg), and proteins were detected by Western blotting with anti-Raf-1 antibody.
Western blot analysis.
Extracted or immunoprecipitated proteins were separated on an SDS-polyacrylamide gel and studied by Western blotting as previously described (26). The primary antibodies were a polyclonal rabbit antiserum recognizing GILZ, a polyclonal rabbit anti-mouse phospho-ERK-1/2, and anti-mouse ERK-1/2 (Cell Signaling), anti-Fos, anti-Jun, anti-Raf-1, monoclonal rat anti-mouse phospho-Raf-1 (series 338; Upstate Biotechnology), anti-MEK, and polyclonal rabbit anti-human phospho-MEK (Ser 217/221; Cell Signaling) antibodies. Secondary antibodies included horseradish peroxidase-labeled antibody (Pierce, Rockford, Ill.). Anti-β-tubulin MAb (Calbiochem, San Diego, Calif.) was used as a control. The antigen-antibody complexes were revealed by enhanced chemiluminescence according to the manufacturer's instructions (SuperSignal Pierce; Pierce).
Molecular modeling. (i) Homology modeling of GILZ.
The three-dimensional (3D) model of human GILZ was constructed by using the structure of DIP, the sleep-inducing peptide immunoreactive peptide (58), and comparative homology modeling and threading approaches. In particular, GILZ and DIP sequences were aligned, and BLOSUM was used as a score matrix with an open gap and gap extension penalties of 10 and 0.05, respectively. A secondary structure with a matching weight of 1 was also used during optimization of the alignment score. The secondary structure of sleep-inducing peptide immunoreactive peptide, DIP (pdb code: 1dip) (58), was retrieved from the pdb file, whereas the secondary structure of GILZ was predicted by the GOR method (11, 21, 40). Only the first conformation in the pdb of DIP was used to build the GILZ model with the program Modeler implemented in Insight II. The final 3D model of the leucine zipper and the C-terminal domains of GILZ was refined by using several cycles of conjugate gradient minimization until a convergence gradient of 0.05 kcal/mol was reached. A high harmonic constraint was applied on the coordinates of all backbone atoms and on the coordinates of the side chain atoms belonging to the conserved residues. Since no statistically significant homology exists for the NH2-terminal region of GILZ (residues 1 to 57), several threading procedures were used to assign a plausible 3D fold. The NH2-terminal domain sequence was submitted to the University of California at Los Angeles folding prediction (17), SAUSAGE (23, 24), and 3D-PSSM (34) servers.
(ii) Docking experiments of GILZ and Raf.
Docking experiments between GILZ and Raf were performed by using the FTDOCK program (19). Briefly, the crystal structure of the Ras-binding domain of C-Raf-1 kinase (pdb code: 1c1y) (47) was docked to GILZ by using the default setting of FTDOCK. The resulting 10,000 solutions were ranked by using a residue pair potential scoring matrix (RPscore [44]) implemented in the FTDOCK distribution. All of the above computations were carried out on an SGI O2 R12000 workstation.
Statistical analysis.
Each experiment was performed at least three times. Representative experiments are shown. Due to the nonnormal distribution of the data, nonparametric tests (Kruskal-Wallis analysis of variance) were adopted for statistical evaluation.
RESULTS
GILZ inhibits anti-CD3-driven IL-2 production and AP-1 transactivation.
IL-2 gene transcription involves the transactivation of AP-1, and the molecular basis of the IL-2 block, in several in vivo and in vitro models of T-cell unresponsiveness, involves a defect in AP-1 transactivation (20, 32, 41, 43, 57, 59). We analyzed the effects of GILZ expression on AP-1 activation and the upstream activation pathway, including Raf, MEK, and ERK.
For this purpose, GILZ cDNA was overexpressed in 3DO cells (11), and stable clones containing empty (pcDNA3) or GILZ-expressing (pcDNA3-GILZ) vectors were prepared. As previously demonstrated (2), pcDNA3-transfected clones, upon activation by anti-CD3 MAb, displayed normal IL-2 production, whereas pcDNA3-GILZ-transfected clones presented low or undetectable IL-2 levels (Table 1). IFN-γ was also tested as a control, and no significant differences were found in GILZ-transfected and control clones (Table 1).
TABLE 1.
Production of IL-2 and IFN-γ by pcDNA3-transfected and pcDNA3-GILZ-transfected clones
| Transfecting plasmid | Mean cytokine concn ± SD
|
|||
|---|---|---|---|---|
| Control cells
|
Anti-CD3 antibody-treated cells
|
|||
| IL-2 (pg/ml) | IFN-γ (ng/ml) | IL-2 (pg/ml) | IFN-γ (ng/ml) | |
| pcDNA3a | -b | - | 745 ± 10 | 8 ± 0.5 |
| pcDNA3-GILZ | - | - | 20 ± 1.5 | 12 ± 1 |
3DO clones containing pcDNA3 or pcDNA3-GILZ were treated for 18 h with adherent anti-CD3 MAb (1 μg/ml). Supernatants were harvested and used in an ELISA.
-, None detected.
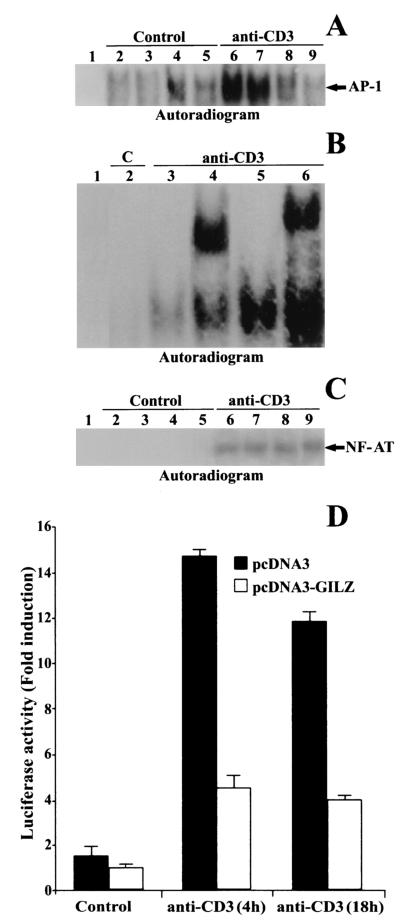
The same clones were used to analyze whether GILZ interferes with AP-1/DNA binding in comparative EMSAs. Nuclear extracts from untreated or anti-CD3-treated clones containing pcDNA3 or pcDNA3-GILZ were prepared. Empty-vector-transfected T-cell clones (PV6) and parental 3DO displayed AP-1 activation upon anti-CD3 stimulation (Fig. 1A, lanes 6 and 7 versus lanes 2 and 3). Clones overexpressing GILZ failed to induce AP-1 activation upon anti-CD3 triggering (Fig. 1A, lanes 8 and 9 versus lanes 4 and 5). The specific competitor (cold AP-1) prevented complex formation (Fig. 1B, lane 3 versus lane 5), and anti-c-Jun and anti-c-Fos antibodies supershifted the complex (Fig. 1B, lanes 4 and 6, respectively). No differences were observed in the pattern of NF-AT transactivation in pcDNA3 and pcDNA3-GILZ clones (Fig. 1C). This is not surprising, because, even though AP-1 is a component of the NF-AT heterotrimeric complex, several models of T-cell functional anergy are characterized by the lack of AP-1 but non-NF-AT transactivation (57).
FIG. 1.
GILZ overexpression inhibits the binding of AP-1 to its DNA motif. (A) EMSA was performed with nuclear extract from untreated and anti-CD3-treated (1 μg/ml, 1 h) 3DO cells (lanes 2 and 6) or pcDNA3-transfected PV6 (lanes 3 and 7), pcDNA3-GILZ-transfected ST7 (lanes 4 and 8), or GIRL-19 (lanes 5 and 9) clones. Lane 1, probe alone. (B) Nuclear extract from untreated or anti-CD3-treated 3DO cells, alone (lane 5) or added with competitor cold probe (lane 3), with anti-c-Jun (lane 4), or with anti-c-Fos antibody (lane 6). Lane 1, labeled probe alone. C, untreated cells. (C) EMSA performed with NF-AT as the probe. Nuclear extract from untreated and anti-CD3-treated (1 μg/ml, 1 h) 3DO cells (lanes 2 and 6) or PV6 (lanes 3 and 7), ST7 (lanes 4 and 8), or GIRL-19 (lanes 5 and 9) clones. Lane 1, probe alone. The results are representative of three experiments. (D) 3DO cells were transfected with the AP-1 luciferase reporter geneand pcDNA3 (solid bar) or with the reporter gene and pcDNA3-GILZ (open bar) and stimulated for 4 and 18 h with plastic-bound anti-CD3 MAb. The values are expressed as the increase (n-fold) in luciferase activity. Each transfection was performed in triplicate. The standard errors were <10%.
The same results were obtained when 3DO cells were cotransfected with an AP-1-dependent reporter gene with or without GILZ cDNA and activated with anti-CD3 MAb. In this experiment, luciferase activity was measured 24 h after transfection in nonstimulated or anti-CD3-triggered cells as a function of AP-1-dependent transcription. The results, expressed as the increase (n-fold) in luciferase activity, of a representative experiment (Fig. 1D) showed that 3DO cells cotransfected with AP-1 reporter and pcDNA3 displayed increased luciferase activity after 4 or 18 h of stimulation with anti-CD3 MAb, whereas cotransfection of pcDNA3-GILZ significantly reduced AP-1 transcriptional activity (P < 0.01) at each time (Fig. 1D).
All of these data indicate that GILZ interferes with AP-1 activation.
GILZ overexpression impairs c-Fos transcription.
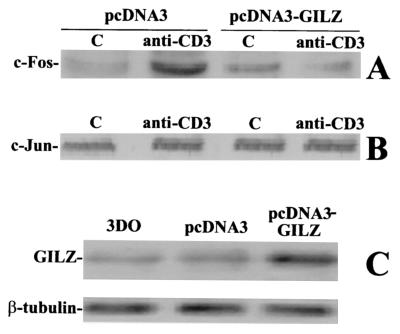
Heterodimeric complex formation and AP-1 activation depend on the Jun and Fos protein levels and thus on their transcriptional control (43). Therefore, we determined whether GILZ interferes with c-Fos and c-Jun transcription by first evaluating the expression of c-Jun and c-Fos proteins in GILZ-overexpressing 3DO clones upon anti-CD3 MAb stimulation. TCR triggering upregulated c-Fos transcription in the pcDNA3-transfected clone but not in the clone overexpressing GILZ (Fig. 2A). The same results were obtained at all times in the time course experiments and with other clones overexpressing GILZ (not shown).
FIG. 2.
GILZ overexpression inhibits c-Fos but not c-Jun trascription. Western blot analysis of c-Fos (A) and c-Jun (B) expression was performed. Nuclear cell lysates (10 μg) from PV6 and GIRL-19, stimulated for 1 h with immobilized anti-CD3 MAb (1 μg/ml), were probed with anti-c-Fos or anti-c-Jun antibodies (1 μg/ml). C, untreated cells. (C) The expression level of endogenous and exogenous GILZ was evaluated by Western blotting with anti-GILZ antiserum.
No c-Jun transcription modulation was observed in either control or GILZ-overexpressing clones (Fig. 2B), suggesting that a constitutive control mechanism for c-Jun transcription is active in this particular experimental model.
The GILZ expression levels of the 3DO cell line and the pcDNA3-transfected and pcDNA3-GILZ-transfected clones are shown in Fig. 2C.
GILZ overexpression inhibits ERK-1/2, MEK, and Raf-1 phosphorylation but not JNK phosphorylation.
The above results indicate that impaired c-Fos protein production could partly explain the decreased transactivation of multimerized AP-1 and suggest a possible new mechanism for GILZ-induced decrease of IL-2 production.
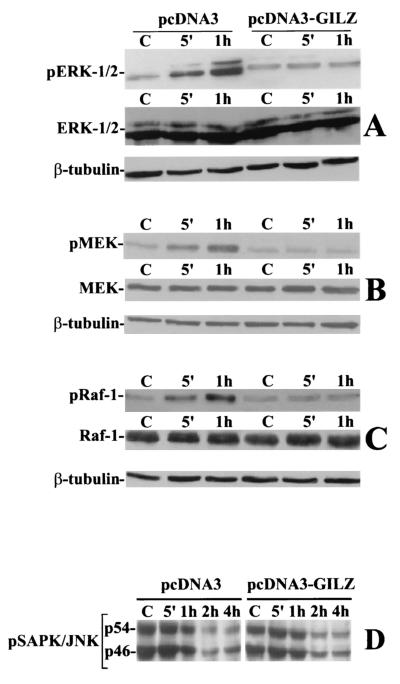
Since AP-1 transactivation (c-Fos transcription and c-Jun phosphorylation) is under the control of the Ras/MAPK pathway (9, 15, 61), we determined whether GILZ interferes with the activation of some members of the MAPK family, such as ERK-1/2. Control clones and GILZ-overexpressing clones were stimulated with anti-CD3 MAb for 5 and 60 min. The results of a representative experiment (Fig. 3A) indicate that the activated control clones displayed an increasing level of ERK-1/2 phosphorylation (the results with one clone that is representative of four that yielded similar results are shown in the figure). GILZ-overexpressing clones did not respond to anti-CD3 triggering. MEK and Raf-1 phosphorylation showed the same behavior (Fig. 3B and C). Moreover, the amount of total ERK-1/2, MEK, and Raf-1 proteins was unchanged (Fig. 3A, B, and C).
FIG. 3.
GILZ overexpression inhibits ERK-1/2, MEK-1/2, and Raf-1 but not JNK phosphorylation. Clones transfected with pcDNA3 (PV6) or pcDNA3-GILZ (GIRL-19) were stimulated for 5 or 60 min with plastic-bound anti-CD3 MAb. Whole-cell lysates were probed with an antibody specific for phosphorylated ERK-1/2 (pERK-1/2) (A), MEK-1/2 (pMEK) (B), or Raf-1 (pRaf-1) (C). Western blots were also performed with anti-ERK-1/2, anti-MEK-1/2, or anti-Raf-1 antibodies to verify that no modulation of protein expression occurred or with β-tubulin to verify that an equivalent amount of proteins was loaded in each lane. PV6 or GIRL-19 was stimulated for the times indicated with plastic-bound anti-CD3 MAb. (D) Whole-cell lysates were probed with an antibody recognizing both phosphorylated forms of JNK: p54 and p46 (pSAPK/JNK). C, untreated cells.
On the other hand JNK, which controls Jun phosphorylation and transcription and whose activation is under the control of the stress-activated MAPK pathway (27), was already phosphorylated in nonstimulated clones (both pcDNA3-transfected and pcDNA3-GILZ-transfected clones). Anti-CD3 MAb triggering did not augment JNK phosphorylation, which, on the contrary, decreased over time (Fig. 3D).
These data indicate that GILZ inhibited ERK-1/2 phosphorylation and the consequent c-Fos transcription and strongly suggest that the failure to synthesize Fos proteins could partly contribute to impair formation of AP-1 heterodimeric complexes and, consequently, AP-1 transactivation.
GILZ interferes with Raf activation by a protein-to-protein interaction mechanism.
It has been shown that the treatment of mast cells with DEX blocked the phosphorylation of Raf-1, MEK, and ERK-2 without affecting Ras activation (53). Since protein-to-protein interaction may have important consequences with regard to protein phosphorylation, activation, and trafficking, we speculated that GILZ could bind proteins of the cascade of MAPKs and inhibit their activation.
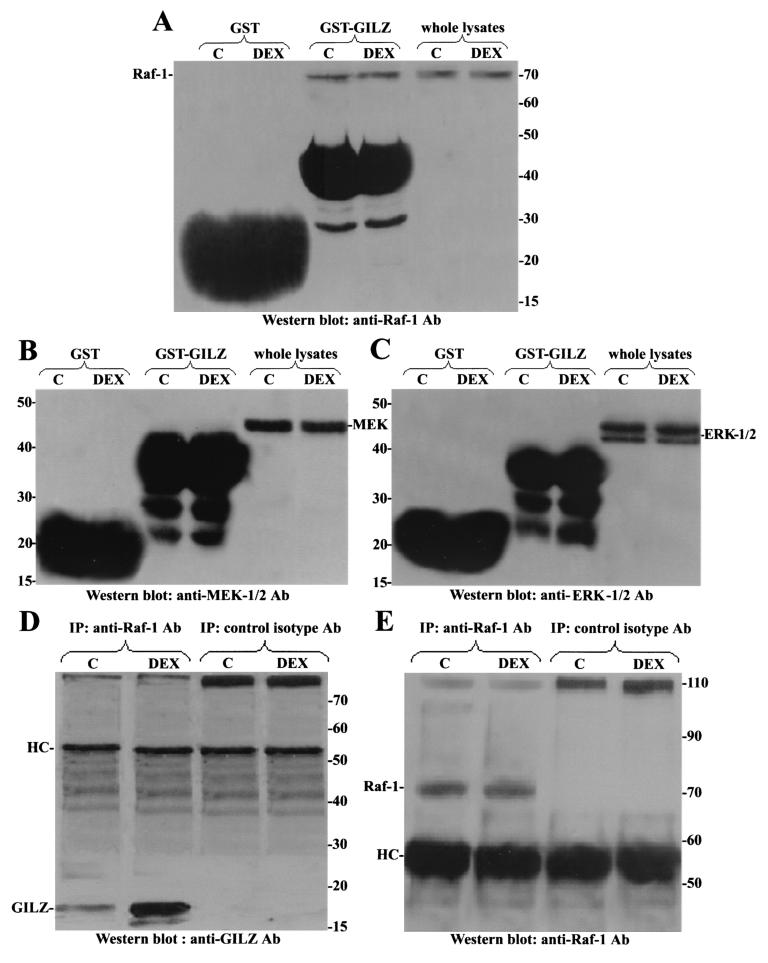
These hypotheses were first experimentally addressed by using murine thymocytes, which upregulate GILZ upon DEX stimulation (11), thus providing a model with the proteins expressed at physiological levels. Murine thymocytes were treated for 6 h with DEX, and cellular lysates were incubated with immobilized GST-GILZ fusion protein. After a washing, the composition of the complex was examined by Western blotting with anti-Raf-1, anti-MEK-1/2, or anti-ERK-1/2 antibody. Figure 4 shows that GST-GILZ fusion protein bound endogenous Raf (Fig. 4A) but not endogenous MEK (Fig. 4B) or ERK (Fig. 4C), suggesting GILZ-Raf-1 interaction, and additional studies examined this possibility further. Thymocyte cell lysates were, in fact, immunoprecipitated with an antibody recognizing Raf-1 protein. Figure 4 shows that anti-Raf antibody, but not a control isotype antibody, immunoprecipitated Raf-1 from untreated and DEX-treated thymocytes (Fig. 4E) but coimmunoprecipitated GILZ mainly from DEX-treated thymocytes (Fig. 4D) in which GILZ was upregulated (11).
FIG. 4.
GILZ interacts with endogenous Raf-1 in mouse thymocytes. Mouse thymocytes were treated for 6 h with DEX (100 nM), and cell lysates were incubated with GST or GST-GILZ beads. Binding of Raf-1 (A), MEK-1/2 (B), and ERK-1/2 (C) was visualized by Western blotting. Whole-cell lysates from thymocytes left untreated or treated with DEX were immunoprecipitated with an anti-Raf-1 or control isotype antibody (4 μg/500 μg of protein). (D and E) Nitrocellulose membrane was probed with an anti-GILZ antiserum (D) and then stripped and reprobed with anti-Raf-1 antibody (E).
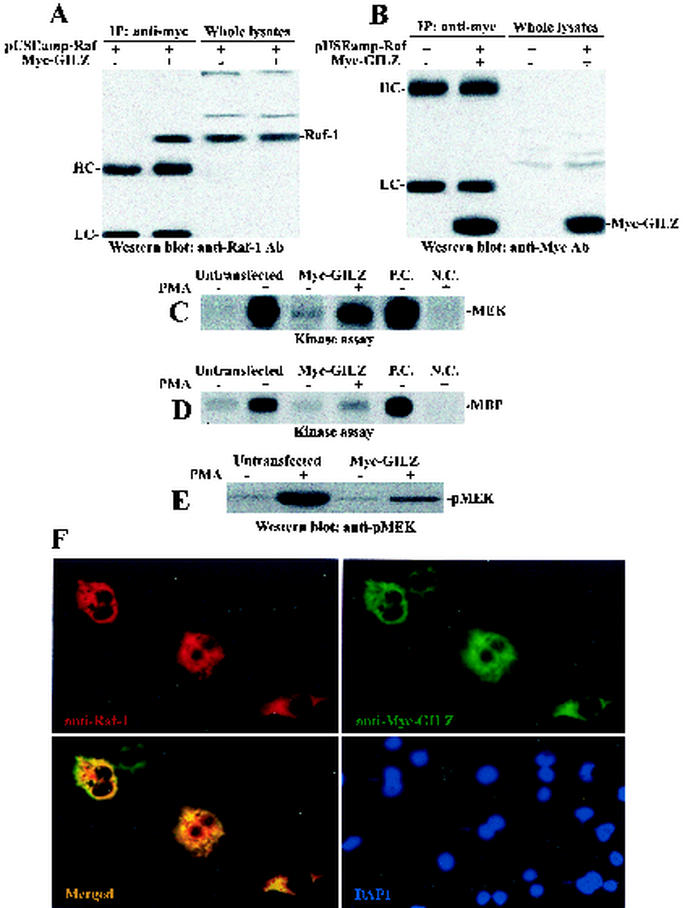
The GILZ-Raf-1 interaction was then examined in COS-7 cells cotransfected with myc-tagged GILZ (myc-GILZ) vector, along with an expression vector carrying Raf-1 (pUSEamp-Raf). Figure 5 shows that anti-myc antibody immunoprecipitates (Fig. 5B) contained immunoreactive Raf-1 in the lysates from cells cotransfected with myc-GILZ and pUSEamp-Raf but not in cells transfected with pUSEamp-Raf alone (Fig. 5A). Whole-cell lysates were analyzed to compare transfected- protein levels.
FIG. 5.
GILZ interacts with Raf-1 in COS-7-transfected cells. COS-7 cells were cotransfected with pUSEamp-Raf-1 (2 μg) and myc-GILZ (2 μg) vectors. Immunoprecipitation was performed with anti-myc antibody (3 μg/500 μg of protein), and immunoreactive proteins were visualized with anti-Raf-1 (A) or anti-myc (B) antibodies. Whole-cell lysates were loaded to control GILZ and Raf-1 expression. Serum-starved COS-7 cells, either untransfected or transfected with myc-GILZ, were treated for 15 min with PMA (10 ng/ml). Raf-1 immunoprecipitates were analyzed for kinase activity in the presence of [γ-32P]ATP by using GST-MEK (C) or GST-MEK, GST-ERK, and MBP (D) as substrates. P.C., positive control performed with 10 U of purified Raf-1 kinase; N.C., negative control performed with Raf-1 immunocomplex from PMA-stimulated cells in the absence of MEK substrate. (E) MEK phosphorylation was also assayed by Western blot with an antibody specific for phosphorylated MEK (pMEK). myc-GILZ-Raf-cotransfected COS-7 cells were immunostained with anti-myc and anti-Raf antibodies. Single staining and superimposed images are shown. (F) Nuclei were visualized by DAPI staining.
To further investigate the impact of GILZ-Raf-1 interaction, we tested whether GILZ interferes with Raf-1 kinase activity. COS-7 cells were left untransfected or were transfected with myc-GILZ and activated with PMA for 15 min. Endogenous Raf-1 was recovered by immunoprecipitation with anti-Raf-1 antibody and assayed for MEK and MBP phosphorylation in the kinase assays. Figure 5C and D shows that the kinase activity of Raf-1 was severely impaired in the myc-GILZ-transfected cells in which transfected GILZ bound endogenous Raf-1 (not shown). The same results were obtained when MEK phosphorylation was tested by Western blotting with anti-pMEK-1/2 antibody (Fig. 5E). Finally, fluorescent microscopy revealed extensive colocalization between Raf-1 and GILZ in myc-GILZ-Raf-cotransfected cells (Fig. 5F).
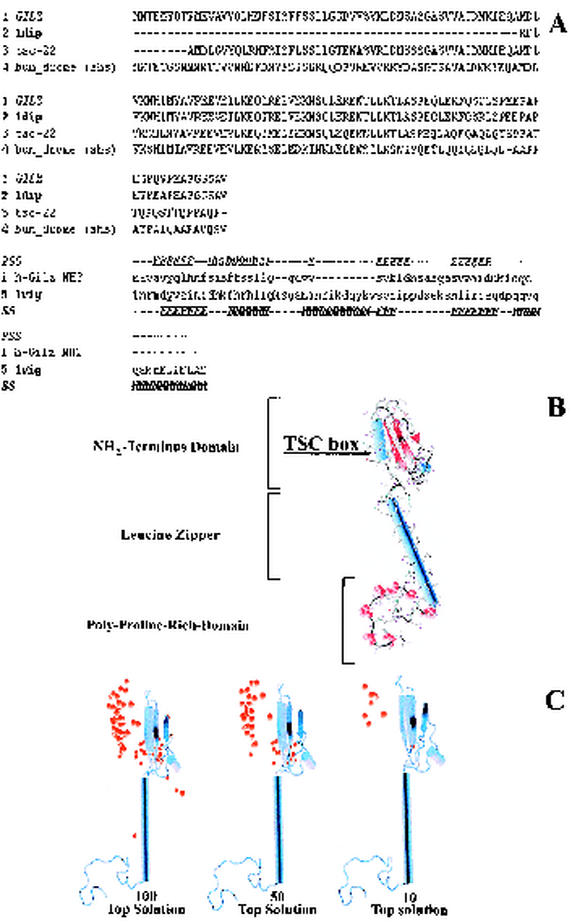
Molecular modeling.
To map the GILZ-Raf-1 binding domains, we constructed a 3D model of human GILZ by the modeling computer analysis and performed docking experiments between GILZ and Raf.
(i) Homology modeling of GILZ.
A 3D model of human-GILZ was constructed by using comparative homology modeling and threading approaches. In particular, the structure of DIP, a member of the TSC family, was used as a template to build the TSC box, leucine zipper, and the C-terminal domains of GILZ (residues 58 to 134). The sequences of GILZ and DIP were aligned by using the module Align123 of Insight II (Fig. 6A).
FIG. 6.
(A) GILZ sequence alignment with a set of TSC family proteins (upper part of diagram) and alignment of NH2-terminal sequence of GILZ to 1vig as proposed by threading server (3DPSSM) (lower part of diagram). PSS indicates the predicted secondary structure for the N-terminal domain. SS indicates the known secondary structure of the library template 1vig. (B) 3D model of human GILZ. α-Helices are represented as light blue cylinders, β-sheets are represented as red ribbons, and proline residues are indicated in red CPK. (C) Molecular modeling of Raf-1-GILZ interaction. Distribution of solutions (i.e., the 100, 50, and 10 top solutions found) of docking experiments of Raf-1 and GILZ; the red balls indicate the center of mass of Raf-1, for each solution, interacting with GILZ.
All methods used (see Materials and Methods) identified the solution 1vig (structure of human vigilin) in the best 10 solutions. Thus, the sequences of 1vig and of the amino-terminal domain of GILZ were aligned as proposed by threading servers and a 3D model of this domain was built by using Modeler (Fig. 6B). The model was refined with several cycles of conjugate gradient minimization until a gradient convergence of 0.05 kcal/mol was reached.
To assemble the whole GILZ protein, the NH2-terminal domain was docked on the remaining part of GILZ. Briefly, residues belonging to the TSC box of GILZ (residues 58 to 68) were added to the model of the NH2-terminal domain by assigning to them the coordinates of the residues constituting the last 1vig helix. Thus, the NH2-terminal domain was docked by fitting its TSC box helix on the respective one of GILZ and taking into account the different spatial dispositions of the TSC box found in the nuclear magnetic resonance conformations of 1dip. Each of the resulting models was evaluated in terms of docked energy. The one with the lowest energy (TSC box disposition of conformer 3 of 1dip) was stored and submitted to energy minimization cycles of conjugate gradient. The minimization cycles were stopped when a convergence gradient of 0.05 kcal/mol was reached. All minimization protocols were carried out by using the Charmm22 force field and by using a distance-dependent function of 1 for the dielectric constant. The geometric accuracy of all models was checked by using the Procheck and Verify3D programs (38, 45).
(ii) Docking experiments of GILZ and Raf-1.
Docking experiments between GILZ and Raf-1 suggested that in all of the top 10 solutions Raf-1 interacted through its RBD with the NH2-terminal domain of GILZ (Fig. 6C).
GILZ interacts with the RBD of Raf through its NH2-terminal domain.
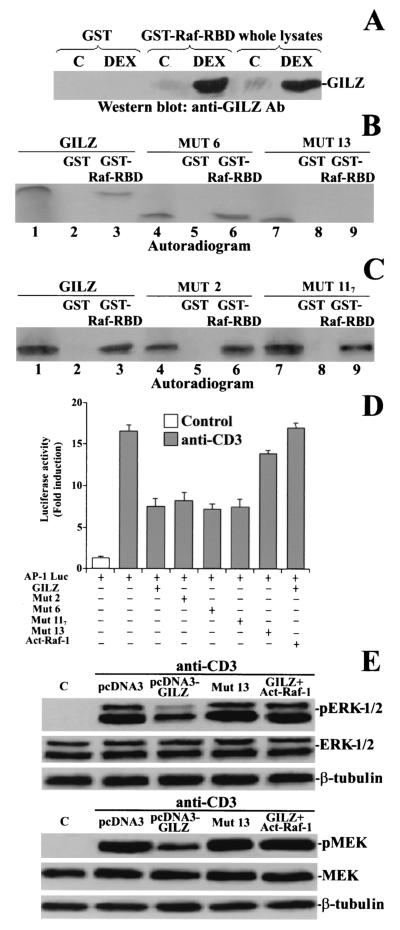
To provide experimental evidence in support of the modeling hypothesis, we used a GST-Raf fusion protein containing a truncated form of Raf (residues 1 to 149) with the binding motif for Ras, the RBD (residues 51 to 131). Protein lysates from untreated and DEX-treated thymocytes were incubated with immobilized GST-Raf-RBD fusion protein and the blotted nitrocellulose membrane was probed with anti-GILZ antibody. Immunoreactive GILZ was retained by immobilized GST-Raf-RBD mainly in DEX-treated thymocytes (Fig. 7A ). Moreover, no aspecific binding was detected with GST-protein alone. As expected, a protein identical in size and immunoreactivity was found in total lysates from DEX-treated thymocytes (Fig. 7A). These results suggest that Raf-1 may interact with GILZ by the same part of the molecule binding Ras.
FIG. 7.
The GST-Raf-RBD interacts with the GILZ amino-terminal region. (A) GST pulldown was performed with GST-Raf-RBD fusion protein corresponding to the human RBD (residues 1 to 149) of Raf or GST alone, attached to glutathione-Sepharose beads as bait and whole-cell lysates from untreated and DEX-treated thymocytes. The membrane was probed with anti-GILZ antiserum. Total lysates from DEX-treated and untreated thymocytes were loaded to control GILZ expression. (B) GST-Raf-RBD fusion protein was incubated for 18 h with the 35S-labeled in vitro-transcribed and translated proteins GILZ (lane 3), mutant 6 (lane 6), and mutant 13 (lane 9). Lane 1, GILZ; lane 2, GST plus GILZ; lane 4, mutant 6; lane 5, GST plus mutant 6; lane 7, mutant 13; lane 8, GST plus mutant 13. (C) GST-Raf-RBD fusion protein was incubated for 18 h with the 35S-labeled in vitro-transcribed and -translated protein GILZ (lane 3) or mutant 2 (lane 6) or mutant 117 (line 9). Lane 1, GILZ; lane 2, GST plus GILZ; lane 4, mutant 2; lane 5, GST plus mutant 2; lane 7, mutant 117; lane 8, GST plus mutant 117. (D) 3DO cells were transfected with the AP-1 luciferase reporter gene, along with GILZ, mutant 2, mutant 6, mutant 117, mutant 13, or GILZ plus activated Raf-1, and then stimulated for 18 h with plastic-bound anti-CD3 MAb. The values are expressed as the increase (n-fold) of luciferase activity compared to that in unstimulated cells. The values of transfected control groups are comparable; only one of them is shown in the figure. Each transfection was performed in triplicate. The standard errors were < 10%. (E) 3DO cells, transfected with the indicated vectors, were stimulated for 1 h with anti-CD3 MAb (1 μg/ml). Phosphorylation of ERK and MEK was visualized by Western blotting.
Since the modeling analysis suggested the N-terminal domain of GILZ as the site of interaction with Raf-RBD, we performed in vitro GST pulldown experiments by using GST-Raf-RBD fusion protein as bait with in vitro-translated GILZ or the deleted forms of GILZ (mutants 6 and 13), which lack, respectively, the COOH-terminal proline-rich and the NH2-terminal domains. Figure 7B shows that the GST-Raf-RBD fusion protein, but not GST alone, bound the GILZ full-length protein (line 3) and the GILZ lacking the COOH-terminal proline-rich region (mutant 6, lane 6) but did not interact with GILZ lacking the NH2-terminal region (mutant 13, lane 9). These results concur with the molecular modeling hypothesis and indicate that Raf-1 binds, through its RBD, the NH2-terminal domain of GILZ.
Finally, since GILZ, like other leucine zipper proteins, could homodimerize by means of its leucine zipper domain (42), we determined whether the homodimeric form of GILZ was required for binding to Raf-1. As shown in Fig. 7C, GST-Raf-RBD retained both in vitro-translated full-length GILZ and in vitro-translated mutants 2 and 117, which are unable to homodimerize (B. Di Marco and C. Riccardi, unpublished data), suggesting that GILZ binds Raf-1 independently of its state of homodimerization.
The functional meaning of these interactions was tested by determining the inhibitory effects of each mutant on the luciferase activity of the AP-1-dependent reporter gene. As shown in Fig. 7D, the luciferase activity, induced by anti-CD3 treatment, was reduced by cotransfection of GILZ or GILZ mutants 2, 6, and 117, all of which are able to bind Raf-1, but the activity was virtually unaffected by the coexpression of mutant 13, which did not interact with Raf-1. These data suggest that the GILZ-Raf-1 interaction modulates AP-1 transcriptional activity.
If GILZ-Raf-1 binding, in sequestering Raf-1 and consequently in inhibiting its activation, were responsible for the impaired AP-1 transcriptional activity, the overexpression of activated Raf would bypass the inhibitory effect. To test this hypothesis, activated Raf-1 was cotransfected, along with GILZ and the AP-1-dependent reporter gene. Figure 7D shows that activated Raf-1 reversed GILZ inhibitory effect on the induction of AP-1 transcriptional activity. Similarly, activated Raf-1 overexpression bypassed the GILZ inhibitory effect on ERK-1/2 and MEK phosphorylation (Fig. 7E). The overexpression of mutant 13 did not inhibit ERK and MEK phosphorylation (Fig. 7E).
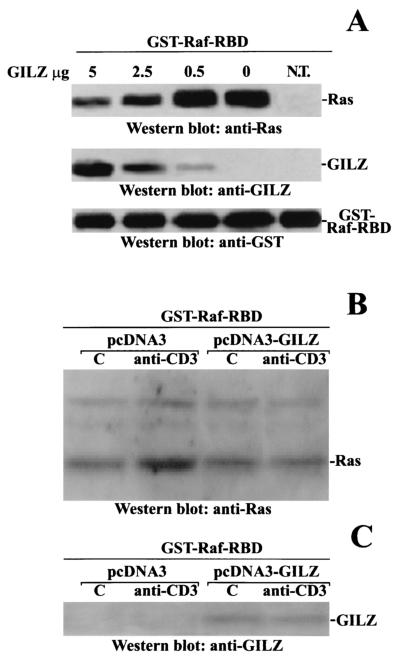
These results indicate that the inhibitory effect of GILZ on MEK-ERK activation depends on GILZ binding to Raf-1 and inhibiting its activation. Therefore, we analyzed the role of GILZ on the Ras-induced Raf-1 activation. We determined whether GILZ competes with Ras for binding to Raf-1, thus interfering with the formation of Ras-Raf-1 complexes. We expressed activated Ras in COS-7 cells and used the GST-Raf-RBD fusion protein as bait in a binding assay. The GST-Raf-RBD-Ras complex was incubated with increasing amounts of purified GILZ. After being washed, the complex was analyzed by Western blotting. The addition of GILZ resulted in GILZ binding and a concomitant displacement of activated Ras from GST-Raf-RBD beads, suggesting that GILZ interferes in Ras-Raf complex formation (Fig. 8A). This may explain the decrease of GST-Raf-RBD-Ras complex in anti-CD3-activated clone overexpressing GILZ when GST-Raf-RBD was used to evaluate the Ras activation (Fig. 8B). This effect might be due to competitive inhibition of GILZ with Ras for binding to Raf-1 (Fig. 8C) rather than to a real decrease in the Ras-GTP level.
FIG. 8.
GILZ interferes with Ras-Raf-1 complex. (A) Activated Ras was expressed in COS-7 cells, and 100 μg of cellular extracts was incubated for 1 h with 2.5 μg of GST-Raf-RBD. The complex was purified by adsorption to glutathione-Sepharose beads, washed, and resuspended in PBS. Purified GILZ was added at the concentrations indicated. After 1 h at 4°C, the GST-Raf-RBD beads were washed and examined for associated proteins by Western blotting with anti-Ras, anti-GILZ, and anti-GST antibodies. N.T., nontransfected cells. Clones transfected with pcDNA3 (PV6) or pcDNA3-GILZ (GIRL-19) were stimulated for 1 h with plastic-bound anti-CD3 MAb. A total of 200 μg of cellular extracts was incubated for 1 h with 2.5 μg of GST-Raf-RBD. (B) Western blotting was performed with anti-Ras antibody. (C) The nitrocellulose membrane was then stripped and reprobed with anti-GILZ antibody.
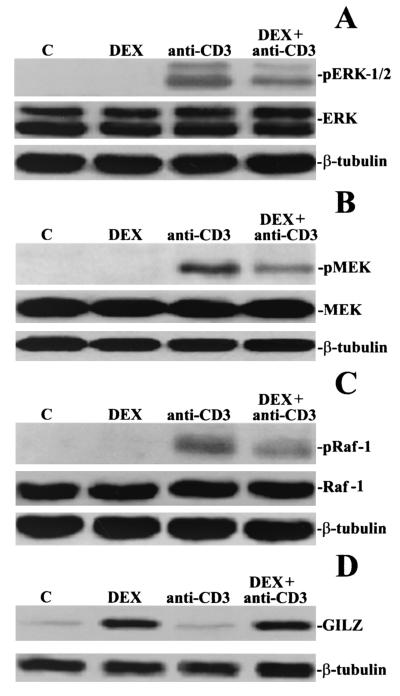
The biological relevance of these data was evaluated by determining whether the induction of GILZ by DEX correlates with the inhibition of anti-CD3-induced signaling. Murine thymocytes were stimulated for 6 h with DEX and then for different times with anti-CD3 MAb (from 30 min to 2 h). Cellular extracts were separated on SDS gels, and MAPK phosphorylation was assayed with the appropriate antibodies. Figure 9 shows that the anti-CD3-activated phosphorylation (1 h) of Raf-1, MEK, and ERK-1/2 was reduced by DEX pretreatment. Similar results were obtained at different times of anti-CD3 MAb treatment (not shown). Overall, these data were consistent with GILZ being a regulatory molecule of the Raf-1/MEK/ERK pathway.
FIG. 9.
DEX inhibits Raf-1, MEK, and ERK-1/2 phosphorylation. Mouse thymocytes left untreated or pretreated for 6 h with DEX (100 nM) were stimulated for 1 h with plastic-bound anti-CD3 MAb. Western blotting was performed with an anti-pERK-1/2 (A), anti-pMEK-1/2 (B), anti-pRaf-1 (C), or anti-GILZ (D) antibody. Western blotting was also performed with an anti-ERK-1/2, anti-MEK-1/2, or anti-Raf-1 antibody to verify that no modulation of protein expression occurred or with β-tubulin to verify that equivalent amounts of proteins were loaded in all lanes.
DISCUSSION
The data presented here indicate that GILZ, by binding to Raf-1, inhibits the MAPK pathway and suggest a new mechanism for GCH transcriptional control of this signaling circuitry.
T-cell activation is characterized by the release of a series of lymphokines that regulate apoptosis, T-cell proliferation, and clonal expansion (16, 20, 25, 59). GCH inhibition of T-cell activation and the consequent immunosuppression are achieved through a combination of genomic and nongenomic mechanisms and also through interference with the MAPK pathway (1, 8, 50, 51, 53, 55, 56, 62). In fact, interaction between GR and MAPK pathway components has been evoked to explain impaired IL-2 gene transcription and the consequent inhibition of cell activation and proliferation in different cells and physiologic settings. For example, recent evidence suggests that Raf-1, 14-3-3, and GR coimmunoprecipitate within the same protein complex in the cytoplasm of rat liver cells, and this provides a plausible explanation of the GCH effect on the Ras-Raf-1 signaling pathway (62). Moreover, other studies with mast cells suggest that DEX inhibits activation of Raf-1, MEK, and ERK but does not affect Ras activation (53). This effect is due to disruption of the Raf-1 multimeric complex without affecting proteins, such as 14-3-3, in this complex. According to this model, the loss of DEX-induced Hsp90 from the Raf-1 complex correlates with inhibition of antigen-stimulated Raf-1 activity and suggests that a loss of Hsp90 and the impairment of Raf activation are linked (8). In addition, GCH increase the expression of MAPK phosphatase 1, and this effect is necessary for GCH-mediated inhibition of ERK-1/2 activation (33). Finally, GR directly binds AP-1, thus inhibiting one of the final steps in the MAPK cascade (56).
In the present study, we demonstrated that an indirect mechanism involving GILZ is responsible for GCH-mediated inhibition of the MAPK pathway, which could then contribute to inhibit T-cell activation. GILZ overexpression in 3DO clones suppressed the Raf pathway and, consequently, interfered with the c-Fos transcription. A direct protein-to-protein interaction, occurring both in vitro and in vivo (Fig. 4, 5, and 7), may be responsible for this inhibitory mechanism. In fact, GILZ fusion protein bound Raf, but not MEK and ERK, in cellular lysates from murine thymocytes and antibody anti-Raf coimmunoprecipitated GILZ, especially in DEX-treated mouse thymocytes (Fig. 4).
Based on several protein motifs, it has been suggested that GILZ belongs to a TSC family (35) of leucine zipper proteins comprising at least five other members (TSC-22, THG-1, KIAA0669, DIP, and shc), which can potentially homodimerize or heterodimerize by means of its leucine zipper pattern. All members of the TSC family share a high degree of homology in the dimerization domain (TSC-22 box and leucine zipper pattern) but have different N-terminal and C-terminal domains. Experimental evidence supports the hypothesis that TSC-22 and THG repressor activity resides in the unconserved N- and C-terminal domains (35). Although GILZ shares a proline-rich region in the C-terminal domain with TSC-22 and THG-1, these domains may have totally different functions. Molecular modeling analysis (Fig. 6) and experimental evidence suggested that GILZ interacts through its NH2-terminal domain with the NH2-terminal portion of Raf corresponding to the Ras-binding side. In fact, the GST-Raf-RBD fusion protein was able to bind GILZ in vitro (Fig. 7B) and in vivo in murine thymocytes (Fig. 7A). Deletion of the GILZ N-terminal region, but not of the proline-rich region, resulted in complete abrogation of binding to Raf-RBD and inhibition of AP-1 transcriptional activity (Fig. 7B and D). In contrast, the homodimerization state of GILZ was not required to bind Raf-1 (Fig. 7C).
Raf is known to bind directly to the GTP-bound form of Ras (13). This Raf-Ras-GTP interaction does not, however, result directly in Raf activation. In fact, the Raf-Ras interaction is required for Raf recruitment and translocation from cytosol to plasma membrane (46). Furthermore, Raf-1 bound in the cytosol to 14-3-3 protein, an arachidonate-selective acyltransferase and putative phospholipase A2. This protein associates with a number of key signaling proteins and cell cycle regulators; its role in MAPK activation is permissive for Raf-1 recruitment and activation, even though it is totally displaced when Ras recruits Raf to the plasma membrane (39). Furthermore, Raf-1 kinase activity and function are regulated both positively and negatively by phosphorylation and dephosphorylation signals (7, 12). If Raf binds GILZ by using the same portion of the molecule that interacts with activated Ras, this raises the question of competition between GILZ and Ras for Raf binding. In fact, one hypothesis might be that GILZ binding Raf creates a steric obstacle for the Ras-Raf association and the consequent activation and recruitment of Raf in the cell membrane that could explain its diminished phosphorylation. Our data suggest that a steric interference of GILZ with Raf-Ras binding is possible (Fig. 8). However, we cannot rule out the possibility that GILZ, rather than acting as a direct competitor for Raf-Ras binding, reduces the affinity of Raf for Ras and again induces an impaired Raf translocation to the cell membrane and consequently diminishes phosphorylation.
Whatever mechanism is triggered by the GILZ-Raf interaction, it is important to note that it inhibits Raf, MEK, and ERK activation and consequently impairs AP-1 activation.
Another mechanism has been proposed to account for the GILZ-induced defect in AP-1 activation. The in vitro interaction of recombinant GILZ with c-Fos and c-Jun inhibits AP-1 binding to its target DNA (42). Although we obtained the same results (not shown), we demonstrate here that this was not the only mechanism of GILZ-induced AP-1 inhibition. The coexistence of multiple mechanisms is common to many biological systems where more than one molecular event can control any one pathway. In the case of GILZ, the redundancy may be due to the need to block the Raf-ERK cascade in order to inhibit not only c-Fos transcription but also the activity of other transcription factors controlled by this pathway. Were this the case, GILZ, as mediator of GCH immunosuppressive and anti-inflammatory activities, would have a larger spectrum of action.
The lack of IL-2 production, which was due, at least in part, to the block of MAPK driven by GILZ overexpression, resembles the state of functional unresponsiveness with impaired IL-2 production and Ras activation, a characteristic of anergic T cells (32, 43, 57). Anergy, one of the responses of the immunity system to continuous antigen challenge, is also one of the T-cell responses to exogenous GCH (10, 49, 52). Therefore, the observation that GILZ triggers the same molecular events caused by GCH and/or repeated exposure to the antigen once again confirms the functional cross talk between TCRs and GRs at various levels of signaling events (30). Moreover, GILZ-mediated inhibition of the Raf-1 pathway suggests yet another mechanism accounting for the anti-inflammatory and immunosuppressive effects of GCH.
Acknowledgments
This work was supported by the Associazione Italiana Ricerca sul Cancro (AIRC).
REFERENCES
- 1.Auphan, N., J. A. DiDonato, C. Rosette, A. Helmberg, and M. Karin. 1995. Immunosuppression by glucocorticoids: inhibition of NF-κB activity through induction of IκB synthesis. Science 270:286-290. [DOI] [PubMed] [Google Scholar]
- 2.Ayroldi, E., G. Migliorati, S. Bruscoli, C. Marchetti, O. Zollo, L. Cannarile, F. D'Adamio, and C. Riccardi. 2001. Modulation of T-cell activation by the glucocorticoid-induced leucine zipper factor via inhibition of nuclear factor κB. Blood 98:743-753. [DOI] [PubMed] [Google Scholar]
- 3.Ayroldi, E., O. Zollo, L. Cannarile, F. D'Adamio, U. Grohmann, D. V. Delfino, and C. Riccardi. 1998. Interleukin-6 (IL-6) prevents activation-induced cell death: IL-2-independent inhibition of Fas/fasL expression and cell death. Blood 92:4212-4219. [PubMed] [Google Scholar]
- 4.Barnes, P. J., and I. Adcock. 1993. Anti-inflammatory actions of steroids: molecular mechanisms. Trends Pharmacol. Sci. 14:436-441. [DOI] [PubMed] [Google Scholar]
- 5.Beato, M. 1991. Transcriptional control by nuclear receptors. FASEB J. 5:2044-2051. [DOI] [PubMed] [Google Scholar]
- 6.Boise, L. H., B. Petryniak, X. Mao, C. H. June, C. Y. Wang, T. Lindsten, R. Bravo, K. Kovary, J. M. Leiden, and C. B. Thompson. 1993. The NFAT-1 DNA binding complex in activated T cells contains Fra-1 and JunB. Mol. Cell. Biol. 13:1911-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chong, H., J. Lee, and K. L. Guan. 2001. Positive and negative regulation of Raf kinase activity and function by phosphorylation. EMBO J. 14:3716-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cissel, D. S., and M. A. Beaven. 2000. Disruption of Raf-1/heat shock protein 90 complex and Raf signaling by dexamethasone in mast cells. J. Biol. Chem. 275:7066-7070. [DOI] [PubMed] [Google Scholar]
- 9.Coffer, P., M. De Jonge, A. Mettouchi, B. Binetruy, J. Ghysdael, and W. Kruijer. 1994. JunB promoter regulation: Ras-mediated transactivation by c-ets-1 and c-ets-2. Oncogene 9:911-921. [PubMed] [Google Scholar]
- 10.Cupps, T. R., and A. S. Fauci. 1982. Corticosteroid-mediated immunoregulation in man. Immunol. Rev. 165:133-155. [DOI] [PubMed] [Google Scholar]
- 11.D'Adamio, F., O. Zollo, R. Moraca, E. Ayroldi, S. Bruscoli, A. Bartoli, L. Cannarile, G. Migliorati, and C. Riccardi. 1997. A new dexamethasone-induced gene of the leucine zipper family protects T lymphocytes from TCR/CD3-activated cell death. Immunity 7:803-812. [DOI] [PubMed] [Google Scholar]
- 12.de Rooij, J., and J. L. Bos. 1997. Minimal Ras-binding domain of Raf1 can be used as an activation-specific probe for Ras. Oncogene 14:623-625. [DOI] [PubMed] [Google Scholar]
- 13.Dhillon, A. S., S. Meikle, Z. Yazici, M. Eulitz, and W. Kolch. 2002. Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J. 21:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diamond, M. I., J. N. Minner, S. K. Yoshinaga, and K. R. Yamamoto. 1990. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science 249:1266-1272. [DOI] [PubMed] [Google Scholar]
- 15.Downward, J., J. D. Graves, P. H. Warne, S. Rayter, and D. A. Cantrell. 1990. Stimulation of p21ras upon T-cell activation. Nature 346:719-723. [DOI] [PubMed] [Google Scholar]
- 16.Durand, D. B., J. P. Shaw, M. R. Bush, R. E. Replogle, R. Belagase, and G. R. Crabtree. 1988. Characterization of antigen receptor response elements within the interleukin-2 enhancer. Mol. Cell. Biol. 8:1715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer, D., and D. Eisenberg. 1996. Protein fold recognition using sequence-derived predictions. Protein Sci. 5:947-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franklin, R. A., A. Tordai, H. Patel, A. M. Gardner, G. L. Johnson, and E. W. Gelfand. 1994. Ligation of the T-cell receptor complex results in activation of the Ras/Raf-1/MEK/MAPK cascade in human T lymphocytes. J. Clin. Investig. 93:2134-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabb, H. A., R. M. Jackson, and M. J. Sternberg. 1997. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J. Mol. Biol. 272:106-120. [DOI] [PubMed] [Google Scholar]
- 20.Garrity, P. A., D. Chen, E. V. Rothenberg, and B. J. Wold. 1994. Interleukin-2 transcription is regulated in vivo at the level of coordinated binding of both constitutive and regulated factors. Mol. Cell. Biol. 14:2159-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibrat, J. F., J. Garnier, and B. Robson. 1987. Further developments of protein secondary structure prediction using information theory: new parameters and consideration of residue pairs. J. Mol. Biol. 198:425-443. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman, G. S. 1993. Immunosuppressive therapy for autoimmune diseases. Ann. Allergy 70:263-274. [PubMed] [Google Scholar]
- 23.Huber, T., and A. E. Torda. 1998. Protein fold recognition without Boltzmann statistics or explicit physical basis. Protein Sci. 7:142-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber, T., and A. E. Torda. 1999. Protein sequence threading, the alignment problem, and a two-step strategy. J. Comput. Chem. 20:1455-1467. [Google Scholar]
- 25.Hughes, C. C., and J. S. Pober. 1996. Transcriptional regulation of the interleukin-2 gene in normal human peripheral blood T cells. Convergence of costimulatory signals and differences from transformed T cells. J. Biol. Chem. 271:5369-5377. [DOI] [PubMed] [Google Scholar]
- 26.Iwata, M., S. Hanaoka, and K. Sato. 1991. Rescue of thymocytes and T-cell hybridomas from glucocorticoid-induced apoptosis by stimulation via the T-cell receptor/CD3 complex: a possible in vitro model for positive selection of the T-cell repertoire. Eur. J. Immunol. 21:643-648. [DOI] [PubMed] [Google Scholar]
- 27.Jacinto, E., G. Werlen, and M. Karin. 1998. Cooperation between Syk and Rac1 leads to synergistic JNK activation in T lymphocytes. Immunity 8:31-41. [DOI] [PubMed] [Google Scholar]
- 28.Jain, J., C. Loh, and A. Rao. 1995. Transcriptional regulation of the IL-2 gene. Curr. Opin. Immunol. 7:333-342. [DOI] [PubMed] [Google Scholar]
- 29.Jain, J., V. E. Valge-Archer, and A. Rao. 1992. Analysis of the AP-1 sites in the IL-2 promoter. J. Immunol. 148:1240-1250. [PubMed] [Google Scholar]
- 30.Jamieson, C. A., and K. R. Yamamoto. 2000. Cross talk pathway for inhibition of gluccocorticoid-induced apoptosis by T-cell receptor signaling. Proc. Natl. Acad. Sci. USA 97:7319-7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabelitz, D., T. Pohl, and K. Pechhold. 1993. Activation-induced cell death (apoptosis) of mature peripheral T lymphocytes. Immunol. Today 14:338-339. [DOI] [PubMed] [Google Scholar]
- 32.Kang, S. M., B. Beverly, A. C. Tran, K. Brorson, R. H. Schwartz, and M. J. Lenardo. 1992. Transactivation by AP-1 is a molecular target of T-cell clonal anergy. Science 257:1134-1138. [DOI] [PubMed] [Google Scholar]
- 33.Kassel, O., A. Sancono, J. Kratzschmar, B. Kreft, M. Stassen, and A. C. Cato. 2001. Glucococorticoids inhibit MAP kinase via increased expression and decreased degradation of MKP-1. EMBO J. 20:7108-7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 35.Kester, H. A., C. Blanchetot, J. den Hertog, P. T. van der Saag, and B. van der Burg. 1999. Transforming growth factor-beta-stimulated clone-22 is a member of a family of leucine zipper proteins that can homo- and heterodimerize and has transcriptional repressor activity. J. Biol. Chem. 274:27439-27447. [DOI] [PubMed] [Google Scholar]
- 36.Kolch, W. 2000. Meaningful relationships: the regulation of the Ras/Raf/Mek/Erk pathway by protein interactions. Biochem. J. 351:289-305. [PMC free article] [PubMed] [Google Scholar]
- 37.Kwak, L. W., and D. L. Longo. 1996. Lymphomas, p. 347-375. In H. M. Pinedo, D. L. Longo, and B. A. Chabner (ed.), Cancer chemotherapy and biological response modifiers. Elsevier Science, Amsterdam, The Netherlands.
- 38.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 39.Luo, Z. J., X. F. Zhang, U. Rapp, and J. Avruch. 1995. Identification of the 14.3.3 ζ domains important for self-association and Raf binding. J. Biol. Chem. 40:23681-23687. [DOI] [PubMed] [Google Scholar]
- 40.Luthy, R., J. U. Bowie, and D. Eisenberg. 1992. Assessment of protein models with three-dimensional profiles. Nature 356:83-85. [DOI] [PubMed] [Google Scholar]
- 41.Maggirvar, S. B., E. W. Harhaj, and S. C. Sun. 1997. Regulation of the interleukin-2 CD28 responsive element by NF-ATp and various NF-κB/Rel transcription factors. Mol. Cell. Biol. 17:2605-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mittellstadt, P. R., and J. D. Ashwell. 2001. Inhibition of AP-1 by the glucocorticoid-inducible protein GILZ. J. Biol. Chem. 276:29603-29610. [DOI] [PubMed] [Google Scholar]
- 43.Mondino, A., C. D. Whaley, D. R. DeSilva, W. Li, M. K. Jenkins, and D. L. Mueller. 1996. Defective transcription of the IL-2 gene is associated with impaired expression of c-Fos, FosB, and JunB in anergic T helper 1 cells. J. Immunol. 157:2048-2057. [PubMed] [Google Scholar]
- 44.Moont, G., H. A. Gabb, and M. J. E. Sternberg. 1999. Use of pair potentials across protein interfaces in screening predicted docked complexes. Proteins 35:364-373. [PubMed] [Google Scholar]
- 45.Morris, A. L., M. W. MacArthur, E. G. Hutchinson, and J. M. Thornton. 1992. Stereochemical quality of protein structure coordinates. Proteins 12:345-364. [DOI] [PubMed] [Google Scholar]
- 46.Morrison, D. K., and R. E. Cutler. 1997. The complexity of Raf-1 regulation. Curr. Opin. Cell Biol. 9:174-179. [DOI] [PubMed] [Google Scholar]
- 47.Nassar, N., G. Horn, C. Herrmann, A. Scherer, F. McCormick, and A. Wittinghofer. 1995. The 2.2 Å crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature 375:554-560. [DOI] [PubMed] [Google Scholar]
- 48.Raff, M. C. 1992. Social controls on cell survival and cell death. Nature 356:397-400. [DOI] [PubMed] [Google Scholar]
- 49.Ramirez, F. 1998. Glucocorticoids induce a Th2 response in vitro. Dev. Immunol. 6:233-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ray, A., and K. E. Prefontaine. 1994. Physical association and functional antagonism between the p65 subunit of transcription factor NF-κB and the glucocorticoid receptor. Proc. Natl. Acad. Sci. USA 91:752-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riccardi, C., G. Cifone, and G. Migliorati. 1999. Glucocorticoid hormone-induced modulation of gene expression and regulation of T-cell death: role of GITR and GILZ, two dexamethasone-induced genes. Cell Death Differ. 12:1155-1163. [DOI] [PubMed] [Google Scholar]
- 52.Riccardi, C., O Zollo, G. Nocentini, S. Bruscoli, A. Bartoli, F. D'Adamio, L. Cannarile, D. Delfino, E. Ayroldi, and G. Migliorati. 2000. Glucocorticoid hormones in the regulation of the cell death. Therapie 55:165-169. [PubMed] [Google Scholar]
- 53.Rider, L. G., N. Hirasawa, F. Santini, and M. A. Beaven. 1996. Activation of the mitogen-activated protein kinase cascade is suppressed by low concentrations of dexamethasone in mast cells. J. Immunol. 157:2374-2380. [PubMed] [Google Scholar]
- 54.Rincon, M. 2001. MAP-kinase signaling pathways in T-cell. Curr. Opin. Immunol. 13:339-345. [DOI] [PubMed] [Google Scholar]
- 55.Scheinman, R. I., P. C. Cogswell, A. K. Lofquist, and A. S. Baldwin, Jr. 1995. Role of transcriptional activation of IkBα in mediation of immunosuppression by glucocorticoids. Science 270:283-286. [DOI] [PubMed] [Google Scholar]
- 56.Schüle, R., P. Rangarajan, S. Kliewer, L. I. Ransone, J. Bolado, N. Yang, I. M. Verma, and R. M. Evans. 1990. Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell 62:1217-1226. [DOI] [PubMed] [Google Scholar]
- 57.Schwartz, R. H. 1997. T-cell clonal anergy. Curr. Opin. Immunol. 9:351-357. [DOI] [PubMed] [Google Scholar]
- 58.Seidel, G., K. Adermann, T. Schindler, A. Ejchart, R. Jaenicke, W. G. Forssmann, and P. Rosch. 1997. Solution structure of porcine delta sleep-inducing peptide immunoreactive peptide A homolog of the shortsighted gene product. J. Biol. Chem. 272:30918-30927. [DOI] [PubMed] [Google Scholar]
- 59.Serfling, E., R. Barthelmas, I. Pleuffer, B. Schenk, S. Zarius, R. Swoboda, F. Mercurio, and M. Karin. 1989. Ubiquitous and lymphocyte-specific factors are involved in the induction of the mouse interleukin-2 gene in T lymphocytes. EMBO J. 8:465-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shimonkevitz, R., J. Kappler, P. Marrack, and H. Grey. 1983. Antigen recognition by H-2-restricted T cells. I. Cell-free antigen processing. J. Exp. Med. 158:303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whitehurst, C. E., and T. D. Geppert. 1996. MEK1 and the extracellular signal-regulated kinases are required for the stimulation of IL-2 gene transcription in T cells. J. Immunol. 156:1020-1029. [PubMed] [Google Scholar]
- 62.Widen, C., J. Zilliacus, J. A. Gustafsson, and A. C. Wikstrom. 2000. Glucocorticoid receptor interaction with 14-3-3 and Raf-1, a proposed mechanism for cross-talk of two signal transduction pathways. J. Biol. Chem. 275:39296-39301. [DOI] [PubMed] [Google Scholar]
- 63.Wlemik, P. H. 1996. Leukemia and myeloma, p. 347-375. In H. M. Pinedo, D. L. Longo, and B. A. Chabner (ed.), Cancer chemotherapy and biological response modifiers. Elsevier Science, Amsterdam, The Netherlands.