Abstract
Cyclin D1, the regulatory subunit for mid-G1 cyclin-dependent kinases, controls the expression of numerous cell cycle genes. A cyclic AMP-responsive element (CRE), located upstream of the cyclin D1 mRNA start site, integrates mitogenic signals that target the CRE-binding factor CREB, which can recruit the transcriptional coactivator CREB-binding protein (CBP). We describe an alternative mechanism for CREB-driven cyclin D1 induction that involves the ubiquitous POU domain protein Oct-1. In the breast cancer cell line MCF-7, overexpression of Oct-1 or its POU domain strongly increases transcriptional activation of cyclin D1 and GAL4 reporter genes that is specifically dependent upon CREB but independent of Oct-1 DNA binding. Gel retardation and chromatin immunoprecipitation assays confirm that POU forms a complex with CREB bound to the cyclin D1 CRE. In solution, CREB interaction with POU requires the CREB Q2 domain and, notably, occurs with CREB that is not phosphorylated on Ser 133. Accordingly, Oct-1 also potently enhances transcriptional activation mediated by a Ser133Ala CREB mutant. Oct-1/CREB synergy is not diminished by the adenovirus E1A 12S protein, a repressor of CBP coactivator function. In contrast, E1A strongly represses CBP-enhanced transactivation by CREB phosphorylated on Ser 133. Our observation that Oct-1 potentiates CREB-dependent cyclin D1 transcriptional activity independently of Ser 133 phosphorylation and E1A-sensitive coactivator function offers a new paradigm for the regulation of cyclin D1 induction by proliferative signals.
The D-type cyclins control progression through the G1 phase of the cell cycle (54) as the regulatory component in the cyclin D/cdk4-6 kinase complex. Mitogens stimulate the transient accumulation of cyclin D1 and thus cyclin/cdk kinase activity by acting at both the transcriptional and posttranscriptional levels (7, 40, 41). Enhanced cyclin D1 expression is a hallmark of many cancers, particularly mammary carcinoma, in which over 50% of primary breast tumors show increased levels of cyclin D1 protein (43). Consistent with this, mice engineered to overexpress cyclin D1 in mammary epithelium develop lethal mammary carcinoma (52). This appears to be directly linked to Ras signaling, since mice bearing a disrupted cyclin D1 locus are resistant to breast cancers induced by the neu and ras oncogenes (55). Thus, mitogenic signaling through cyclin D1 represents a key step in the control of mammary cell proliferation in vivo.
Transcriptional regulation of cyclin D1 can be reproduced in transient-transfection assays with a promoter fragment that spans the sequences from −973 to +110 relative to the mRNA initiation site (20). Depending upon the cell line used, from one to several elements in this region have been implicated in promoter control (2, 8, 17, 20, 23, 24, 33, 44, 49, 53). These include established or potential binding sites for the transcription factors AP1, Ets-1, NF-κB, SP-1, TCF/LEF, Oct-1, ATF-2, and CREB. Many of these factors are activated by mitogenic signals, although with kinetics that paradoxically does not correspond to those of cyclin D1 transcriptional induction. Thus, the true regulatory events governing cyclin D1 transcriptional control in mid-G1 have yet to be elucidated.
The cyclic AMP-responsive element (CRE), located upstream of the mRNA start site, has a key role in both basal and induced cyclin D1 expression (4, 8, 28, 33). The CRE-binding protein CREB is an essential component in its activity, either alone or in association with ATF-1 or ATF-2. Numerous signaling pathways lead to phosphorylation of the CREB/ATF complex, which in turn promotes recruitment of the coactivator CREB-binding protein (CBP) and thereby potentiates transcription (reviewed in reference 30). However, CREB phosphorylation peaks within an hour after mitogen stimulation and then progressively diminishes over several hours, i.e., prior to the window of cyclin D1 transcriptional activity. Moreover, overexpression of p300 but not CBP stimulates cyclin D1 reporter gene expression through the AP1 site (2). Thus, the CREB/CRE complex appears to function through an alternative mechanism in the cyclin D1 promoter.
We wondered whether the ubiquitous protein Oct-1 might cooperate with CREB in regulating cyclin D1 expression. Oct-1 has been implicated in transcriptional control of both housekeeping and tissue-specific genes, such as histone H2B, small nuclear RNAs, and immunoglobulins, primarily via its homeodomain (14, 32, 46). This domain spans amino acids 280 to 440 and contains a bipartite DNA binding motif termed the POU domain, which comprises two conserved regions, the POU-specific and POU-homeo domains (21, 47). Both contain a helix-turn-helix motif that mediates both DNA binding and protein-protein interactions with a wide range of basal and activating transcription factors (6, 16, 25, 27, 29, 35, 45, 56, 57).
Therefore, we investigated if Oct-1 might also cooperate with CREB to activate the cyclin D1 promoter in the breast cancer cell line MCF-7. Oct-1 strongly increases CREB-driven transcriptional induction of a cyclin D1 reporter gene, an effect that is dependent upon the POU domain but independent of the putative Oct-1 binding site in the cyclin D1 promoter. This synergy is specific for CREB and reflects an interaction between POU and CREB bound to the cyclin D1 CRE. Solution binding studies show that complexes contain unphosphorylated CREB and localize the interaction region in CREB to the Q2 transactivation domain near the C terminus. Consistent with these observations, functional cooperation between Oct-1 and CREB does not require CREB phosphorylation on Ser 133 and, accordingly, is independent of CBP, in direct contrast to CREB activation through protein kinase A (PKA)-mediated phosphorylation on Ser 133. These data reveal the existence of a novel cyclin D1 regulatory element that consists of an Oct-1-CREB complex bound to the CRE near the transcriptional start site in the cyclin D1 promoter. Notably, this complex functions independently of normal mitogenic signaling to CREB and offers new insights into the transcriptional regulation of the key cell cycle regulator cyclin D1.
MATERIALS AND METHODS
Construction of reporter genes and expression vectors.
The human cyclin D1 promoter deletions and mutations were engineered by PCR and cloned upstream of the luciferase reporter gene in pXP2. The GAL4-Luc reporter plasmid was described previously (22) (provided by J. Licht). All constructs and mutations were confirmed by sequencing. The expression vectors encoding full-length Oct-1 or the Pou domain (46) (provided by W. Herr), ATF-1, full-length CREB, dominant negative A-CREB (provided by C. Vinson) (1), and mutant CREB S133A (15) (provided by B. Lutz) in pCDNA3 have been described previously. CREB deletion mutants were generated by PCR and cloned in pCDNA3. The Rous sarcoma virus-wild-type CBP (RSV-CBPwt) expression vector was kindly provided by R. Kwok. Cytomegalovirus (CMV) E1A 12S (51), GAL4-CREB, GAL4-CREB S133A (22) (provided by J. Licht), and GAL4-Nur77 (37) have been described previously. The PKA expression vector was obtained from Stratagene (Amsterdam, The Netherlands).
Reporter assays and cell culture.
Human MCF-7 breast cancer cells (obtained from D. Chalbos, INSERM U540, Montpellier, France) were maintained in Dulbecco's modified Eagle's medium with 10% (vol/vol) fetal calf serum and 1% penicillin-streptomycin. Prior to transfection, MCF-7 cells were seeded at a density of 106 cells/well in six-well dishes. In transient-expression studies, cells were transfected with the FuGEN6 transfection reagent (Roche Diagnostics, Mannheim, Germany) as described by the manufacturer. Transfections were performed with a constant ratio of 1 μg of DNA to 3 μl of FuGEN6. The transfection mixes contained 400 ng of luciferase reporter construct, 100 ng of expression vector, 100 to 300 ng of A-CREB expression vector, 20 ng of PKA expression vector, 150 to 300 ng of E1A 12S expression vector, and 1 ng of CMV-Renilla luciferase or 10 ng of Rous sarcoma virus-Renilla luciferase reporter control plasmid (Promega). The culture medium was changed after 6 h, and luciferase activity was determined after an additional 24 h.
The effect of an expression vector was compared with the effect of an equal amount of vector cassette. Treatment with forskolin (25 μM) was performed as indicated in the legend to Fig. 5, and results were compared with dimethyl sulfoxide vehicle treatment. Luciferase activity was determined following the protocol supplied with the dual luciferase kit (Promega) with an AutoLumat LB 950 (EG&G Berthold, Bad Willdbad, Germany). The luciferase measurements are represented as the mean ± standard error of the mean of three experiments, each performed in duplicate.
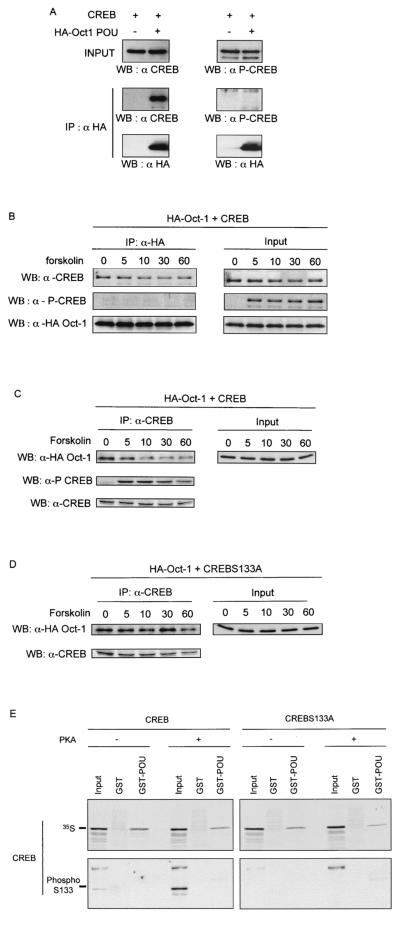
FIG. 5.
Oct-1 interacts with unphosphorylated CREB. (A) MCF-7 cells were transfected with pCMV-HA-POU and pCMV-CREB as indicated. HA-Oct-1 POU was immunoprecipitated (IP) from total cell lysates with anti-HA polyclonal antiserum (HA). Binding of CREB and phospho-CREB was detected by immunoblotting (WB) with anti-CREB (α CREB) or anti-P-Ser 133-CREB (α P-CREB) polyclonal antiserum. (B) The same experiment as described for A except that cells were induced with forskolin for the indicated times (in minutes) prior to cell lysis. (C) The same experiment as in B except that the immunoprecipitation (IP) was performed with anti-CREB polyclonal antiserum and Oct-1 binding was revealed by immunoblotting with anti-HA polyclonal antiserum (WB). (D) Identical to C except that anexpression vector for the S133A CREB mutant was transfected. (E) In vitro-translated, 35S-labeled wild-type or S133A mutant CREB was incubated in vitro with ATP alone (−) or together with PKA (+). Subsequently the 35S-labeled proteins were incubated with glutathione-agarose beads bearing either the GST-Oct-1 POU domain fusion protein (GST-POU) or GST alone. In the upper panel, bound 35S-labeled proteins were visualized by autoradiography of dried SDS-PAGE gels. In the lower panel, the binding of phospho-S133A CREB was detected by immunoblotting as described for A. Input represents 10% of the in vitro-transcribed-translated proteins used in the incubation.
Northern blot analysis.
Total RNA was isolated with Trizol reagent as recommended by the manufacturer (Life Technologies). RNA samples were fractionated on an agarose gel and transferred overnight onto a nylon filter. The next day, RNA was cross-linked with a UV cross-linker (Stratagene). For detection of cyclin D1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs, blots were hybridized overnight at 42°C in 50% formamide-5× SSC (1× SSC is 0.15 M sodium citrate)-1× PE (50 mM Tris-HCl [pH 7.5], 0.1% sodium pyrophosphate, 1% sodium dodecyl sulfate [SDS],25% polyvinylpyrrolidone, 0,25% Ficoll, 5 mM EDTA)-150 μg of salmon sperm DNA. All the probes were generated with a random-primed labeling kit (Life Technologies) in the presence of [α-32P]dCTP, 3,000 Ci/mmol (NEN-Dupont). Washes were performed twice in 2× SSC-0.1% SDS for 10 min at room temperature and then twice in 0.1× SSC-0.1% SDS for 30 min at 65°C for detection of all mRNAs.
In vitro binding assay.
Glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli strain BL21. Bacteria were grown to mid-log phase, induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside, grown for 3 h at 37°C, and collected by centrifugation. Bacteria were washed and resuspended in BugBuster protein extraction reagent (Novagen), as recommended by the manufacturer, and lysed by sonication, and the supernatant was clarified by centrifugation at 25,000 × g for 30 min. GST fusion proteins were isolated by incubation of the lysate with 50 μl of glutathione-agarose beads (Sigma) for 1 h at 4°C. Beads were collected by centrifugation and washed three times in ice-cold phosphate-buffered saline. An aliquot of beads was boiled in 2× SDS loading buffer, separated by electrophoresis through a 12% polyacrylamide gel, and Coomassie stained to analyze bound proteins.
GST fusion proteins (2 μg) immobilized on glutathione-agarose beads were used for each pulldown assay. GST fusion proteins were incubated in interaction buffer (40 mM KCl, 0,1 mM EDTA, 20 mM Tris-HCl [pH 8], 0,1% Nonidet P-40, 10% glycerol). Binding assays were conducted in 1 ml of interaction buffer with 10 μl of in vitro-transcribed-translated [35S]methionine-labeled protein (Promega). CREB and CREB mutant proteins were phosphorylated with PKA (New England Biolabs) as described by the manufacturer. After a 1-h incubation, beads were collected by centrifugation, washed three times in interaction buffer, and boiled in 2× SDS loading buffer. Bound proteins were separated by electrophoresis through a polyacrylamide gel, which was transferred onto a nitrocellulose filter and exposed to film at room temperature overnight.
Coimmunoprecipitation and immunoblotting.
For immunoprecipitation from MCF-7 cells, 3 × 106 cells were seeded 1 day prior to transfection in 140-cm dishes. The cells were transfected with the indicated combination of expression vectors (see the figure legends) by FuGEN6 transfection reagent (Roche Diagnostics, GmbH, Mannheim, Germany)-mediated transfection with 10 μg of total DNA. Cells were harvested 48 h posttransfection and lysed for 20 min at 4°C in 1.5 ml of extraction buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 2 mM dithiothreitol, Boehringer complete protease inhibitor cocktail, 1% each Sigma phosphatase inhibitor cocktails I and II). Cell lysates were clarified by centrifugation at 18,000 × g for 15 min at 4°C.
Hemagglutinin (HA) fusion proteins were immunoprecipitated with 5 μg of affinity-purified anti-HA mouse monoclonal antibody (sc-7392; Santa Cruz Biotechnology, Santa Cruz, Calif.), or 5 μg of control antibody during an overnight incubation with 10 μl of a 50% slurry of protein A-agarose (Roche Molecular Biochemicals). Immune complexes were collected by slow-speed centrifugation, washed three times in extraction buffer, and boiled in 2× SDS loading buffer, and denatured proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were transferred to a nitrocellulose filter (Schleicher and Schuell Protran BA 83 nitrocellulose transfer membrane), which was blocked in 5% nonfat milk-150 mM NaCl-50 mM Tris-HCl [pH 8.0]-0.05% Tween 20.
Immunoblots were performed with rabbit anti-HA antibody at 2 μg/ml (Zymed), anti-CREB rabbit polyclonal antibody diluted 1:1,000 (Ozyme/CST, St. Quentin Yvelines, France), or anti-phospho-Ser 133 CREB rabbit polyclonal antibody diluted 1:1,000 (Ozyme/CST, St. Quentin Yvelines, France). Immune complexes were visualized by enhanced chemiluminescence (Roche Diagnostics GmbH). Membranes were stripped with Re-Blot Plus-Trial (Chemicon International) according to the manufacturer's recommendations when sequentially immunoblotted for phospho-CREB and total CREB.
Chromatin immunoprecipitation.
Approximately 6 × 106 transfected MCF-7 cells were grown on 10-cm dishes and cross-linked by addition of formaldehyde (to 1% final concentration) to attached cells. Cross-linking was allowed to proceed at room temperature for 10 min and was terminated with glycine (final concentration, 0.125 M). Cells were washed with phosphate-buffered saline and scraped. Cells were collected by centrifugation and incubated at 4°C for 20 min in buffer containing 10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, and Boehringer complete protease inhibitor cocktail. Nuclei were collected by centrifugation at 4,000 rpm for 5 min, resuspended in sonication buffer (50 mM HEPES [pH 7.5], 140 mM NaCl, 1% Triton X-100, and Boehringer complete protease inhibitor cocktail) and incubated at 4°C for 30 min. Samples were sonicated on ice to an average length of 500 to 1,000 bp and then microcentrifuged at 14,000 rpm.
Chromatin was incubated with 10 μl of agarose conjugate antibody (HA, sc-7392 AC) in sonication buffer containing 1 mg of salmon sperm DNA per ml and 1 mg of bovine serum albumin per ml at 4°C overnight. Immunoprecipitates were washed two times with sonication buffer, two times with sonication buffer containing 500 mM NaCl, two times in TE buffer containing 250 mM LiCl and 0,5% NP-40, and two times in TE buffer. Pellets were resuspended in 100 μl of TE and incubated at 55°C for 3 h with 10 μg each of RNase A and proteinase K. Cross-links were reversed by incubating samples at 65°C overnight. Samples were purified on NucleoSpin extract (Macherey-Nagel), eluted in 50 μl of TE buffer, and assayed by semiquantitative PCR.
PCR mixtures contained 3 μl of immunoprecipitate or total input, 10 pmol of each primer, 1× Thermophilic buffer (Qiagen), 0.2 mM each dATP, dCTP, dGTP, and dTTP, 1.25 U of HotStarTaq DNA polymerase (Qiagen), and 1 μCi of [α-32P]dCTP in a total volume of 50 μl. After 30 cycles of amplification, PCR products were electrophoresed by 5% PAGE and visualized by autoradiography of the dried gels. Previous control experiments established that these PCR conditions are within the linear range of amplification. Each experiment was performed at least three times.
Electrophoretic mobility shift assays.
Binding reactions were performed with 4 μl of in vitro-transcribed-translated protein produced by coupled transcription-translation in reticulocyte extracts (Promega) and/or 1 μg of GST or GST-POU fusion protein in a 20-μl volume containing 20 mM HEPES (pH 7.8), 2 mM MgCl2, 5% glycerol, 30 ng of poly(dI-dC)-poly(dI-dC), and 20 fmol of a 32P-labeled oligonucleotide probe spanning the CRE (underlined) of the cyclin D1 promoter (5′-CAACAGTAACGTCACACGGAC-3′). After incubation for 30 min at room temperature, the sample were subjected to electrophoresis on 5% PAGE in 0.5× Tris-borate-EDTA at 15 mA for 2 to 3 h at room temperature. Complexes were revealed by autoradiography of the dried gels at room temperature and at −70°C with intensifying screens.
RESULTS
Oct-1 increases cyclin D1 promoter activity by a CREB binding site-mediated mechanism.
To examine the ability of Oct-1 to control cyclin D1 expression, we transiently transfected human MCF-7 breast cancer cells with expression vectors encoding the human Oct-1 protein or its POU domain alone, since the latter mediates many activities attributed to Oct-1. The POU domain activated a luciferase reporter gene driven by the cyclin D1 promoter (Fig. 1A).
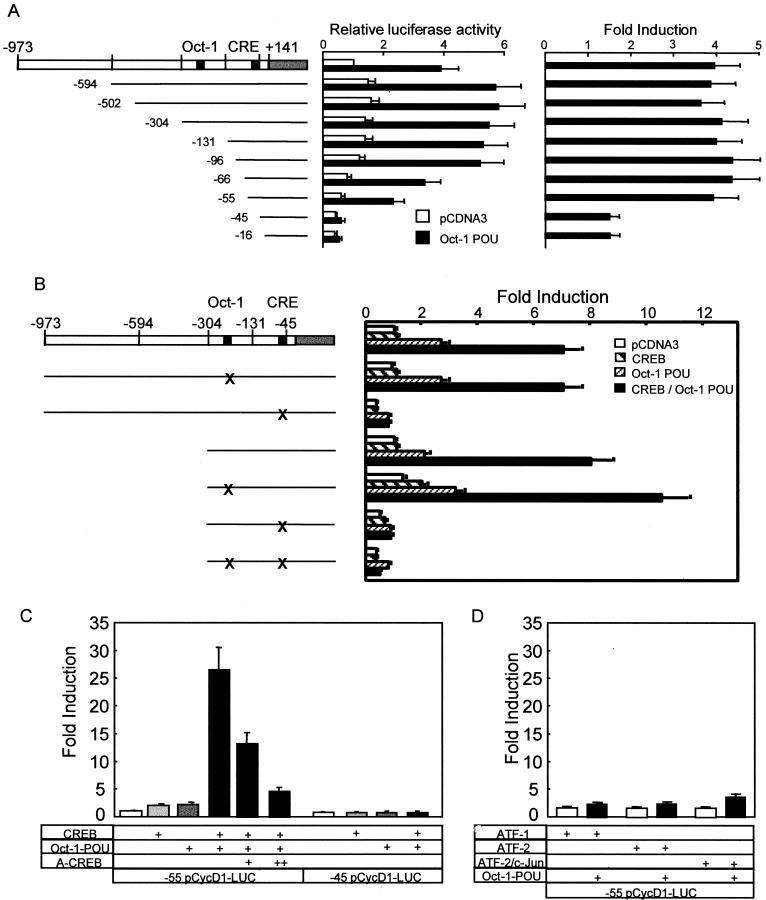
FIG. 1.
Oct-1 POU domain-induced cyclin D1 promoter activity is dependent on CREB and the CRE. (A) MCF-7 cells were cotransfected with expression vectors for the POU domain of Oct-1 or empty vector (pCDNA3) with 5′ deletion mutants in the cyclin D1 promoter. The relative luciferase activity was standardized to that observed with pCDNA3 and the full-length promoter. The error bars represent the standard error of the mean. Each experiment was performed at least three times in duplicate (left panel). The right panel shows the induction for each 5′ deletion mutant, calculated by dividing the luciferase activity generated by the POU domain by that observed with pCDNA3 for each deletion mutant. (B) MCF-7 cells were transfected with expression vectors for the POU domain of Oct-1, CREB, or empty vector (pCDNA3), together with a wild-type or mutant cyclin D1-luciferase reporter construct, as indicated schematically to the left. X indicates the mutated site in the promoter. The induction is presented relative to the activity with pCDNA3 and the full promoter, and the error bars represent the standard error of the mean. (C) Cyclin D1 reporters with 5′ deletions to −55 and −45 were transfected with the indicated expression vectors and analyzed as described for B; 100 ng (+) and 300 ng (++) of the dominant negative A-CREB expression vector were cotransfected. (D) Oct-1-POU was coexpressed with ATF-1, ATF-2, and c-Jun as well as the cyclin D1 (−55/+141)-luciferase reporter, as indicated below the bars.
To map the DNA sequences involved, we examined the effect of the POU domain on the activity of several cyclin D1 promoter 5′ deletion mutants (Fig. 1A). Deletion from −973 to −55 did not affect induction, although this removed the Oct-1 consensus sequence located between positions −304 and −131. Similarly, mutation of the Oct-1 consensus sequence in the cyclin D1 promoter did not block induction by the POU domain (Fig. 1B), implicating another promoter element. This was identified by deletion of another 10 bp, which removed the cyclic AMP-responsive element (CRE) and diminished transcription in the presence of both Oct-1-POU and an empty expression vector (Fig. 1A). When corrected for the latter, basal, POU-independent activation, these data revealed a striking decrease upon removal of the CRE (Fig. 1A, right panel). Consistent with this, mutation of the CRE site in the context of either the −973 or a −304 cyclin D1 reporter construct abolished activation of the promoter by the Oct-1 POU domain (Fig. 1B).
Since Oct-1 POU factor cannot bind the cyclin D1 CRE sequence directly (data not shown), we examined whether this effect might involve CRE-binding protein (CREB). While transfection of a CREB expression vector did not activate the cyclin D1 reporter gene in the absence of signal-dependent activation, CREB coexpression with Oct-1 POU led to a strong induction (Fig. 1B, C). This was specific for CREB, since Oct-1 POU failed to augment the weak level of transactivation observed upon expression of ATF-1, ATF-2, or ATF-2 together with c-Jun (Fig. 1D). Moreover, synergistic activation by CREB and Oct-1 POU together was suppressed by mutation or deletion of the CRE but not the Oct-1 consensus sequence (Fig. 1B, C), as well as by cotransfection of A-CREB (1), a dominant-negative version that prevents the basic region of CREB from binding to DNA (Fig. 1C). These results showed that the Oct-1 POU domain activated the cyclin D1 promoter indirectly through a mechanism involving CREB binding to the CRE located between promoter sequences −55 and −45.
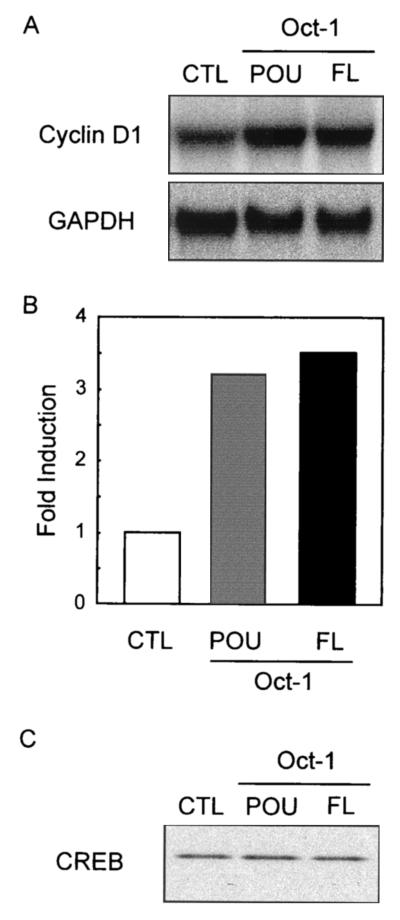
We then tested the effect of wild-type Oct-1 and POU expression on the endogenous cyclin D1 gene. Transient transfection of either expression vector led to an increase in cyclin D1 mRNA relative to GAPDH, indicating that wild-type Oct-1 or the POU domain alone is sufficient to activate the cyclin D1 gene under noninduced conditions (shown in Fig. 2A and quantified in Fig. 2B). Notably, endogenous CREB protein levels were unchanged in the presence of either expression vector (Fig. 2C).
FIG. 2.
Oct-1 overexpression increases endogenous cyclin D1 mRNA levels. (A) MCF-7 cells were transfected with either pCDNA3 (CTL), the Oct-1 full-length expression vector (FL), or the POU domain expression vector (POU). RNA was isolated 48 h posttransfection and analyzed by Northern blotting and hybridization with probes for cyclin D1 and GAPDH. (B) The hybridization signals were quantified with a phosphorimager, and the induction was calculated by standardizing to the GAPDH signal. (C) Endogenous CREB protein was detected by immunoblotting with anti-CREB polyclonal antiserum.
Oct-1-dependent CREB activity results from direct interaction between these factors.
To avoid potential complications due to endogenous CREB/ATF proteins interacting with the CRE, we coexpressed a chimeric protein containing CREB fused to the GAL4 DNA-binding domain (GAL4-CREB) and a luciferase reporter gene driven by GAL4 DNA-binding sites. GAL4-CREB increased luciferase activity 5-fold, and coexpression with full-length Oct-1 or the POU domain led to a 40-fold increase in reporter activity (Fig. 3A). Oct-1 alone did not induce the GAL4 reporter gene, nor did the GAL4 DNA-binding domain (GAL4 1 to 147), either alone or together with Oct-1 (Fig. 3A). Oct-1 coexpression also did not enhance transcriptional activity mediated by Nur77 (GAL4-Nur77), a transcription factor belonging to the orphan receptor family. These results strongly suggested that Oct-1 was a specific coactivator of CREB, implicating a direct interaction between these two proteins.
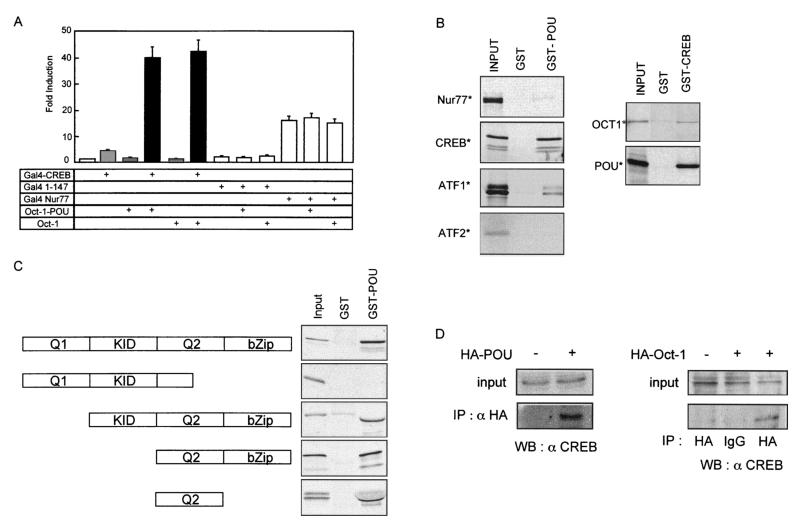
FIG. 3.
CREB interacts in vitro and in cellulo with Oct-1 independently of DNA binding. (A) MCF-7 cells were transfected as indicated with expression vectors for the GAL4 DNA-binding domain alone (GAL4 1-147), the GAL4 DNA-binding domain fused to CREB (GAL4-CREB) or Nur77 (GAL4-Nur77), full-length Oct-1, the Oct-1 POU domain, or pCDNA3 (first bar). All transfections contained the reporter pG5tk-Luc, which contains five binding sites for GAL4 fused to the thymidine kinase minimal promoter and the firefly luciferase gene. The induction is presented relative to the activity with pCDNA3, and the error bars represent the standard error of the mean. (B) The in vitro-translated, 35S-labeled proteins indicated to the left of each panel were incubated with purified recombinant GST-Oct-1 POU domain (GST-POU), GST-CREB, or GST alone prebound to glutathione-agarose beads. Bound 35S-labeled proteins were visualized by autoradiography of dried SDS-PAGE gels. Input represents 10% of the in vitro-transcribed-translated proteins used in the incubation. (C) The diagram to the left shows the domains of CREB protein present in the in vitro-translated, 35S-labeled proteins that were incubated with GST-POU or GST alone as described for B. Input represents 10% of the in vitro-transcribed-translated proteins. (D) MCF-7 cells were transfected with pCMV-HA-Oct-1 or pCMV-HA-POU, as indicated. The HA fusion proteins were immunoprecipitated (IP) from total cell lysates with anti-HA polyclonal antiserum (HA) or nonspecific rabbit IgG (IgG). Binding of endogenous CREB to the immunoprecipitated protein was detected by immunoblotting (WB) with anti-CREB polyclonal antiserum. Input represents 5% of the total cell lysate.
Physical interactions between CREB and Oct-1 were analyzed in vitro by pulldown experiments performed with GST fusion proteins. Glutathione-Sepharose beads bearing recombinant GST or a GST-Oct-1-POU domain fusion protein were incubated with 35S-labeled CREB, ATF-1, ATF-2, or Nur77, produced by coupled transcription-translation in a reticulocyte lysate system. CREB bound to the GST-Oct-1-POU fusion and not to the GST control (Fig. 3B). ATF-1 interacted more weakly with GST-Oct-1-POU, whereas neither ATF-2 nor Nur77 bound to the GST proteins (Fig. 3B). In the reciprocal experiment, GST-CREB bound to in vitro-expressed, 35S-labeled Oct-1 or POU domain but not to GST alone (Fig. 3B). Thus, the preferential interaction of Oct-1 with CREB could explain their functional synergy on the cyclin D1 promoter.
To map which part of CREB mediated its interaction with the POU domain, we generated a series of mutants lacking different functional domains (Fig. 3C). After coupled in vitro transcription-translation, the 35S-labeled proteins were tested in pulldown assays with GST or GST-Oct-1 POU. Deletion of the C-terminal portion of the Q2 domain and the entire bZip domain prevented interaction with GST-Oct-1 POU (Fig. 3C, second panel), indicating that these two regions were necessary for binding. The region required for binding actually mapped to the Q2 domain itself, since deletion of the KID and bZip domains did not significantly affect in vitro interactions (Fig. 3C). Thus, the transactivation domain Q2 was sufficient to confer selective binding to Oct-1 POU in vitro.
We used coimmunoprecipitation experiments to test for a similar interaction in vivo (Fig. 3D). HA-tagged Oct-1 or POU domain was expressed in MCF-7 cells and immunoprecipitated from cellular lysates with an HA-specific antibody. Endogenous CREB coprecipitated with both proteins but not with nonspecific immunoglobulin (IgG) or in the absence of HA-Oct-1 expression (Fig. 3D). This indicates that multiprotein complexes containing both CREB and Oct-1 exist in vivo and that the POU domain of Oct-1 is sufficient for complex formation.
Oct-1 POU domain interacts with CREB bound to the cyclin D1 CRE.
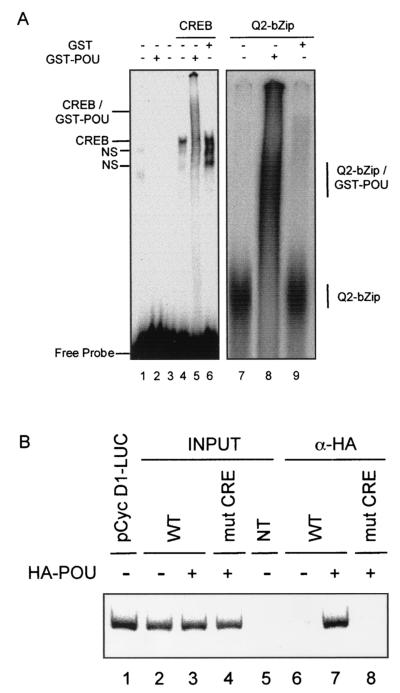
The transfection and solution interaction results led us to test for interactions between the Oct-1 POU domain and CREB complexed to the cyclin D1 CRE. These used GST or GST-Oct-1 POU, along with CREB generated by coupled in vitro transcription-translation. Binding reactions containing various combinations of proteins were performed with a 32P-labeled oligonucleotide derived from the cyclin D1 promoter region spanning the CRE and analyzed by native PAGE. CREB formed a specific complex on the cyclin D1 CRE probe that was not observed in control reactions (Fig. 4A). GST-Oct-1 POU did not bind to the cyclin D1 CRE on its own (lane 2) but, together with CREB, generated a series of smeared, slower-migrating complexes that were not found with GST alone (compare lanes 5 and 6). More importantly, GST-Oct-1 POU gave rise to similar, more distinct slower-migrating complexes with a minimal form of CREB containing just the bZip and Q2 domains (lane 8) that were not seen with GST alone (lane 9). This indicates that CREB can recruit the Oct-1 POU domain to the cyclin D1 CRE in vitro via the same region implicated in solution interactions between these two proteins.
FIG. 4.
Oct-1 POU binds to a CREB-cyclin D1 CRE complex in vitro and in transfected cells. (A) Binding reactions were assembled with a 32P-labeled oligonucleotide spanning the cyclin D1 CRE and the following proteins, as indicated above the lanes: unprogrammed reticulocyte lysate (lane 1); recombinant, full-length CREB (lanes 4 to 6) or the Q2-bZip region of CREB (lanes 7 to 9), produced with programmed reticulocyte lysates; and recombinant GST (lanes 6 and 9) or GST-Oct-1 POU (lanes 2, 5, and 9), purified from bacteria. After electrophoresis on native PAGE, complexes were visualized by autoradiography of the dried gels. The right panel shows the relevant portion of the autoradiogram. Lane 3 shows the probe alone under binding conditions. The composition of the different complexes is indicated to the side of the two panels; NS, nonspecific complexes. (B) Chromatin immunoprecipitation analysis of Oct-1 POU interaction with the cyclin D1 reporter gene in transfected MCF-7 cells. Cells were cotransfected with pCMV-HA-POU (lanes 3, 4, 7, and 8) and either a wild-type −131 pCycD1-LUC reporter gene (WT, lanes 2, 3, 6, and 7) or the same reporter bearing point mutations in the CRE (mut CRE, lanes 4 and 8). After cross-linking, nuclei were purified from transfected cells and sonicated, and after cross-link reversal, an aliquot of chromatin was immunoprecipitated with anti-HA monoclonal antiserum (α-HA, lanes 6 to 8). The cyclin D1 CRE was detected by PCR with primers specific to the cyclin D1-luciferase reporter plasmid in the presence of a trace amount of [α-32P]dCTP. Input indicates PCRs that contained chromatin prior to immunoprecipitation. NT contained chromatin from nontransfected cells (lane 5), while lane 1 shows amplification of the purified plasmid −131 pCycD1-LUC. The panel shows the autoradiogram of the dried gel and is representative of several independent experiments analyzed by 32P incorporation or ethidium bromide staining.
We then used chromatin immunoprecipitation experiments to demonstrate that this complex forms on the cyclin D1 CRE in transfected MCF-7 cells, where it is clearly functional (see Fig. 1). Cells were transfected with an expression vector for the HA-tagged Oct-1 POU domain and a −131 cyclin D1 reporter gene that contains either a wild-type or mutated CRE but lacks the consensus Oct-1 binding site. DNA was purified from chromatin prior to or after immunoprecipitation with an HA-specific antibody and analyzed by PCR. Similar levels of the wild-type and mutant reporter plasmid were present in the total chromatin fraction from the transfected cells (Fig. 4A, lanes 2 to 4). However, only chromatin containing the wild-type reporter gene was coimmunoprecipitated with HA-Oct-1 POU (compare lanes 7 and 8). Notably, no signal was observed in the absence of HA-Oct-1 POU (lane 6) or in nontransfected cells (lane 5). Thus, the Oct-1 POU domain is complexed with the cyclin D1 reporter gene via the CRE in transfected cells. Taken together with the analyses above, these data strongly suggest that Oct-1 affects cyclin D1 promoter activity through a protein-protein complex with CREB bound to the cyclin D1 CRE.
Oct-1 interacts with the unphosphorylated form of CREB.
A variety of intracellular signaling pathways, notably one involving PKA, phosphorylate CREB on Ser 133, thereby enhancing transcriptional activation via the recruitment of CBP (reviewed in reference 30). To test whether Oct-1 interaction with CREB was influenced by CREB phosphorylation, CREB was expressed alone or together with HA-tagged Oct-1 POU domain in MCF-7 cells that were treated with forskolin to activate PKA. This led to CREB phosphorylation on Ser 133, as detected by immunoblotting with antibodies specific for phospho-CREB (Fig. 5A). Remarkably, phospho-CREB did not coprecipitate with HA-Oct-1 POU, in contrast to CREB unmodified on Ser 133 (Fig. 5A).
To confirm this observation, MCF-7 cells transfected with expression vectors for HA-Oct-1 POU and CREB were subjected to prolonged serum starvation and then treated with forskolin for different times (Fig. 5B). Once again, the phospho-Ser 133 form of CREB did not coprecipitate with HA-Oct-1, although phosphorylated Ser 133-CREB (P-Ser 133-CREB) was readily detectable in the cell lysates. In contrast, CREB associated with Oct-1 at all time points, and forskolin treatment caused a weak decrease in this binding (Fig. 5B). This decrease in Oct-1-CREB association after PKA activation was more striking in the reverse experiment, where HA-Oct-1 association was assayed after immunoprecipitation with CREB-specific antibodies (Fig. 5C). Coexpression of CREB bearing a Ser 133 to Ala mutation (CREB S133A), which cannot be phosphorylated on this residue, also coprecipitated with Oct-1 and showed no decrease upon forskolin stimulation (Fig. 5D).
This suggested that CREB phosphorylation might interfere with its binding to Oct-1. To test this, we assayed the interaction of in vitro-translated, 35S-labeled CREB phosphorylated with PKA with GST-Oct-1 POU. PKA strongly increased the level of CREB phosphorylation on Ser 133 (input lanes, Fig. 5E, lower left panel); remarkably, we could detect no binding of phosphorylated CREB to GST-Oct-1-POU by immunoblotting (Fig. 5E, lower left panel) or direct 32P labeling (data not shown), while the 35S-labeled CREB in the same reaction mix bound to the immobilized POU domain (Fig. 5E, upper left panel). As expected, CREB S133A was insensitive to PKA and interacted with GST-Oct-1 POU (Fig. 5E, right panel). These data demonstrate that CREB phosphorylated on Ser 133 is excluded from binding to the Oct-1 POU domain in vitro.
Oct-1-enhanced CREB transcriptional activity is independent of Ser 133 phosphorylation and insensitive to E1A inhibition of CBP/p300.
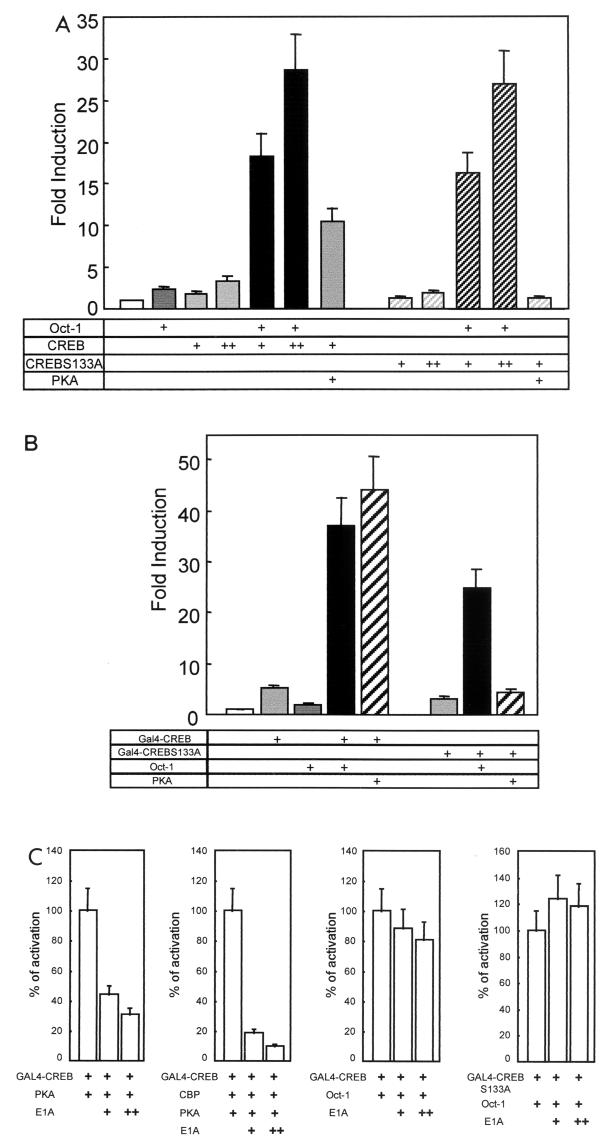
Recruitment of CBP to P-Ser 133-CREB strongly increased its capacity to activate transcription. Our results suggested that Oct-1 is a cofactor that also targeted CREB in its unphosphorylated form. To test this functionally, we analyzed the activity of CREB S133A in transient-transfection assays. CREB S133A cooperated with Oct-1 to activate the cyclin D1 reporter gene indistinguishably from wild-type CREB (Fig. 6A). On the other hand, CREB S133A did not respond to PKA, which did activate wild-type CREB, yielding a 10-fold induction of the cyclin D1 reporter.
FIG.6.
Oct-1 synergy with CREB is independent of Ser 133 phosphorylation or E1A-sensitive CBP coactivation. (A) MCF-7 cells were transfected with the (−55/+141) cyclin D1-luciferase reporter and, as indicated, with Oct-1, 100 ng (+) or 200 ng (++) of CREB, 100 ng (+) or 200 ng (++) of CREB S133A, or PKA expression vector. The induction is presented relative to the activity with pCDNA3 (bar 1), and the error bars represent the standard error of the mean. (B) MCF-7 cells were transfected with expression vectors for the GAL4 DNA-binding domain fused to wild-type CREB or the S133A mutant, pCMV-Oct-1, and PKA expression plasmid, as indicated. All transfections contained the reporter pG5tk-Luc, which contains five binding sites for GAL4 fused to the thymidine kinase minimal promoter and the firefly luciferase gene. The induction is presented relative to the activity with pCDNA3 (bar 1), and the error bars represent the standard error of the mean. (C) MCF-7 cells were transfected essentially as described (B), except that 150 ng (+) or 300 ng (++) of CBP or E1A expression vectors was included in the transfection, as indicated. The bars represent the luciferase activity as a percentage of that observed in the first lane of each graph, and the error bars present the standard error of the mean.
The role of Oct-1 as a cofactor of CREB was reconfirmed with the GAL4 reporter system. Overexpression of GAL4-CREB alone induced a 5-fold activation of the GAL4 reporter gene, and Oct-1 enhanced induction to nearly 40-fold, which is nearly the level observed with PKA-activated CREB (Fig. 4A and 6B). Oct-1 also synergized with GAL4-CREB S133A, which was insensitive to PKA, as expected (Fig. 6B).
These data show that Oct-CREB synergy is not dependent upon CREB phosphorylation on Ser 133, suggesting that p300/CBP in not required for this effect. To unequivocally rule out this possibility, we added an expression vector for the adenovirus E1A protein to our transfection assays. E1A binds p300 and CBP and blocks their capacity to coactivate transcription (reviewed in reference 19). Accordingly, E1A inhibited PKA-dependent CREB activation, which is mediated by CBP recruitment (Fig. 6C, panel 1). E1A had an even more striking effect when an expression vector for CBP was included in the transfection assay (Fig. 6C, panel 2). In direct contrast, E1A had no effect on Oct-1-driven synergy with GAL4-CREB wild-type (Fig. 6C, panel 3) or, more importantly, GAL4-CREB S133A (Fig. 6C, panel 4). These results strongly suggest that Oct-1 cooperates with CREB to activate the cyclin D1 promoter through a direct interaction. This does not require CREB phosphorylation on Ser 133 and is independent of E1A-sensitive p300/CBP coactivator function.
DISCUSSION
CREB phosphorylation on Ser 133 is tightly linked to mitogen-driven induction of gene expression through the recruitment of the coactivator CBP to promoters (reviewed in reference 30). Indeed, PKA-catalyzed phosphorylation of CREB enhanced transcription of the cyclin D1 reporter via the CRE immediately upstream of the transcript start site in our assay system, transiently transfected MCF-7 cells. Nevertheless, the rapid appearance and subsequent decay of P-Ser 133-CREB (18, 50) precedes the transcriptional activation of cyclin D1 in vivo and therefore seems unlikely to play a prominent role in regulating the cyclin D1 promoter. Instead, our data indicate that a complex involving Oct-1 and CREB can strongly activate transcription of a cyclin D1 reporter gene independently of CREB phosphorylation on Ser 133. Furthermore, overexpression of the adenovirus E1A 12S protein, which binds to p300/CBP and thereby inhibits coactivation (reviewed in reference 19), did not block Oct-1/CREB synergy while strongly suppressing transcription mediated by PKA-phosphorylated CREB. Thus, cooperation between Oct-1 and CREB does not require p300/CBP activities repressed by E1A binding. In fact, this synergy may not directly require CBP, since it was also not stimulated by overexpression of CBP.
This activity of Oct-1 thus bears striking resemblance to that of the LIM-only transcriptional coactivators (12, 13). This tissue-specific family of proteins potentiate transactivation by CREB family members independently of their phosphorylation state or of CBP. Given its ubiquitous expression, Oct-1 may represent a functional homologue of LIM-only proteins that is active in all cell types.
The stimulatory effect of Oct-1 on the cyclin D1 promoter was specific for CREB, either directly bound to the CRE in the cyclin D1 promoter or in the context of a fusion with the GAL4 DNA-binding domain. It also required the POU domain of Oct-1 but not its binding site in the cyclin D1 promoter, instead functioning through an association with CREB. Consistent with this functional interaction, we detected complexes between these two proteins in GST pulldown assays, in gel retardation experiments, and in transfected cells that involve the cyclin D1 CRE, Oct-1 POU domain, and CREB, within which the Q2 domain was sufficient for in vitro association. Since the CREB KID domain was not necessary, it is not surprising that complex formation did not depend on CREB activation by phosphorylation on Ser 133, which actually appeared to exclude CREB from interacting with Oct-1. This was not observed in the functional assays and could be an artifact arising from a protein, possibly CBP, interacting preferentially with phospho-Ser 133-CREB in solution and interfering with Oct-1 binding.
Unlike the KID domain, the Q2 domain of CREB can activate transcription constitutively (39). In vitro it recruits TFIID (11, 34, 42) through interactions with hTAF(II)130 and thereby strongly stimulates transcription on naked templates (10, 26) but not on chromatin-bound templates (3), where it is repressed. Moreover, the Q2/TFIID complex is not sufficient to activate a CRE-driven reporter gene under unstimulated conditions, indicating that it requires additional components to function. In the case of the cyclin D1 promoter, our results indicate that Oct-1 is such a cofactor, acting through an interaction between CREB and the Oct-1 POU domain. Notably, this does not involve the DNA binding functions of this domain but rather protein-protein interactions.
Oct-1 can also interact with components of the basal transcription machinery, namely TFIIB (35) and TATA binding protein (56). Via binding to TFIID and TFIIB, the Oct-1/CREB complex could facilitate transcriptional activation without the participation of a coactivator. Other protein-DNA complexes would then be implicated in ensuring stable coactivator binding to the cyclin D1 promoter in both a constitutive and signaling-dependent manner, leading to full transcriptional induction. This has been documented for AP1 (2), but could also involve NF-κB, SP1, E2F, ATF-2, and TCF/LEF, many of which interact with p300 or CBP upon mitogenic signal-driven phosphorylation (5, 9, 31, 36, 48).
Overexpression of the cyclin D1 protein is found in many human cancers and is particularly linked to oncogenic transformation in mammary epithelium (43, 52). Here we show that Oct-1 is a potentially important cofactor in regulating cyclin D1 expression in a cell line derived from a human breast cancer and thus is potentially linked to transformation. Interestingly, overexpression of the Oct-1 POU domain in the mouse thymus led to the development of thymic lymphomas, indicating that Oct-1 can be oncogenic (38). This raises the possibility that, under appropriate conditions, Oct-1 makes an important contribution to aberrant transcriptional activation of cyclin D1 that underlies the onset and progression of cancer.
Acknowledgments
This work was supported by grants from the French Association pour la Recherche sur le Cancer (ARC), numbers 9276 (R.A.H.), 5890 (R.A.H.), and 5961 (A.P.).
The first two authors contributed equally to the results of this work.
We thank W. Herr, J. Licht, C. Vinson, and B. Lutz for generous gifts of materials. We are grateful to A. Blangy, M. Weill, and P. Fort for helpful discussions and A. LeCam for critical reading of the manuscript.
REFERENCES
- 1.Ahn, S., M. Olive, S. Aggarwal, D. Krylov, D. D. Ginty, and C. Vinson. 1998. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol. Cell. Biol. 18:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albanese, C., M. D'Amico, A. T. Reutens, M. Fu, G. Watanabe, R. J. Lee, R. N. Kitsis, B. Henglein, M. Avantaggiati, K. Somasundaram, B. Thimmapaya, and R. G. Pestell. 1999. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J. Biol. Chem. 274:34186-34195. [DOI] [PubMed] [Google Scholar]
- 3.Asahara, H., B. Santoso, E. Guzman, K. Du, P. A. Cole, I. Davidson, and M. Montminy. 2001. Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB. Mol. Cell. Biol. 21:7892-7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier, F., R. J. Lee, A. C. Taylor, R. G. Pestell, and P. LuValle. 1999. Identification of the cyclin D1 gene as a target of activating transcription factor 2 in chondrocytes. Proc. Natl. Acad. Sci. USA 96:1433-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billon, N., D. Carlisi, M. B. Datto, L. A. van Grunsven, A. Watt, X. F. Wang, and B. B. Rudkin. 1999. Cooperation of Sp1 and p300 in the induction of the CDK inhibitor p21WAF1/CIP1 during NGF-mediated neuronal differentiation. Oncogene 18:2872-2882. [DOI] [PubMed] [Google Scholar]
- 6.Chandran, U. R., B. S. Warren, C. T. Baumann, G. L. Hager, and D. B. DeFranco. 1999. The glucocorticoid receptor is tethered to DNA-bound Oct-1 at the mouse gonadotropin-releasing hormone distal negative glucocorticoid response element. J. Biol. Chem. 274:2372-2378. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, M., V. Sexl, C. J. Sherr, and M. F. Roussel. 1998. Assembly of cyclin D-dependent kinase and titration of p27Kip1 regulated by mitogen-activated protein kinase kinase (MEK1). Proc. Natl. Acad. Sci. USA 95:1091-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Amico, M., J. Hulit, D. F. Amanatullah, B. T. Zafonte, C. Albanese, B. Bouzahzah, M. Fu, L. H. Augenlicht, L. A. Donehower, K. Takemaru, R. T. Moon, R. Davis, M. P. Lisanti, M. Shtutman, J. Zhurinsky, A. Ben-Ze'ev, A. A. Troussard, S. Dedhar, and R. G. Pestell. 2000. The integrin-linked kinase regulates the cyclin D1 gene through glycogen synthase kinase 3beta and cAMP-responsive element-binding protein-dependent pathways. J. Biol. Chem. 275:32649-32657. [DOI] [PubMed] [Google Scholar]
- 9.Duyndam, M. C., H. van Dam, P. H. Smits, M. Verlaan, A. J. van der Eb, and A. Zantema. 1999. The N-terminal transactivation domain of ATF-2 is a target for the cooperative activation of the c-jun promoter by p300 and 12S E1A. Oncogene 18:2311-2321. [DOI] [PubMed] [Google Scholar]
- 10.Felinski, E. A., J. Kim, J. Lu, and P. G. Quinn. 2001. Recruitment of an RNA polymerase II complex is mediated by the constitutive activation domain in CREB, independently of CREB phosphorylation. Mol. Cell. Biol. 21:1001-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felinski, E. A., and P. G. Quinn. 1999. The CREB constitutive activation domain interacts with TATA-binding protein-associated factor 110 (TAF110) through specific hydrophobic residues in one of the three subdomains required for both activation and TAF110 binding. J. Biol. Chem. 274:11672-11678. [DOI] [PubMed] [Google Scholar]
- 12.Fimia, G. M., D. De Cesare, and P. Sassone-Corsi. 1999. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature 398:165-169. [DOI] [PubMed] [Google Scholar]
- 13.Fimia, G. M., D. De Cesare, and P. Sassone-Corsi. 2000. A family of LIM-only transcriptional coactivators: tissue-specific expression and selective activation of CREB and CREM. Mol. Cell. Biol. 20:8613-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher, C., N. Heintz, and R. G. Roeder. 1987. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell 51:773-781. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, G. A., and M. R. Montminy. 1989. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59:675-680. [DOI] [PubMed] [Google Scholar]
- 16.Gstaiger, M., O. Georgiev, H. van Leeuwen, P. van der Vliet, and W. Schaffner. 1996. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 15:2781-2790. [PMC free article] [PubMed] [Google Scholar]
- 17.Guttridge, D. C., C. Albanese, J. Y. Reuther, R. G. Pestell, and A. S. Baldwin, Jr. 1999. NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 19:5785-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagiwara, M., A. Alberts, P. Brindle, J. Meinkoth, J. Feramisco, T. Deng, M. Karin, S. Shenolikar, and M. Montminy. 1992. Transcriptional attenuation following cAMP induction requires PP-1-mediated dephosphorylation of CREB. Cell 70:105-113. [DOI] [PubMed] [Google Scholar]
- 19.Hagmeyer, B. M., P. Angel, and H. van Dam. 1995. Modulation of AP-1/ATF transcription factor activity by the adenovirus-E1A oncogene products. Bioessays 17:621-629. [DOI] [PubMed] [Google Scholar]
- 20.Herber, B., M. Truss, M. Beato, and R. Muller. 1994. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene 9:2105-2107. [PubMed] [Google Scholar]
- 21.Herr, W., R. A. Sturm, R. G. Clerc, L. M. Corcoran, D. Baltimore, P. A. Sharp, H. A. Ingraham, M. G. Rosenfeld, M. Finney, G. Ruvkun, et al. 1988. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev. 2:1513-1516. [DOI] [PubMed] [Google Scholar]
- 22.Houvras, Y., M. Benezra, H. Zhang, J. J. Manfredi, B. L. Weber, and J. D. Licht. 2000. BRCA1 physically and functionally interacts with ATF-1. J. Biol. Chem. 275:36230-36237. [DOI] [PubMed] [Google Scholar]
- 23.Hulit, J., T. Bash, M. Fu, F. Galbiati, C. Albanese, D. R. Sage, A. Schlegel, J. Zhurinsky, M. Shtutman, A. Ben-Ze'ev, M. P. Lisanti, and R. G. Pestell. 2000. The cyclin D1 gene is transcriptionally repressed by caveolin-1. J. Biol. Chem. 275:21203-21209. [DOI] [PubMed] [Google Scholar]
- 24.Joyce, D., B. Bouzahzah, M. Fu, C. Albanese, M. D'Amico, J. Steer, J. U. Klein, R. J. Lee, J. E. Segall, J. K. Westwick, C. J. Der, and R. G. Pestell. 1999. Integration of Rac-dependent regulation of cyclin D1 transcription through a nuclear factor-κB-dependent pathway. J. Biol. Chem. 274:25245-25249. [DOI] [PubMed] [Google Scholar]
- 25.Kakizawa, T., T. Miyamoto, K. Ichikawa, A. Kaneko, S. Suzuki, M. Hara, T. Nagasawa, T. Takeda, J. Mori, M. Kumagai, and K. Hashizume. 1999. Functional interaction between Oct-1 and retinoid X receptor. J. Biol. Chem. 274:19103-19108. [DOI] [PubMed] [Google Scholar]
- 26.Kim, J., J. Lu, and P. G. Quinn. 2000. Distinct cAMP response element-binding protein (CREB) domains stimulate different steps in a concerted mechanism of transcription activation. Proc. Natl. Acad. Sci. USA 97:11292-11296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kutoh, E., P. E. Stromstedt, and L. Poellinger. 1992. Functional interference between the ubiquitous and constitutive octamer transcription factor 1 (OTF-1) and the glucocorticoid receptor by direct protein-protein interaction involving the homeo subdomain of OTF-1. Mol. Cell. Biol. 12:4960-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, R. J., C. Albanese, R. J. Stenger, G. Watanabe, G. Inghirami, G. K. Haines, 3rd, M. Webster, W. J. Muller, J. S. Brugge, R. J. Davis, and R. G. Pestell. 1999. pp60v-src induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways. A role for cAMP response element-binding protein and activating transcription factor-2 in pp60v-src signaling in breast cancer cells. J. Biol. Chem. 274:7341-7350. [DOI] [PubMed] [Google Scholar]
- 29.Luo, Y., and R. G. Roeder. 1995. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol. Cell. Biol. 15:4115-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayr, B., and M. Montminy. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell. Biol. 2:599-609. [DOI] [PubMed] [Google Scholar]
- 31.Morris, L., K. E. Allen, and N. B. La Thangue. 2000. Regulation of E2F transcription by cyclin E-Cdk2 kinase mediated through p300/CBP co-activators. Nat. Cell Biol. 2:232-239. [DOI] [PubMed] [Google Scholar]
- 32.Murphy, S., J. B. Yoon, T. Gerster, and R. G. Roeder. 1992. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol. Cell. Biol. 12:3247-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagata, D., E. Suzuki, H. Nishimatsu, H. Satonaka, A. Goto, M. Omata, and Y. Hirata. 2001. Transcriptional activation of the cyclin D1 gene is mediated by multiple cis-elements, including SP1 sites and a cAMP-responsive element in vascular endothelial cells. J. Biol. Chem. 276:662-669. [DOI] [PubMed] [Google Scholar]
- 34.Nakajima, T., C. Uchida, S. F. Anderson, J. D. Parvin, and M. Montminy. 1997. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 11:738-747. [DOI] [PubMed] [Google Scholar]
- 35.Nakshatri, H., P. Nakshatri, and R. A. Currie. 1995. Interaction of Oct-1 with TFIIB. Implications for a novel response elicited through the proximal octamer site of the lipoprotein lipase promoter. J. Biol. Chem. 270:19613-19623. [DOI] [PubMed] [Google Scholar]
- 36.Perkins, N. D., L. K. Felzien, J. C. Betts, K. Leung, D. H. Beach, and G. J. Nabel. 1997. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science 275:523-527. [DOI] [PubMed] [Google Scholar]
- 37.Philips, A., S. Lesage, R. Gingras, M. H. Maira, Y. Gauthier, P. Hugo, and J. Drouin. 1997. Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Mol. Cell. Biol. 17:5946-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin, X. F., Y. Luo, H. Suh, J. Wayne, Z. Misulovin, R. G. Roeder, and M. C. Nussenzweig. 1994. Transformation by homeobox genes can be mediated by selective transcriptional repression. EMBO J. 13:5967-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinn, P. G. 1993. Distinct activation domains within cAMP response element-binding protein (CREB) mediate basal and cAMP-stimulated transcription. J. Biol. Chem. 268:16999-17009. [PubMed] [Google Scholar]
- 40.Ravenhall, C., E. Guida, T. Harris, V. Koutsoubos, and A. Stewart. 2000. The importance of ERK activity in the regulation of cyclin D1 levels and DNA synthesis in human cultured airway smooth muscle. Br. J. Pharmacol. 131:17-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rimerman, R. A., A. Gellert-Randleman, and J. A. Diehl. 2000. Wnt1 and MEK1 cooperate to promote cyclin D1 accumulation and cellular transformation. J. Biol. Chem. 275:14736-14742. [DOI] [PubMed] [Google Scholar]
- 42.Saluja, D., M. F. Vassallo, and N. Tanese. 1998. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol. Cell. Biol. 18:5734-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 44.Shtutman, M., J. Zhurinsky, I. Simcha, C. Albanese, M. D'Amico, R. Pestell, and A. Ben-Ze'ev. 1999. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 96:5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strubin, M., J. W. Newell, and P. Matthias. 1995. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell 80:497-506. [DOI] [PubMed] [Google Scholar]
- 46.Sturm, R. A., G. Das, and W. Herr. 1988. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 2:1582-1599. [DOI] [PubMed] [Google Scholar]
- 47.Sturm, R. A., and W. Herr. 1988. The POU domain is a bipartite DNA-binding structure. Nature 336:601-604. [DOI] [PubMed] [Google Scholar]
- 48.Sun, Y., F. T. Kolligs, M. O. Hottiger, R. Mosavin, E. R. Fearon, and G. J. Nabel. 2000. Regulation of beta-catenin transformation by the p300 transcriptional coactivator. Proc. Natl. Acad. Sci. USA 97:12613-12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tetsu, O., and F. McCormick. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422-426. [DOI] [PubMed] [Google Scholar]
- 50.Wadzinski, B. E., W. H. Wheat, S. Jaspers, L. F. Peruski, Jr., R. L. Lickteig, G. L. Johnson, and D. J. Klemm. 1993. Nuclear protein phosphatase 2A dephosphorylates protein kinase A-phosphorylated CREB and regulates CREB transcriptional stimulation. Mol. Cell. Biol. 13:2822-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, H. G., P. Yaciuk, R. P. Ricciardi, M. Green, K. Yokoyama, and E. Moran. 1993. The E1A products of oncogenic adenovirus serotype 12 include amino-terminally modified forms able to bind the retinoblastoma protein but not p300. J. Virol. 67:4804-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, T. C., R. D. Cardiff, L. Zukerberg, E. Lees, A. Arnold, and E. V. Schmidt. 1994. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 369:669-671. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe, G., C. Albanese, R. J. Lee, A. Reutens, G. Vairo, B. Henglein, and R. G. Pestell. 1998. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol. Cell. Biol. 18:3212-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 55.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]
- 56.Zwilling, S., A. Annweiler, and T. Wirth. 1994. The POU domains of the Oct-1 and Oct2 transcription factors mediate specific interaction with TBP. Nucleic Acids Res. 22:1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zwilling, S., H. Konig, and T. Wirth. 1995. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 14:1198-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]