Abstract
Streptococcus suis is an important cause of infectious diseases in young pigs. Little is known about the virulence factors or protective antigens of S. suis. Recently, we have identified two proteins of the arginine deiminase system (ADS) of S. suis, which were temperature induced and expressed on the streptococcal surface (N. Winterhoff, R. Goethe, P. Gruening, M. Rohde, H. Kalisz, H. E. Smith, and P. Valentin-Weigand, J. Bacteriol. 184:6768-6776, 2002). In the present study, we analyzed the complete ADS of S. suis. Due to their homologies to the recently published S. gordonii ADS genes, the genes for arginine deiminase, ornithine carbamoyl-transferase, and carbamate kinase, which were previously designated adiS, octS, and ckS, respectively, were renamed arcA, arcB, and arcC, respectively. Our data revealed that arcA, arcB, and arcC of the S. suis ADS are transcribed from an operon (arcABC operon). Additionally, putative ADS-associated genes were cloned and sequenced which, however, did not belong to the arcABC operon. These were the flpS gene upstream of the arcABC operon with homology to the flp transcription regulator of S. gordonii and the arcD, arcT, arcH, and argR genes downstream of the arcABC operon with high homologies to a putative arginine-ornithine antiporter, a putative dipeptidase of S. gordonii, a putative β-N-acetylhexosaminidase of S. pneumoniae, and a putative arginine repressor of S. gordonii, respectively. The transcriptional start point of the arcABC operon was determined, and promoter analysis provided evidence that multiple factors contribute to the regulation of the ADS. Thus, a putative binding site for a transcription regulator of the Crp/Fnr family, an ArgR-binding site, and two cis-acting catabolite response elements were identified in the promoter-operator region of the operon. Consistent with this, we could demonstrate that the ADS of S. suis is inducible by arginine and reduced O2 tension and subject to carbon catabolite repression. Furthermore, comparing an arcA knockout mutant in which expression of the three operon-encoded proteins was abolished with the parental wild-type strain showed that the arcABC operon of S. suis contributes to survival under acidic conditions.
Streptococcus suis is a major cause of meningitis, septicemia, bronchopneumonia, and sudden death in young pigs. As a zoonotic agent, S. suis infects humans, causing meningitis and septicemia (2, 9, 13). Very little is known about the pathogenesis, virulence factors, and protective antigens of S. suis. The serotype 2 polysaccharide capsule of S. suis is the only virulence factor proven so far (10). However, in S. suis 35 different capsular serotypes have been described, and virulence differs among the serotypes and strains of the same serotype. Serotype 2 S. suis is considered the most prevalent capsular type in diseased pigs (1, 25), and the capsule of S. suis serotype 2 has been shown to play an essential role in pathogenesis (46). The muramidase-released protein (52), the extracellular protein factor (52), suilysin (26, 30), adhesins (49), and the fibronectin- and fibrinogen-binding protein (17) have been described as virulence-associated factors. However, virulent strains lacking virulence factors have been isolated from diseased pigs (25), indicating that as-yet-unknown virulence factors exist.
We have previously identified two cell wall-associated proteins of S. suis that were induced by a temperature shift from 32°C or 37°C to 42°C (53). Amino-terminal sequence analysis of the two proteins indicated homologies to an ornithine carbamoyl-transferase (OCT) and the streptococcal acid glycoprotein from S. pyogenes, an arginine deiminase (AD) recently proposed as a putative virulence factor (15, 16). Cloning and sequencing of the respective genes of S. suis, adiS (80.2% homology to the sagP gene), and octS (81.2% homology to an OCT of S. pyogenes) and their adjacent upstream and downstream regions revealed that they were clustered together with two additional open reading frames (ORFs). The first ORF (orf-2) showed 59.8% homology to a gene encoding a hypothetical cytosolic protein, and the second ORF (ckS) showed 70.1% homology to a carbamate kinase of S. pyogenes. Thus, we concluded that the genes adiS, octS, and ckS make up the AD system (ADS) of S. suis (53).
The ADS catalyzes the conversion of arginine to ornithine, ammonia, and carbon dioxide and concomitantly generates 1 mol of ATP per mol of arginine consumed. The ADS is widely distributed among prokaryotic organisms, including halobacteria, Pseudomonas spp., Bacillus spp., lactic acid bacteria, and oral streptococci (7, 14, 22, 36, 43, 55). It comprises three major enzymes, the AD, the OCT, and the carbamate kinase. The genes encoding the ADS are commonly organized as an operon. In addition to the genes encoding the catalytic activities, a number of genes can be found associated with the ADS gene cluster. These include regulatory genes belonging to different families of transcriptional regulators, not-yet-characterized genes, and genes encoding putative transport proteins. The gene arrangement and regulation and the biological role of the ADS differ among species (3, 7, 18, 19, 22, 35, 57). In some bacteria, such as Pseudomonas aeruginosa and Bacillus licheniformis, the ADS permits growth under anaerobic conditions (18, 36). In others, such as oral streptococci (20) and Lactobacillus sakei (55), expression of the ADS is under the control of carbon catabolite repression (CCR) and its expression is increased in the presence of arginine (19, 55). In oral streptococci and S. pyogenes, the ADS seems to provide protection against acidic stress by the production of ammonia (7, 15). Furthermore, it has been shown that the AD of S. pyogenes is involved in adhesion to and invasion of epithelial cells (15, 37). Little is known about the regulation of the ADS in S. pyogenes and other pathogenic bacteria.
In this study, we characterized the ADS of S. suis with respect to its genetic organization and regulation and putative functions.
MATERIALS AND METHODS
Materials, bacterial strains, and growth conditions.
If not stated otherwise, all chemicals were purchased from Sigma (Munich, Germany). Previously described S. suis strains I9841/1 (1) and 10 (46) were grown in Todd-Hewitt broth (THB; Oxoid, Wesel, Germany) and subcultured on Columbia blood agar base (Oxoid) containing 6% (vol/vol) sheep blood overnight at 37°C. For microaerophilic and anaerobic growth conditions, bacteria were inoculated into freshly autoclaved THB and cultured in jars with respective gas delivery envelopes (CampyGen or AnaeroGen; Oxoid, Wesel, Germany) overnight at 37°C. The influence of different glucose concentrations on AD activity was analyzed with bacteria grown in Trypticase soy broth (TSB; Difco) supplemented with different glucose concentrations (0.125 to 1%). To examine the influence of glucose, galactose, and arginine on AD activity, the bacteria were grown in a low-carbohydrate tryptone-vitamin (TV)-based minimal medium (5) supplemented with the respective sugar at 0.2 or 2%, with or without 1% arginine. For temperature stress experiments, S. suis was cultured as described previously (53).
Escherichia coli strains were grown in Luria-Bertani broth and subcultured on Luria-Bertani agar plates. If required, antibiotics were added at the following concentrations: spectinomycin at 100 μg/ml (S. suis) and 50 μg/ml (E. coli), ampicillin at 100 μg/ml (E. coli), and kanamycin at 25 μg/ml (E. coli).
DNA techniques and sequence analysis.
Chromosomal S. suis DNA was prepared according to standard procedures as described by Sambrook and Russell (44). Plasmid preparations were performed with Plasmid Kits from Macherey and Nagel (Dueren, Germany) according to the manufacturer's instructions.
The up- and downstream regions of the arcABC operon were cloned by PCR and sequenced (Seqlab, Goettingen, Germany). The specific primer sequences are available on request. Sequence analyses were performed with the HUSAR server (Heidelberg Unix Sequence Analysis Resource).
RNA preparation.
Total RNA was prepared by the method of Chomczynski and Sacchi (12), with some modifications. Briefly, S. suis cultures were stress treated for 1 h at 42°C (53) or incubated at 37°C to late exponential growth phase. Ten milliliters of each culture was chilled on ice and then centrifuged for 10 min at 14,000 × g and 4°C. The bacterial pellet was suspended in 600 μl denaturation buffer (4 M guanidinium thiocyanate, 25 mM sodium citrate, pH 7, 0.5% Sarkosyl, 100 mM 2-mercaptoethanol) with 400 mg zirconium beads. The cells were disrupted by bead beating for 2 × 3 min with intermediate cooling on ice. Cell debris and zirconium beads were removed by centrifugation, and the supernatant was transferred into a new 2-ml polypropylene tube. The preparation was then continued exactly as described by Chomczynski and Sacchi (12). RNA was treated with DNase I (Roche, Mannheim, Germany) for 30 min at 37°C, followed by phenol extraction and precipitation.
Primer extension.
The transcriptional start point of the AD operon was determined by primer extension analysis as described by Sambrook and Russell (44). Briefly, 20 μg DNase I-treated RNA from temperature-induced S. suis cultures was incubated with 100,000 cpm of [γ-32P]dATP-labeled primer PE (5′-CTGCCTCTCATTTCCTCTTTACAT-3′) for 15 min at 58°C and reverse transcribed at 42°C for 1 h. The extension product was analyzed on a denaturing 6% polyacrylamide gel containing 6 M urea. Sequencing was performed with the Reader Sequencing Kit (Fermentas, St. Leon-Rot, Germany) according to the manufacturer's instructions with plasmid pBAD and primer PE.
cDNA synthesis and PCR.
For cDNA synthesis, 2 μg of DNase I-treated total RNA from S. suis grown at 37°C was reverse transcribed with primers specific for flpS (5′-AAACATGGCTAACCGTTTCC-3′), arcA (5′-GTAGCAAATGGATCCCGTGT-3′), arcC (5′-TTTGCATGCAGAGTCATCAAT-3′), and arcD (5′-GCAAGATGATGGCTACACCA-3′) and 100 U Superscript II (Invitrogen, Groningen, The Netherlands) as described by the manufacturer. PCRs for gene-specific and intragenic products were performed with 2 U Taq polymerase (Invitrogen) and specific oligonucleotide primer pairs according to the manufacturer's instructions. Primer pair 1 (P1 [5′-GGAGTTTTGATGATAAGCAACG-3′] and P2 [5′-AAACATGGCTAACCGTTTCC-3′]) and primer pair 2 (P3 [5′-TGGTTCGAGAGGACCAGTTT-3′] and P2) were used in combination with the flpS cDNA. PCRs with the arcA cDNA were performed with primer pair 3 (P4 [5′-ATGAGAGGAGGAAAGCGTA-3′] and P5 [5′-TCATGCTCTTTCTGTGCATCTT-3′]) and primer pair 4 (P6 [5′-GCAACTGAAGCAGGATGG-3′] and P7 [5′-CCTGCCTCTCATTTCCTCTTT-3′]). Primer pairs 4, 5 (P8 [5′-TTTGCTCAAGCTCTTCGTGA-3′] and P9 [5′-ACCTTCAGCCAAGGCAACTA-3′]), and 6 (P10 [5′-GTACTCGTGCTGCCTTCACA-3′] and P11 [5′-CTGCCAAACATTGAGAAGCA-3′]) were used in the PCR with the arcC cDNA, and primer pairs 7 (P12 [5′-AAATATTATAGGAGGTTTTTGCGATG-3′] and P13 [5′-GCAAGATGATGGCTACACCA-3′]) and 8 (P13 and P14 [5′-TCATTGCAAAGGCTAAGCAG-3′]) were used in combination with the arcD cDNA. PCR conditions consisted of 2 min of initial denaturation at 94°C; 30 cycles of 1 min at 94°C, 1 min at 57 to 60°C, and 1 min at 72°C; and a final elongation for 10 min at 72°C.
Production and purification of recombinant proteins.
Recombinant ArcA, ArcB, and ArcC were produced as His tag fusions in E. coli with the QIAexpress paired pREP4-pQE plasmid system (QIAGEN, Hilden, Germany). Genes were amplified with deleted translation initiation codons by PCR from chromosomal DNA with respective specific oligonucleotide primers containing restriction sites (arcAKpnI [5′-GGGGTACCTCAAACCATCCAATTCAT-3′], arcAHindIII [5′-CCCAAGCTTTTAGATGTCTTCACGTTC-3′], arcBKpnI [5′-GGGGTACCACAAACGTATTTAAAGGT-3′], arcBHindIII [5′-CCCAAGCTTTACACACGTGGAACAAAT-3′], arcCBamHI [5′-CGGGATCCTGTAAGCGATTACTATTT-3′], and arcCHindIII [5′-CCCAAGCTTCAACGATCATACAGCTGC-3′]). PCR products were cloned first into the pCR2.1 TA cloning vector (Invitrogen). The KpnI-HindIII arcA and arcB fragments and the BamHI-HindIII arcC fragments were subsequently subcloned into correspondingly digested pQE vectors and verified by sequencing (Seqlab). Resulting plasmids pQE32arcA, pQE30arcB, and pQE30arcC were introduced into E. coli strain M15(pREP4) (QIAGEN). After isopropyl-β-d-thiogalactopyranoside (IPTG) induction, proteins were overexpressed as N-terminal hexahistidyl derivates (His6-ArcA, His6-ArcB, and His6-ArcC) and isolated by Ni2+-nitrilotriacetic acid affinity chromatography according to the manufacturer's instructions. Polyclonal rabbit antisera against the purified proteins were obtained from Seqlab.
Construction of gene-specific knockout mutants.
Inactivation of the arcABC operon was achieved by insertion of a spectinomycin resistance cassette into the arcA gene. A 2,840-nucleotide (nt) PCR fragment (positions −965 to +1875 according to the transcriptional start point) comprising the arcA gene and adjacent regions was amplified by PCR from chromosomal DNA of S. suis strain I9841/1 with oligonucleotide primers 5′-TGGTTCGAGAGGACCAGTTT-3′ and 5′-TTGTCAATGTCTGGCACCAT-3′. The PCR was performed in a total volume of 50 μl containing 100 ng chromosomal DNA of S. suis and 2 U Taq polymerase (Invitrogen) as described by the manufacturer. PCR conditions were an initial 2 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 57°C, and 3 min at 72°C; and a final elongation for 10 min at 72°C. The PCR product was cloned into the pGEM-T Easy vector (Promega, Mannheim, Germany), resulting in plasmid pGEMAD. The DNA insert was then subcloned as an ApaI-SacI fragment into the pBluescriptII SK+ vector (Stratagene, Heidelberg, Germany), resulting in plasmid pBAD. The internal ClaI-EcoRV fragment (positions +35 to +341) was replaced with the ClaI-SmaI fragment of pICspc (39, 46) containing the spectinomycin resistance gene, resulting in pBADspc. The EcoRI fragment from plasmid pBADspc, containing the inserted spectinomycin resistance cassette and the arcA-adjacent regions, was inserted into the pIC vector to gain plasmid pICADspc. Electrotransformation of S. suis was done as described by Smith et al. (47), with 1 μg DNA of plasmid pICADspc and a Bio-Rad Gene Pulser apparatus (Bio-Rad, Munich, Germany). Pulses were performed with settings of 25 μF, 2.5 kV, and 200 Ω and a time constant of 4.5 to 4.6 ms. After electroporation, 1 ml THB-0.3 M sucrose was added to the cells. Bacteria were incubated for 2 h at 37°C and plated on agar plates containing selective antibiotics. The arcABC mutant strain was grown under aerobic conditions in THB-1% yeast extract (THY) and subcultured on Columbia blood agar base containing 6% (vol/vol) horse blood and spectinomycin (100 μg/ml) overnight at 37°C. Control of the arcABC mutant strain was performed by PCR with oligonucleotide primers for arcB (5′-GGGGTACCACAAACGTATTTAAAGGT-3′ and 5′-CCCAAGCTTTACACACGTGGAACAAAT-3′), arcC (5′-CGGGATCCTGTAAGCGATTACTATTT-3′ and 5′-CCCAAGCTTCAACGATCATACAGCTGC-3′), and spc (5′-TGGTACCGTGGAATCATCCT-3′ and 5′-GGAGAAGATTCAGCCACTGC-3′) and primers adjacent to the insertion site of the spc resistance gene cassette (5′-AGCGAGAGCAGTTTGCTACC-3′ and 5′-TCATGCTCTTTCTGTGCATCTT). The PCR was performed as described above. Southern analyses were performed with BamHI-cleaved genomic DNA according to standard protocols (44).
Immunoblot analysis.
Immunoblot analysis of whole-cell lysates was performed as previously described (53), with specific anti-ArcA, anti-ArcB, and anti-ArcC antisera at a final dilution of 1:200.
Determination of AD activity.
AD activity from whole-cell lysates was assayed as described previously (53). Results were expressed as nanomoles of citrulline produced per hour per milligram of streptococcal protein.
Survival of S. suis under acidic conditions.
Experiments were performed as described previously (4). Briefly, S. suis cultures were grown overnight in TSB medium and harvested by centrifugation. Cells were suspended in 20 mM Na2HPO4-1 mM MgCl2-25 mM arginine-HCl adjusted to pH 4, 5, 6, or 7. Controls were incubated at pH 4 without arginine-HCl. Suspensions were incubated for up to 4 h at 37°C. The number of viable bacteria was quantified by plating on THY. Results were expressed as percent survival compared to the CFU of the initial inoculum.
RESULTS
Genetic organization and characterization of the ADS gene cluster.
While our first report on the ADS of S. suis was in press (53), the ADS of S. gordonii was published by Dong et al. (20). Subsequent sequence analysis of the S. suis ADS genes revealed high homologies to their analogues in S. gordonii. Hence, the genes for AD, ornithine carbamoyl-transferase, and carbamate kinase, previously designated adiS, octS, and ckS, respectively, were renamed arcA, arcB, and arcC, respectively. Further sequence analysis of ORF2 revealed similarities to an acetyltransferases of S. pyogenes and S. agalactiae. However, due to a weak Shine-Dalgarno box and a start codon within the AD coding sequence, we assume that this ORF is not expressed in S. suis. Therefore, this ORF was not assigned a specific name.
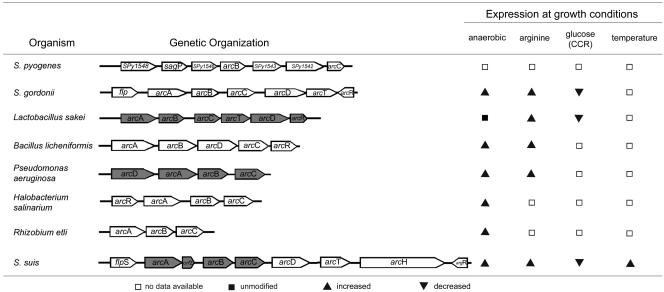
The genes of the ADS are generally organized in an operon-like structure (56). Many ADS operons are associated with genes encoding an arginine-ornithine antiporter and a transcription regulator of the Crp/Fnr or ArgR/AhrC family (3, 15, 22, 33, 35, 36) (see also Fig. 6). Therefore, we cloned and sequenced the up- and downstream flanking regions of the arcA, arcB, and arcC gene cluster of S. suis. Sequence analysis confirmed an additional ORF of 696 nt upstream of the arcA gene. The predicted protein sequence shared 43% identity with a transcription regulator of S. pyogenes (AE014160) belonging to the family of Crp/Fnr transcription regulators and also a homology of 36% to the ArcR protein of B. licheniformis (CAB95946). Alignment with the partial genome of S. gordonii (www.tigr.org) revealed the highest sequence identity to S. gordonii flp (64%). Analysis of the predicted amino acid sequence revealed a putative CAP effector domain in the N-terminal region of the protein and a helix-turn-helix motif in the C-terminal region, which might be responsible for DNA binding. Due to the homology to the Flp regulator protein of S. gordonii, this ORF was named flpS. Upstream of the flpS gene, two additional ORFs were identified; both putative proteins showed homologies of up to 78% to conserved and hypothetical proteins SP1565 (AAK756562) and spr1422 (E98049), respectively, of S. pneumoniae that have not been further characterized. Downstream of arcC, analyses revealed four further ORFs. The first one (1,502 nt) was named arcD due to a homology of 70% to an arginine-ornithine antiporter of S. gordonii (AAN65258). The second, a 1,358-nt ORF, was named arcT because of a homology of 67 to 71% to arcT of S. gordonii (AF534569). The third ORF, with a size of 4,259 nt, showed a homology of up to 69% to a β-N-acetylhexosaminidase of S. pneumoniae (L36923). The last ORF, 473 nt, showed a homology of 70% to arcR of S. gordonii (AF534569). Consistent with what has been described in S. gordonii (22), strong similarities were found to regulatory proteins involved in arginine metabolism; e.g., the pI of the first 100 amino acids was basic (9.25), while the pI of the last ones was acidic (3.82). Furthermore, a typical serine-arginine motif essential for DNA binding was found in the N-terminal region of the ArgR sequence, while conserved amino acid residues important for arginine binding were detected at positions 103 (alanine), 125, and 126 (aspartic acid). Thus, this ORF was named argR.
FIG. 6.
Comparison of the genetic organization and regulation of known ADSs. The ADSs of S. pyogenes (15), S. gordonii (19, 20), L. sakei (55), B. licheniformis (35), P. aeruginosa (22), H. salinarum (43), R. etli (18), and S. suis are depicted. Gray gene symbols represent genes with confirmed operon structures.
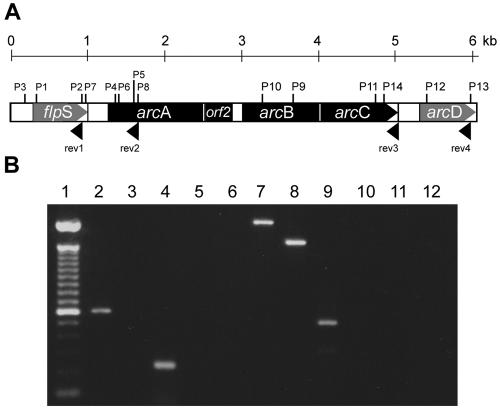
Next, we determined which genes of the ADS cluster of S. suis were organized in an operon. For this, RNA isolated from S. suis cultures grown at 37°C was reverse transcribed with oligonucleotide primers specific for the flpS gene, the arcA gene, the arcC gene, and the arcD gene (Fig. 1A). Reverse transcription (RT)-PCR analyses were performed with the respective gene-specific primer pairs. For each cDNA, at least one primer pair consisted of a gene-specific oligonucleotide primer and an additional primer pair specific for the indicated 5′ adjacent genes. As shown in Fig. 1B, all gene-specific RT-PCRs generated the expected PCR products (lanes 2, 4, 7, 8, and 9). However, only the PCR fragments for the intergenic regions between arcA and arcB and between arcB and arcC were amplified (lanes 7 and 8), indicating that these genes compose the arcABC operon of S. suis, whereas genes flpS and arcD appeared to be located outside the operon.
FIG. 1.
The arcABC operon of S. suis consists of the arcA, arcB, and arcC genes. (A) Schematic representation of the positions of the primer sequences used for RT (rev1 to rev3) and the primers used for PCR (P1 to P14) as described in Materials and Methods. (B) Ethidium bromide-stained gel from the RT-PCR analysis of mRNA of S. suis grown in THB at 37°C. Lane 1, molecular weight marker; lane 2, flpS intragenic PCR fragment (603 bp, primer pair 1); lane 3, flpS upstream region 3′-end flpS PCR fragment (primer pair 2); lane 4, arcA intragenic PCR fragment (229 bp, primer pair 3); lanes 5 and 6, flpS-arcA gene intergenic PCR fragment (primer pair 4 each); lane 7, arcA-arcB intergenic PCR fragment (2,132 bp, primer pair 5); lane 8, arcB-arcC intergenic PCR fragment (1,621 bp, primer pair 6); lane 9, arcD intragenic PCR fragment (527 bp, primer pair 7); lane 10, arcC-arcD intergenic PCR fragment (primer pair 8); lanes 11 and 12, control reactions with mRNA without RT and double-distilled H2O, respectively.
Analysis of the promoter region and the 5′ untranslated region of the arcABC operon.
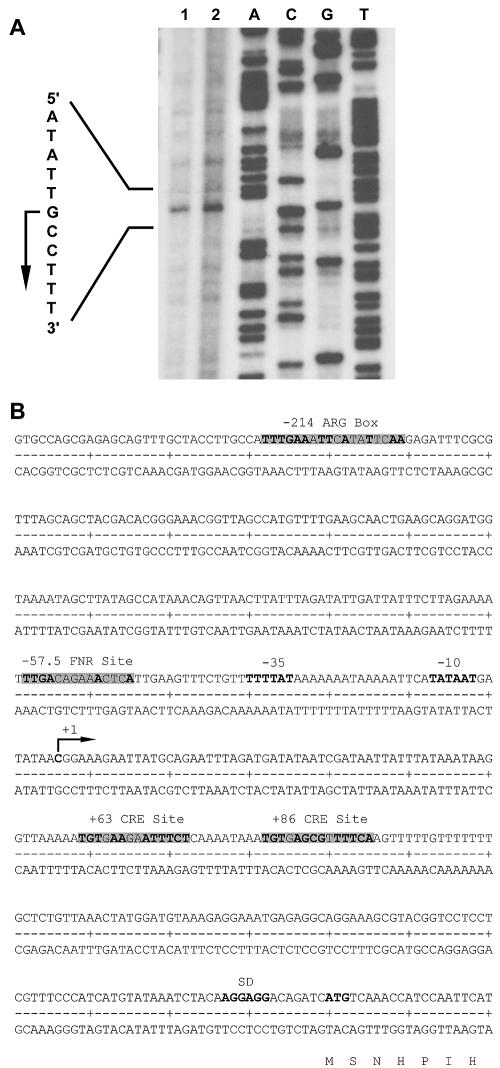
We determined the transcriptional start point of the arcABC operon by primer extension analysis with RNA isolated from temperature stress-induced S. suis cultures (Fig. 2A). The transcriptional start point was identified 214 nt upstream of the arcA start codon, which we identified previously by N-terminal sequencing (53). Sequence analysis of the promoter region (Fig. 2B) revealed a −10 region, TATAAT, that matched the typical consensus sequence and a −35 region, TTTTAT, 17 nt upstream of the −10 region which, however, shared only two bases (bold letters) with the typical consensus sequence TTGACA.
FIG. 2.
Analysis of the promoter region and the 5′ untranslated region of the arcABC operon. (A) Primer extension experiments with 10 μg (lane 1) and 15 μg (lane 2) of heat-induced (42°C) RNA of S. suis strain I9841/1. (B) Sequence of the promoter region of the S. suis arcABC operon. The transcriptional start point (arrow), the predicted −10 and −35 boxes, the predicted ARG box, the predicted Fnr-binding site, the predicted cre-binding sites, and the predicted Shine-Dalgarno (SD) sequence of arcA are indicated.
Based on the homologies of the flpS gene to the family of Crp/Fnr transcription regulators and to ArcR homologous proteins, we analyzed the promoter sequence for possible Crp/Fnr DNA-binding sites. A putative binding site for a transcription regulator of the Crp/Fnr family was identified at position −57.5 upstream of the transcriptional start point. This sequence, TTGACAGAAACTCA, showed homologies (bold letters) to the Fnr consensus region of E. coli, TTGAT-N1-N4-ATCAA (21).
In other bacteria, the ADS is inducible by arginine via activation through ArgR. Therefore, we investigated the region upstream of the promoter of S. suis arcA with regard to E. coli ArgR-binding sites, ANTGAATAATTATTCAN or TNTGAATTTAAATTCAN, respectively (34). A potential binding site for ArgR (TTTGAAATTCATATTCAA) was identified at positions −196 to −214 from the transcriptional start point.
In oral streptococci (20) and L. sakei (55), expression of the ADS is under the control of CCR. In AT-rich gram-positive bacteria, CCR is mediated by trans-acting catabolite control protein A (CcpA). CcpA binds to cis-acting catabolite response elements (cre) in the presence of favored carbohydrate sources to repress the expression of catabolic genes and operons. Genes repressed by CcpA generally contain cre boxes (TGWAARCGYTWNCW) within and downstream of the promoter (28, 29, 50). We identified two potential CcpA-dependent cre sites at +63 (TGTGAAGAATTTCT) and +86 (TGTGAGCGTTTTCA) with bases matching the consensus (bold letters) within the untranslated region of the S. suis arcABC operon (Fig. 2B).
Regulation of the ADS by arginine, reduced oxygen tension, and carbohydrate catabolite repression.
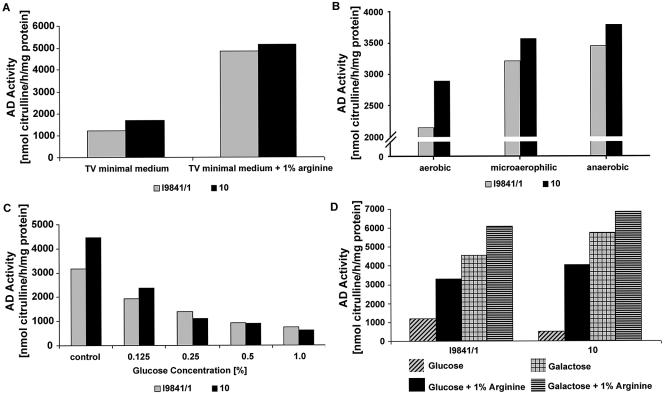
The presence of a putative ArgR-binding site, a possible Fnr-binding site, and the two potential cre-binding sites suggested that the ADS of S. suis is transcriptionally controlled by a multitude of regulator mechanisms. Thus, we examined the regulation of the ADS by variation of growth conditions. First, we determined whether expression of the ADS is induced by arginine (ArgR regulated) and under anaerobic growth conditions (flpS regulated). We measured the AD activities of S. suis strains I9841/1 and 10 grown in TV minimal medium in the presence and absence of 1% arginine. As shown in Fig. 3A, the AD activities of both strains tested were substantially enhanced when the bacteria were grown in the presence of arginine, indicating that the ADS is inducible by arginine. In addition, we tested the AD activities of S. suis grown under aerobic, microaerophilic, and anaerobic culture conditions. Results revealed that the increase in AD activity is correlated with a decrease in O2 tension (Fig. 3B).
FIG. 3.
Regulation of the ADS by arginine, reduced oxygen tension, and carbohydrate catabolite repression. (A) AD activities of S. suis serotype 2 strains grown in TV medium with or without 1% arginine (open bars, strain I9841/1; filled bars, strain 10). (B) AD activities of S. suis serotype 2 strains grown in THB medium under aerobic, microaerophilic, or anaerobic conditions (open bars, strain I9841/1; filled bars, strain 10). (C) AD activities of S. suis serotype 2 strains grown in TSB with increasing amounts of glucose (open bars, strain I9841/1; filled bars, strain 10). (D) S. suis serotype 2 strains grown in TV medium with or without 1% arginine and different carbohydrate sources (glucose and galactose). Results of representative experiments are shown. AD activities were determined as described in Materials and Methods. Each experiment was repeated at least twice.
The presence of two potential cre-binding sites prompted us to examine whether the ADS of S. suis is subject to CCR. Thus, bacteria were grown in glucose-free TSB medium and in medium supplemented with increasing concentrations of glucose. As shown in Fig. 3C, 0.125% glucose reduced AD activity by approximately 45%. AD activities were also determined for S. suis grown in TV minimal medium on various other energy sources (Fig. 3D). We expected higher levels of AD activity in the presence of sugars, e.g., galactose, which were not repressive of AD expression in other streptococci (20, 27). Accordingly, higher levels of AD activity were obtained from S. suis cultures grown in the presence of 0.2% galactose than from cells grown in the presence of 0.2% glucose. Although the presence of glucose had a repressive effect on AD expression, addition of 1% arginine to the growth medium seemed to partially abolish this effect, which was reflected by an increase in AD activity (Fig. 3D). Supplementation of glucose or galactose to a final concentration of 2% induced CCR in S. suis cultures, but addition of 1% arginine seemed to abolish the effect, most notably for cultures grown in the presence of galactose (data not shown).
Role of the ADS in survival of S. suis under acidic conditions.
Ammonium production by the ADS has been shown to protect bacteria from acidic stress (38). To study the role of the ADS of S. suis in an acidic environment, we constructed an arcA knockout mutant. The 5′ end of the arcA gene and its nontranslated region (positions +35 to +341 bp) were replaced with a spectinomycin (spc) resistance gene cassette. The resulting mutant strain was confirmed by PCR (Fig. 4A) and Southern analyses (Fig. 4B) and designated 10ΔarcA. Phenotypically, the mutant strain was examined via immunoblot analysis with antisera raised against the recombinant ArcA, ArcB, and ArcC proteins of S. suis (Fig. 4B). Whole-cell lysates of S. suis strains I9841/1, 10, and 10ΔarcA grown at 37°C were prepared and, after SDS-PAGE, blotted onto nitrocellulose membranes. The membranes were probed with anti-ArcA, anti-ArcB, and anti-ArcC antisera, respectively. As shown in Fig. 4C, the mutant strain did not express any of the proteins of the arcABC operon, indicating that expression of the complete operon was abolished by insertion of the spc resistance gene cassette. Accordingly, there was no detectable AD activity in the mutant strain compared to the wild-type strain (2,762.91 nmol citrulline/h/mg protein).
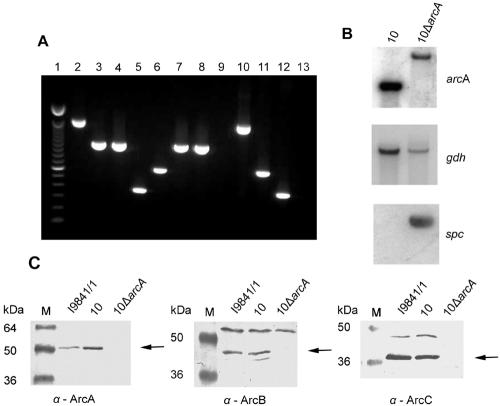
FIG. 4.
Construction of an arcA knockout mutant. (A) Control of strain 10ΔarcA in comparison with wild-type strain 10 by PCR. An ethidium bromide-stained agarose gel with the PCR DNA fragments is shown. Amplification was performed with chromosomal DNAs of strain 10ΔarcA (lanes 2 to 5) and wild-type strain 10 (lanes 6 to 9). ΔarcA-containing plasmid pICADspc (lane 10), wild-type arcA-containing plasmid pBAD (lane 11), and spectinomycin donor plasmid pICspc (lane 12) were used as positive controls, and double-distilled H2O (lane 13) was used as a negative control. Lane 1, molecular weight marker; lanes 2, 6, and 10, amplification reaction to verify insertion of the spc resistance gene cassette (1,609-bp fragment, successful insertion; 614-bp fragment, no insertion); lanes 3 and 7, arcB-specific DNA fragment (1,039 bp); lanes 4 and 8, arcC-specific DNA fragment (1,027 bp); lanes 5, 9, and 12, spc cassette-specific DNA fragment (361 bp). (B) Southern analysis of wild-type strain 10 and mutant strain 10ΔarcA with probes specific for the arcA, gdh (41), and spc genes. (C) Immunoblot analyses with whole-cell lysates of S. suis strains I9841/1, 10, and 10ΔarcA (lane M, molecular size marker). Specific antisera against the ArcA, ArcB, and ArcC (α-ArcA to -C) proteins were used at a 1:200 dilution. The specific protein bands are indicated by arrows.
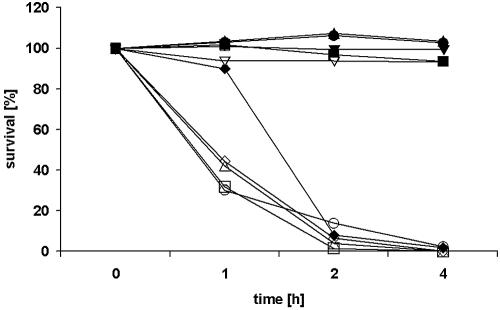
To analyze the ability of the ArcABC mutant strain to survive under acidic conditions, the mutant and parental strains were exposed to pH values of 4, 5, 6, and 7 in the presence or absence 25 mM l-arginine, respectively. Viable streptococci were quantified by serial platings on THY agar, and results were expressed as percent survival (CFU count at the indicated time point compared to that of the initial inoculum). Both the mutant and wild-type strains were sensitive to acidic pH in the absence of l-arginine (Fig. 5). In the presence of l-arginine, the wild-type strain survived under all pH conditions tested, whereas the ArcABC mutant strain was almost completely killed after a 2-h exposure to acidic pH (pH 4 to 6; Fig. 5). The viability of the mutant strain was not affected at pH 7. The decreased viability of the ArcABC mutant strain indicated that the arcABC operon of S. suis contributes to survival under acidic conditions.
FIG. 5.
Survival of S. suis in an acidic environment. Wild-type strain 10 (filled symbols) and mutant strain 10ΔarcA (open symbols) were grown to stationary phase and harvested, and equal numbers of bacteria were suspended in 20 mM Na2HPO4-1 mM MgCl2-25 mM l-arginine-HCl at pH 4 (□), pH 5 (▵), pH 6 (○), and pH 7 (▿). Buffer without l-arginine-HCl served as a control (⋄). The bacteria were incubated at 37°C, and after 1, 2, and 4 h, the number of viable organisms was determined by plating on THY agar. Results are presented as percent survival (CFU count at the indicated time point compared to that of the initial inoculum). Results of a representative experiment of three are shown.
DISCUSSION
The ADS is a multienzyme pathway that catalyzes the conversion of arginine to ornithine, ammonia, and CO2, with concomitant production of ATP (56). Due to these properties, ADS-expressing bacteria can overcome oxygen and nutrient starvation and are able to tolerate acidic environments (8, 38). Apart from the genes encoding the catalytic activities, other associated genes were found within the ADS gene cluster (Fig. 6). These genes include regulatory genes belonging to different families of transcription regulators, not-yet-characterized genes, and genes encoding putative transport proteins. Furthermore, the genes encoding the ADS are commonly organized as an operon. However, the gene arrangement, regulation, and physiological role of the ADS differs among species (Fig. 6) (3, 7, 18, 19, 22, 35, 57). The ADS is widely distributed among prokaryotes, and the primary structures of the enzymes involved in the AD pathway have been reasonably conserved throughout evolution. To date, ADS genes have been identified and sequenced in bacteria and archaea; no ADS genes or ADS enzyme activity has been reported for higher eukaryotes (56). In the amitochondriate parasitic protist Giardia intestinalis, the first two proteins of the ADS pathway, AD and OCT, were shown to be among 16 immunodominant proteins, underlining the importance of the ADS pathway in this parasite (42). The relevance of the ADS in pathogenic bacteria, however, has been described only for S. pyogenes. Here the ADS can be considered as a virulence factor. A protein originally designated streptococcal acid glycoprotein and originally reported to inhibit proliferation of human epidermoid carcinoma cells and human peripheral blood-derived mononuclear cells was ultimately identified as an AD that additionally contributed to acidic survival (15, 16, 54). However, the structure and regulation of the ADS in S. pyogenes have not been analyzed in detail.
Recently, we found that genes adiS, octS, and ckS (renamed arcA, arcB, and arcC, respectively) make up the ADS of S. suis. Interestingly, we could show that ArcA and ArcB were murein associated and that ArcA was expressed on the cell surface (53). Furthermore, we found that a natural, nonencapsulated, ArcA-deficient S. suis strain showed reduced survival after entry into epithelial cells compared to a natural, nonencapsulated, ArcA-expressing S. suis strain (4), emphasizing the putative roles of ArcA and ArcB as virulence factors of S. suis.
In the present study, we examined the structure and regulation of the ADS in S. suis. We identified an additional ORF upstream of the arcA gene named flpS with homologies to a Crp/Fnr transcription regulator of S. pyogenes, the ArcR protein of B. licheniformis, and flp of S. gordonii, which was required for expression of the ADS under anaerobic induction (19). The two additional ORFs upstream of the flpS gene encoded hypothetical proteins that seem not to be functionally associated with the ADS of S. suis. Downstream of arcC, further ORFs were identified which seem to be associated with the arcABC operon. The first one, with high homology to an arginine-ornithine antiporter of S. gordonii, was named arcD and followed by an Xaa-His dipeptidase (arcT) that shared high homologies with S. gordonii arcT (20). The putative β-N-acetylhexosaminidase (arcH), with high homology to an S. pneumoniae β-N-acetylhexosaminidase, has not been described in ADSs of other bacteria and seems to be a special feature of the ADS of S. suis. All these genes appear to be associated with the arcABC operon of S. suis.
Most of the ADSs are highly regulated (3, 20, 36, 57). Their genetic regulation, however, seems to be only partially conserved among organisms (Fig. 6). In P. aeruginosa (33) and B. licheniformis (35), the ADS is expressed under anaerobic conditions via regulators of the Crp/Fnr family. In all ADS-expressing bacteria, induction by Crp/Fnr can be further enhanced in the presence of arginine by ArgR, the transcriptional regulator of the arginine operon (33). In some bacteria, such as lactic acid bacteria (55) and oral streptococci (19), expression of the ADS is under the control of CCR and is inducible by arginine. In the present study, we demonstrated that all these regulation mechanisms apply to the ADS of S. suis.
The ADS of S. suis is induced under microaerophilic and anaerobic growth conditions. Oxygen-dependent regulation of the ADS is probably supported by the presence of the flpS gene, which has homologies to the family of Crp/Fnr transcription regulators and to the ArcR homologue protein of B. licheniformis, upstream of the arcABC operon. Furthermore, the promoter sequence of the arcABC operon contains a possible Crp/Fnr DNA-binding site at position −57.5 from the transcriptional start point of the arcA gene with homologies to the Fnr consensus region of E. coli (21). The members of the Fnr family have been proven to be responsible for anaerobic gene regulation in many gram-negative and some gram-positive bacteria (48). These regulators have been shown to be linked to ADS regulation. For instance, P. aeruginosa and B. licheniformis utilize the ADS exclusively under anaerobic conditions via regulators of the Crp/Fnr family (33, 35). The position of flpS of S. suis within the ADS gene cluster is similar to that of flp of S. gordonii, immediately upstream of arcA (19). The predicted amino acid sequence of FlpS of S. suis revealed a putative CAP effector domain in the N-terminal region of the protein and a helix-turn-helix motif in the C-terminal region; both structures are characteristic of Crp/Fnr regulator proteins (6, 48). In typical Fnr regulators, four conserved cysteine residues are usually found (48). As in the Fnr-like proteins of S. gordonii (19), Lactobacillus casei (24), and Lactococcus lactis (45), two cysteine residues have been found in the predicted amino acid sequence of FlpS of S. suis. Thus, FlpS seems to be more closely related to the ArcR regulators found in other ADS-expressing gram-positive bacteria (3, 35, 57) that have more Crp-like properties and are not typical FeS proteins (32).
We further found that the ADS of S. suis is inducible by arginine. In B. licheniformis and S. gordonii, the AD pathway is activated by the arginine regulators ArgR and ArcR, respectively, in the presence of arginine by binding to conserved ARG boxes. These boxes are located upstream of the promoters apart from the binding sites for Crp/Fnr-type proteins (20, 36). Accordingly, we identified a potential binding site for ArgR at positions −196 to −214 from the transcriptional start point of the arcABC operon, suggesting that the ADS of S. suis is regulated similarly to B. licheniformis and S. gordonii. Furthermore, we identified a putative regulator involved in arginine metabolism named ArgR.
Expression of the ADS in oral streptococci (20) and L. sakei (55) is under the control of CCR. In both bacteria, AD expression is induced in the presence of arginine and repressed by glucose. In AT-rich gram-positive bacteria, CCR is mediated by CcpA. CcpA binds to cis-acting catabolite response elements (cre) in the presence of favored carbohydrate sources to regulate the expression of catabolic genes and operons (50). Sequences resembling Ccp boxes were identified in a number of genes involved in the utilization of secondary carbon sources. In these genes, the target sequence for glucose repression is located within or downstream of the promoter region (28, 50). In the untranslated region of the S. suis arcA gene, downstream of the transcriptional start point, two potential CcpA-dependent cre sites were identified at positions +63 and +86, suggesting that CcpA contributes to CCR in S. suis. CcpA contributes to CCR in S. gordonii (19). However, since other pathways, such as CcpB (11), CcpC (31), and the phosphotransferase system, are known to be involved in CCR of some gram-positive bacteria (23, 40, 51), participation of CcpA in CCR of S. suis has to be confirmed by further studies.
Nonetheless, the presence of a putative ARG box, a possible Fnr-binding site, and two potential cre-binding sites suggests that the ADS of S. suis is transcriptionally controlled by a multitude of regulator proteins.
Initially, we detected the ADS of S. suis by identification of two cell wall-associated proteins that were induced by a temperature shift from 32°C or 37°C to 42°C (53). Interestingly, temperature-induced ADS expression has not been reported for other bacteria. Even though ArcA and AcrB are extracellularly expressed, the S. suis ADS is similar to the ADSs of other, closely related, species. However, there are significant differences in transcriptional organization structure, as emphasized by the presence of ArcH. Elucidation of the temperature regulation and the detailed molecular mechanisms of ADS regulation awaits further studies.
The ADS can be considered a system that protects oral streptococci and S. pyogenes against acidic stress (7, 15). Therefore, we examined the role of the arcABC operon of S. suis regarding survival in an acidic environment by a pH sensitivity test in which we compared an ArcABC mutant strain with its parental strain. The mutant strain was extremely sensitive to low pH, independent of the presence of 25 mM l-arginine, indicating that the ADS facilitates S. suis survival in acidified environments. These results were in good agreement with our previous data demonstrating that a natural, nonencapsulated arcA-negative S. suis strain, in contrast to a nonencapsulated arcA-positive S. suis strain, was able neither to survive in acidic vacuoles inside human epithelial cells nor to resist exposure to low pH in the presence of arginine (4).
In conclusion, our results show that the ADS enables S. suis to overcome oxygen and nutrient starvation and to tolerate acidic environments. Thus, the ADS might facilitate S. suis survival within the different niches of the host and thereby probably contributes to S. suis pathogenesis. Future investigations have to clarify the importance of the ADS for S. suis infections.
Acknowledgments
We gratefully acknowledge H. E. Smith (Division of Infectious Diseases and Food Chain Quality, Institute for Animal Science and Health, Lelystad, The Netherlands) for generously supplying the pICspc plasmid and S. suis strain 10. The sequence data were produced by the Streptococcus suis Sequencing Group at the Sanger Institute and can be obtained from ftp://ftp.sanger.ac.uk//pub/pathogens/ss/.
This study was supported by the Deutsche Forschungsgemeinschaft (DFG), Bonn, Germany (SFB 578 and GRK 745).
REFERENCES
- 1.Allgaier, A., R. Goethe, H. J. Wisselink, H. E. Smith, and P. Valentin-Weigand. 2001. Relatedness of Streptococcus suis isolates of various serotypes and clinical backgrounds as evaluated by macrorestriction analysis and expression of potential virulence traits. J. Clin. Microbiol. 39:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arends, J. P., and H. C. Zanen. 1988. Meningitis caused by Streptococcus suis in humans. Rev. Infect. Dis. 10:131-137. [DOI] [PubMed] [Google Scholar]
- 3.Barcelona-Andres, B., A. Marina, and V. Rubio. 2002. Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J. Bacteriol. 184:6289-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benga, L., R. Goethe, M. Rohde, and P. Valentin-Weigand. 2004. Non-encapsulated strains reveal novel insights in invasion and survival of Streptococcus suis in epithelial cells. Cell. Microbiol. 6:867-881. [DOI] [PubMed] [Google Scholar]
- 5.Burne, R. A., Z. T. Wen, Y. Y. Chen, and J. E. Penders. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 7.Casiano-Colon, A., and R. E. Marquis. 1988. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl. Environ. Microbiol. 54:1318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Champomier Verges, M. C., M. Zuniga, F. Morel-Deville, G. Perez-Martinez, M. Zagorec, and S. D. Ehrlich. 1999. Relationships between arginine degradation, pH and survival in Lactobacillus sakei. FEMS Microbiol. Lett. 180:297-304. [DOI] [PubMed] [Google Scholar]
- 9.Chanter, N., P. W. Jones, and T. J. Alexander. 1993. Meningitis in pigs caused by Streptococcus suis—a speculative review. Vet. Microbiol. 36:39-55. [DOI] [PubMed] [Google Scholar]
- 10.Charland, N., J. Harel, M. Kobisch, S. Lacasse, and M. Gottschalk. 1998. Streptococcus suis serotype 2 mutants deficient in capsular expression. Microbiology 144(Pt. 2):325-332. [DOI] [PubMed] [Google Scholar]
- 11.Chauvaux, S., I. T. Paulsen, and M. H. Saier, Jr. 1998. CcpB, a novel transcription factor implicated in catabolite repression in Bacillus subtilis. J. Bacteriol. 180:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 13.Clifton-Hadley, F. A., and T. J. Alexander. 1980. The carrier site and carrier rate of Streptococcus suis type II in pigs. Vet. Rec. 107:40-41. [DOI] [PubMed] [Google Scholar]
- 14.Cunin, R., N. Glansdorff, A. Pierard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Degnan, B. A., M. C. Fontaine, A. H. Doebereiner, J. J. Lee, P. Mastroeni, G. Dougan, J. A. Goodacre, and M. A. Kehoe. 2000. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 68:2441-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Degnan, B. A., J. M. Palmer, T. Robson, C. E. Jones, M. Fischer, M. Glanville, G. D. Mellor, A. G. Diamond, M. A. Kehoe, and J. A. Goodacre. 1998. Inhibition of human peripheral blood mononuclear cell proliferation by Streptococcus pyogenes cell extract is associated with arginine deiminase activity. Infect. Immun. 66:3050-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Greeff, A., H. Buys, R. Verhaar, J. Dijkstra, L. van Alphen, and H. E. Smith. 2002. Contribution of fibronectin-binding protein to pathogenesis of Streptococcus suis serotype 2. Infect. Immun. 70:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Hooghe, I., C. Vander Wauven, J. Michiels, C. Tricot, P. de Wilde, J. Vanderleyden, and V. Stalon. 1997. The arginine deiminase pathway in Rhizobium etli: DNA sequence analysis and functional study of the arcABC genes. J. Bacteriol. 179:7403-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong, Y., Y. Y. Chen, and R. A. Burne. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J. Bacteriol. 186:2511-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong, Y., Y. Y. Chen, J. A. Snyder, and R. A. Burne. 2002. Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl. Environ. Microbiol. 68:5549-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eiglmeier, K., N. Honore, S. Iuchi, E. C. Lin, and S. T. Cole. 1989. Molecular genetic analysis of FNR-dependent promoters. Mol. Microbiol. 3:869-878. [DOI] [PubMed] [Google Scholar]
- 22.Gamper, M., A. Zimmermann, and D. Haas. 1991. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J. Bacteriol. 173:4742-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gosalbes, M. J., V. Monedero, and G. Perez-Martinez. 1999. Elements involved in catabolite repression and substrate induction of the lactose operon in Lactobacillus casei. J. Bacteriol. 181:3928-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gostick, D. O., J. Green, A. S. Irvine, M. J. Gasson, and J. R. Guest. 1998. A novel regulatory switch mediated by the FNR-like protein of Lactobacillus casei. Microbiology 144:705-717. [DOI] [PubMed] [Google Scholar]
- 25.Gottschalk, M., and M. Segura. 2000. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet. Microbiol. 76:259-272. [DOI] [PubMed] [Google Scholar]
- 26.Gottschalk, M. G., S. Lacouture, and J. D. Dubreuil. 1995. Characterization of Streptococcus suis capsular type 2 haemolysin. Microbiology 141(Pt. 1):189-195. [DOI] [PubMed] [Google Scholar]
- 27.Griswold, A., Y. Y. Chen, J. A. Snyder, and R. A. Burne. 2004. Characterization of the arginine deiminase operon of Streptococcus rattus FA-1. Appl. Environ. Microbiol. 70:1321-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henkin, T. M. 1996. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol. Lett. 135:9-15. [DOI] [PubMed] [Google Scholar]
- 29.Hueck, C. J., and W. Hillen. 1995. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol. Microbiol. 15:395-401. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs, A. A., P. L. Loeffen, A. J. van den Berg, and P. K. Storm. 1994. Identification, purification, and characterization of a thiol-activated hemolysin (suilysin) of Streptococcus suis. Infect. Immun. 62:1742-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jourlin-Castelli, C., N. Mani, M. M. Nakano, and A. L. Sonenshein. 2000. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J. Mol. Biol. 295:865-878. [DOI] [PubMed] [Google Scholar]
- 32.Korner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27:559-592. [DOI] [PubMed] [Google Scholar]
- 33.Lu, C. D., H. Winteler, A. Abdelal, and D. Haas. 1999. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 181:2459-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maas, W. K. 1994. The arginine repressor of Escherichia coli. Microbiol. Rev. 58:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maghnouj, A., A. A. Abu-Bakr, S. Baumberg, V. Stalon, and C. Vander Wauven. 2000. Regulation of anaerobic arginine catabolism in Bacillus licheniformis by a protein of the Crp/Fnr family. FEMS Microbiol. Lett. 191:227-234. [DOI] [PubMed] [Google Scholar]
- 36.Maghnouj, A., T. F. Sousa Cabral, V. Stalon, and C. Vander Wauven. 1998. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor ArgR. J. Bacteriol. 180:6468-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marouni, M. J., E. Ziomek, and S. Sela. 2003. Influence of group A streptococcal acid glycoprotein on expression of major virulence factors and internalization by epithelial cells. Microb. Pathog. 35:63-72. [DOI] [PubMed] [Google Scholar]
- 38.Marquis, R. E., G. R. Bender, D. R. Murray, and A. Wong. 1987. Arginine deiminase system and bacterial adaptation to acid environments. Appl. Environ. Microbiol. 53:198-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsh, J. L., M. Erfle, and E. J. Wykes. 1984. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene 32:481-485. [DOI] [PubMed] [Google Scholar]
- 40.Martin-Verstraete, I., J. Stulke, A. Klier, and G. Rapoport. 1995. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J. Bacteriol. 177:6919-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okwumabua, O., J. S. Persaud, and P. G. Reddy. 2001. Cloning and characterization of the gene encoding the glutamate dehydrogenase of Streptococcus suis serotype 2. Clin. Diagn. Lab. Immunol. 8:251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palm, J. E., M. E. Weiland, W. J. Griffiths, I. Ljungstrom, and S. G. Svard. 2003. Identification of immunoreactive proteins during acute human giardiasis. J. Infect. Dis. 187:1849-1859. [DOI] [PubMed] [Google Scholar]
- 43.Ruepp, A., and J. Soppa. 1996. Fermentative arginine degradation in Halobacterium salinarium (formerly Halobacterium halobium): genes, gene products, and transcripts of the arcRACB gene cluster. J. Bacteriol. 178:4942-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Scott, C., J. R. Guest, and J. Green. 2000. Characterization of the Lactococcus lactis transcription factor FlpA and demonstration of an in vitro switch. Mol. Microbiol. 35:1383-1393. [DOI] [PubMed] [Google Scholar]
- 46.Smith, H. E., M. Damman, J. van der Velde, F. Wagenaar, H. J. Wisselink, N. Stockhofe-Zurwieden, and M. A. Smits. 1999. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 67:1750-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, H. E., H. J. Wisselink, U. Vecht, A. L. Gielkens, and M. A. Smits. 1995. High-efficiency transformation and gene inactivation in Streptococcus suis type 2. Microbiology 141(Pt. 1):181-188. [DOI] [PubMed] [Google Scholar]
- 48.Spiro, S. 1994. The FNR family of transcriptional regulators. Antonie Leeuwenhoek 66:23-36. [DOI] [PubMed] [Google Scholar]
- 49.Tikkanen, K., S. Haataja, and J. Finne. 1996. The galactosyl-(α1-4)-galactose-binding adhesin of Streptococcus suis: occurrence in strains of different hemagglutination activities and induction of opsonic antibodies. Infect. Immun. 64:3659-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Titgemeyer, F., and W. Hillen. 2002. Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek 82:59-71. [PubMed] [Google Scholar]
- 51.Tobisch, S., J. Stulke, and M. Hecker. 1999. Regulation of the lic operon of Bacillus subtilis and characterization of potential phosphorylation sites of the LicR regulator protein by site-directed mutagenesis. J. Bacteriol. 181:4995-5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vecht, U., H. J. Wisselink, M. L. Jellema, and H. E. Smith. 1991. Identification of two proteins associated with virulence of Streptococcus suis type 2. Infect. Immun. 59:3156-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winterhoff, N., R. Goethe, P. Gruening, M. Rohde, H. Kalisz, H. E. Smith, and P. Valentin-Weigand. 2002. Identification and characterization of two temperature-induced surface-associated proteins of Streptococcus suis with high homologies to members of the arginine deiminase system of Streptococcus pyogenes. J. Bacteriol. 184:6768-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshida, J., S. Takamura, and M. Nishio. 1998. Characterization of a streptococcal antitumor glycoprotein (SAGP). Life Sci. 62:1043-1053. [DOI] [PubMed] [Google Scholar]
- 55.Zuniga, M., M. Champomier-Verges, M. Zagorec, and G. Perez-Martinez. 1998. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J. Bacteriol. 180:4154-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuniga, M., G. Perez, and F. Gonzalez-Candelas. 2002. Evolution of arginine deiminase (ADI) pathway genes. Mol. Phylogenet. Evol. 25:429-444. [DOI] [PubMed] [Google Scholar]
- 57.Zuniga, M., M. del Carmen Miralles, and G. Perez-Martinez. 2002. The product of arcR, the sixth gene of the arc operon of Lactobacillus sakei, is essential for expression of the arginine deiminase pathway. Appl. Environ. Microbiol. 68:6051-6058. [DOI] [PMC free article] [PubMed] [Google Scholar]