Abstract
Recently, we identified a pyruvate fermentation pathway in Pseudomonas aeruginosa sustaining anaerobic survival in the absence of alternative anaerobic respiratory and fermentative energy generation systems (M. Eschbach, K. Schreiber, K. Trunk, J. Buer, D. Jahn, and M. Schobert, J. Bacteriol. 186:4596-4604, 2004). Anaerobic long-term survival of P. aeruginosa might be essential for survival in deeper layers of a biofilm and the persistent infection of anaerobic mucus plaques in the cystic fibrosis lung. Proteome analysis of P. aeruginosa cells during a 7-day period of pyruvate fermentation revealed the induced synthesis of three enzymes involved in arginine fermentation, ArcA, ArcB, and ArcC, and the outer membrane protein OprL. Moreover, formation of two proteins of unknown function, PA3309 and PA4352, increased by factors of 72- and 22-fold, respectively. Both belong to the group of universal stress proteins (Usp). Long-term survival of a PA3309 knockout mutant by pyruvate fermentation was found drastically reduced. The oxygen-sensing regulator Anr controls expression of the PPA3309-lacZ reporter gene fusion after a shift to anaerobic conditions and further pyruvate fermentation. PA3309 expression was also found induced during the anaerobic and aerobic stationary phases. This aerobic stationary-phase induction is independent of the regulatory proteins Anr, RpoS, RelA, GacA, RhlR, and LasR, indicating a currently unknown mechanism of stationary-phase-dependent gene activation. PA3309 promoter activity was detected in the deeper layers of a P. aeruginosa biofilm using a PPA3309-gfp (green fluorescent protein gene) fusion and confocal laser-scanning microscopy. This is the first description of an Anr-dependent, anaerobically induced, and functional Usp-like protein in bacteria.
Pseudomonas aeruginosa is a highly adaptable bacterium that colonizes various environmental niches. It is also a leading opportunistic pathogen in human infections. P. aeruginosa is the dominant pathogen causing chronic respiratory infections of cystic fibrosis (CF) patients. This results in progressive lung damage and is the major cause of morbidity and mortality in CF patients (43). Recent data indicate that anaerobic conditions play an important role during persistent infection of the CF lung. P. aeruginosa forms biofilm-like microcolonies in the CF lung mucus embedded in an anaerobic environment (28, 47). Under these conditions, nitrate serves as an alternative electron acceptor sustaining growth under anaerobic conditions. Moreover, nitrate seems to favor the formation of more robust anaerobic biofilms (20, 50). Biofilm formation protects P. aeruginosa in hostile environments like the CF lung from the immune response of the host and leads to high antibiotic tolerance of the cells (7, 25, 41). A recent publication shows that oxygen-restricted cells in deeper layers of a colony biofilm are highly resistant to antibiotics due to stationary-phase conditions caused by oxygen depletion (4).
P. aeruginosa has a limited potential to survive in an anaerobic environment. Growth is supported by denitrification with nitrate or nitrite (5, 8, 51). Moreover, arginine sustains moderate anaerobic growth via fermentation (44). Recently, we have shown that pyruvate—one of the most abundant metabolites in all cells—allows anaerobic long-term survival of P. aeruginosa (13). However, in contrast to denitrification and arginine fermentation, pyruvate fermentation does not sustain anaerobic growth. We identified three enzymes essential for pyruvate fermentation: phosphotransacetylase (Pta), acetate kinase (AckA), and a lactate dehydrogenase (LdhA). The operon encoding Pta and AckA is induced in response to oxygen limitation in dependence of the anaerobic regulatory protein Anr (13). The ackA-pta locus in Escherichia coli is induced in response to starvation during aerobic conditions (31). Pyruvate fermentation in P. aeruginosa might also play an important role as a general endogenous survival metabolism in response to energy starvation.
Here, we report the investigation of the physiological basis of pyruvate fermentation in P. aeruginosa starting with a proteome approach via two-dimensional (2D) gel electrophoresis. We identified two anaerobically induced Usp-type stress proteins (PA3309 and PA4352) and investigated the role of PA3309 during pyruvate fermentation and biofilm growth in more detail. In contrast to E. coli or Usp-type proteins investigated in Mycobacteria (33), our data indicate the presence of Usp-type proteins in P. aeruginosa, which are produced in response to oxygen limitation under the control of the oxygen-sensing regulator Anr. We found evidence for induction of the PA3309 gene in the stationary phase by an Anr-independent unknown regulator.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. For standard molecular biology protocols, E. coli and P. aeruginosa strains were grown in LB medium as described before (13). Antibiotics were used at the following concentrations for P. aeruginosa: carbenicillin at 500 μg/ml, gentamicin at 200 μg/ml, and tetracycline at 100 μg/ml.
TABLE 1.
Strains of P. aeruginosa and E. coli and plasmids used in this study
| Bacterial strain or plasmid | Genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Wild type | 11 |
| PAO6261 | PAO1 Δanr | 49 |
| PAO-MW20 | PAO1 rpoS::aacC1 Gmr | 46 |
| PAO6251 | PAO1 ΔarcDABC | 17 |
| PAO6281 | PAO1 gacA::Ω-Sp/Sm | 36 |
| KS06 | PAO1 attB::(PPA3309-lacZ) | This study |
| KS08 | PAO6261 attB::(PPA3309-lacZ) | This study |
| KS10 | PAO-MW20 attB::(PPA3309-lacZ) | This study |
| KS11 | PAO1 attB::(mini-CTX-lacZ) | This study |
| KS15 | PAO1 attB::(PPA3309-gfp) | This study |
| KS16 | PAO1 ΔPA3309::aacC1 Gmr | This study |
| KS17 | PAO1 ΔPA3309 | This study |
| KS27 | PAO1 attB::(mini-CTX2) | This study |
| KS28 | KS17 attB::(mini-CTX2) | This study |
| KS29 | KS17 attB::(pKS13) | This study |
| KS31 | PAO1 rhlR attB::(PPA3309-lacZ) | This study |
| KS32 | PAO1 lasR attB::(PPA3309-lacZ) | This study |
| KS33 | PAO6281 attB::(PPA3309-lacZ) | This study |
| KS35 | PAO1 ΔrelA | This study |
| KS37 | PAO1 ΔrelA attB::(PPA3309-lacZ) | This study |
| NB015 | PAO1 ΔPA4352 | This study |
| E. coli | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara leu) 7697 galU galK λ−rpsL nupG | Gibco BRL (Invitrogen) |
| S17 λpir | pro thi hsdR+ Tpr Smr; chromosome::RP4-2 Tc::Mu-Km::Tn7/λpir | 9 |
| SM10 | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu (Kmr) | 9 |
| Plasmids | ||
| pEX18Ap | AproriT+sacB+; gene replacement vector with MCS from pUC18 | 23 |
| Mini-CTX-lacZ | Tcr; promoterless lacZ gene | 2 |
| Mini-CTX2 | Tcr; integration vector | 24 |
| pSB219.9A | Gmr; lasR::Gm suicide vector | 1 |
| pSB224.10A | Tcr; rhlR::Tc suicide vector | 1 |
| pPS858 | Apr Gmr; source of gentamicin cassette | 23 |
| pFLP2 | Apr; source of FLP recombinase | 23 |
| pMH305 | Apr Gmr; source of gfp cassette | Morten Hentzer |
| pKS08 | Apr Gmr; pMH305 with 526 bp of PA3309 promoter between EcoRI and BamHI | This study |
| pKS09 | Tcr; mini-CTX2 with NotI fragment liberated from pKS08 containing PA3309 promoter fused to gfp | This study |
| pKS10 | Apr Gmr; pEX18Ap with 723-bp promoter of PA3309, Gmr-gfp fragment from pPS858 and 625 bp downstream of coding region of PA3309 between EcoRI and SalI | This study |
| pKS13 | Tcr; mini-CTX2 with 1,200-bp PCR fragment covering entire PA3309 gene and 455 bp of putative promoter region and 289 bp of downstream region between EcoRI and HindIII | This study |
| pKS15 | Tcr; mini-CTX-lacZ containing 492-bp fragment of putative promoter region of PA3309 gene between EcoRI and BamHI | This study |
| pKS18 | Apr Gmr; pEX18Ap with 796-bp promoter of relA (PA0934), Gmr-gfp fragment from pPS858, and 824 bp of the 3′ coding region of relA between EcoRI and HindIII | This study |
| pNB007 | Apr Gmr; pEX18Ap with 596-bp promoter PA4352, Gmr-gfp fragment from pPS858, and 617 bp downstream of coding region of PA4352 between SacI and HindIII | This study |
Pyruvate fermentation and anaerobic energy starvation experiments.
Pyruvate fermentation experiments with P. aeruginosa were performed with a slightly modified protocol. Instead of OS medium, we used a potassium phosphate (100 mM)-buffered LB medium at a pH value of 7.4. P. aeruginosa was grown aerobically up to an optical density at 578 nm (OD578) of 0.3. Cells were shifted to anaerobic conditions by transferring the culture to rubber-stoppered bottles and immediate addition of 40 mM pyruvate. Incubation was carried out at 37°C without shaking. Viable cell counts were determined as described before (13). Formation of fermentation products and consumption of pyruvate were analyzed using high-performance liquid chromatography analysis as outlined previously (13). Control experiments revealed no consumption of pyruvate during aerobic growth in LB medium up to an OD578 of 0.3.
For anaerobic long-term energy starvation experiments, 40 ml LB medium was inoculated with 104 P. aeruginosa cells/ml and incubated under anaerobic conditions for 20 days. Cell numbers increased up to 4 × 108 cells/ml within 24 h. After 24 h, cells faced severe energy starvation, resulting in a dramatic reduction in the number of cells/ml.
Construction and testing of the promoter-lacZ reporter gene fusions.
Chromosomal promoter-lacZ reporter gene fusions were constructed using the mini-CTX-lacZ vector. A 492-bp PCR product, covering the region from 450 bp upstream and 26 bp downstream of the translational start of the PA3309 gene, was generated using primer Pa-3309-for (5′-CGGAATTCGCCATGGACGAGGAACTG-3′) and Pa-3309-rev (5′-CGGGATCCTCCACGGCTACCAGAATG-3′). Pa-3309-for contained an EcoRI restriction site (underlined) at the 5′ end, and Pa-3309-rev contained a restriction site for BamHI (underlined) also at its 5′ end. The EcoRI- and BamHI-digested PCR product was cloned into the EcoRI and BamHI sites of mini-CTX-lacZ to generate pKS15. Transfer of pKS15 in P. aeruginosa was carried out by a diparental mating using E. coli S17 λ-pir as donor. The CTX integrase of pKS15 promoted integration of the vector into the attB site of the P. aeruginosa genome. The vector was transferred into PAO1, and the anr, rpoS, rhlR, lasR, gacA, and relA mutant strains to generate the P. aeruginosa strains KS06, KS08, KS10, KS31, KS32, KS33, and KS37, respectively (see also Table 1). In these mutant strains, parts of the mini-CTX-lacZ vector containing the tetracycline resistance cassette were deleted using a flippase (FLP) recombinase encoded on the pFLP2 plasmid. Reporter gene fusion assays were performed as outlined before in detail (13, 37). The obtained activities were given in Miller units (29).
Construction of a PPA3309-gfp fusion.
We used a transcriptional PPA3309-gfp (green fluorescent protein [GFP] gene) reporter gene fusion to monitor the promoter activity of PA3309 in biofilms. A 526-bp fragment of the PA3309 promoter region without the translational start codon was amplified by PCR with primers oKS16 (5′-GGAATTCGCCATGGACGAGGAACTG-3′), containing an EcoRI restriction site at the 5′ end, and oKS17 (5′-CGGGATCCAAGGTGTCCCTCCAGAGTG-3′), with a BamHI site. The PCR product was cloned into pMH305 upstream of gfp to generate pKS08. The PPA3309-gfp reporter gene fusion was liberated via NotI digestion and ligated into mini-CTX2 to generate pKS09. This vector was transferred and integrated into the P. aeruginosa genome using the procedure described above. The resulting strain was named KS15.
Construction of P. aeruginosa ΔPA3309, ΔPA4352, ΔrelA, rhlR:Tcr, and lasR::Gmrmutants.
Unmarked gene deletion mutants were obtained using the well-established strategies based on sacB counter selection and FLP recombinase excision (23). First a suicide vector, pKS10, was constructed to replace the PA3309 gene with a gentamicin cassette. After transfer of this vector into P. aeruginosa, a double-crossover mutant was obtained by sacB-based counterselection. The resulting mutant, KS16, was verified by Southern blot analysis. Finally, FLP recombinase encoded on the pFLP2 plasmid removed the FRT-flanked gentamicin cassette to generate KS17. To construct the suicide vector pKS10, the BamHI-digested gentamicin resistance cassette of pPS858 was cloned between two PCR fragments of the PA3309 gene in the multiple cloning site of pEX18Ap. The two PCR fragments contained DNA homologous to the upstream and downstream areas of the PA3309 gene. A 723-bp fragment containing the upstream promoter region of the PA3309 gene was amplified using primers oKS10 (5′-CGGAATTCGAACAAGGCGCTGAAG-3′), with an EcoRI restriction site at the 5′ end, and oKS11 (5′-CGCGGATCCAACTTCAAGGACACTGTA-3′), with a BamHIsite. The primers oKS12 (5′-CGCGGATCCTCCGGTCCTGCTGGT-3′), with a BamHI restriction site, and oKS13 (5′-ACGCGTCGACACGCCATCATCGTCCT-3′), with a SalI restriction site, were used for the amplification of 625 bp of the PA3309 downstream region.
The unmarked ΔPA4352 mutant (NB015) was generated by the same strategy described above using the suicide vector pNB007. In this suicide vector, the following PCR fragments flanked the gentamicin resistance cassette: The 596-bp fragment of the upstream region of the PA4352 gene was amplified using primers oNB01 (5′-CGAGCTCTACGGCGACTTCGTCAAGG-3′), with a SacI restriction site, and oNB02 (5′-CGGGATCCAAGCGGATGCTTCGGACT-3′), with a BamHI site. The primers oNB03 (5′-CGCGGATCCCTTCCGCCGCGCGCTGA-3′), with a BamHI site, and oNB04 (5′-CCCAAGCTTCCCTGGCGCCGCTGACC-3′), with a HindIII site, amplified 617 bp of the corresponding downstream region of PA4352.
The ΔrelA mutant KS35 was constructed by the same strategy described above. For construction of the suicide vector pKS18, the primers oKS40 (5′-GGAATTCGGCCAGTGCATTGCTGTTG-3′), with an EcoRI restriction site at the 5′ end, and oKS41 (5′-CGGGATCCTTACCACGGTGCGCGTAG-3′), with a BamHI site, amplified 796 bp of the putative promoter region of the relA gene. The primers oKS42 (5′-CGCGGATCCCGAGCAGGTCGAGATCA-3′), with a BamHI site, and oKS43 (5′-CCCAAGCTTTGGGCAGTTGCGAGACG-3′), with a HindIII site, were used to amplify 824 bp of the 3′ end of relA.
The two quorum-sensing mutants KS31 (rhlR::Tcr) and KS32 (lasR::Gmr) were constructed as described previously (1) using KS06 as the parent strain.
Plasmid construction for complementation of the PA3309 knockout mutant.
For construction of the complementation plasmid for the chromosomal PA3309 knockout mutant, a 1,200-bp PCR product covering 455 bp of the PA3309 promoter region, the PA3309 gene, and 289 bp downstream of PA3309 was amplified using primers oKS14 (5′-CGGAATTCGCCATGGACGAGGAACTG-3′), with an EcoRI restriction site at the 5′ end, and oKS15 (5′-CCAAGCTTATCCACGTGCCGATGGTC-3′), with a HindIII site. The product was digested with EcoRI and HindIII and ligated into mini-CTX2 to generate pKS13. The vector was transferred into P. aeruginosa KS17 to generate KS29 by a diparental mating with E. coli S17 λpir and integrated at the attB site in the genome as described above. As a control, the empty mini-CTX2 vector was integrated into the genome of PAO1 and KS17 to generate KS27 and KS28, respectively.
Proteomic analysis.
To prevent changes of the cellular protein pattern during cell harvesting and cell extract preparation, the culture was mixed with a double volume of ice-cold potassium phosphate-buffer (0.1 M; pH 7.4) and allowed to cool for 20 min. Cells were centrifuged at 8,000 × g for 30 min at 4°C and washed twice with potassium phosphate buffer. Cells were resuspended in a small volume of potassium phosphate buffer. The protein concentration in whole-cell suspensions was determined using the bicinchoninic acid protein assay (Sigma, Taufkirchen, Germany). Cells were disrupted by incubation of a 360-μl culture aliquot with 150 μl NaOH for 1 h at 70°C. For protein isolation, we modified a protocol described previously (19) and extracted proteins directly from whole cells with phenol and a subsequent acetone precipitation. The precipitated proteins were solubilized in sample buffer consisting of 7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 50 mMdithiothreitol, and 2% ampholytes (Bio-Lyte; Bio-Rad, Munich, Germany). The protein concentration was determined in the sample buffer using the PlusOne 2D Quant kit (Amersham Biosciences, Freiburg, Germany). 2D gel electrophoresis was performed using immobilized pH gradient (IPG) strips 11 or 17 cm in length covering two different pH ranges (pH 4.7 to 5.9 or 5 to 8) (IPG Ready Strips; Bio-Rad, Munich, Germany). For the narrow pH range, the IPG strips (11 cm) were rehydrated overnight in rehydration buffer containing 50 μg of protein. Isoelectric focusing (IEF) was carried out at 20°C under mineral oil in the PROTEAN IEF cell (Bio-Rad, Munich, Germany) for a total of 35,000 V · h. The focused IPG strips were reduced for 15 min in a sodium dodecyl sulfate (SDS) equilibration solution containing 15 mM dithiothreitol and afterwards alkylated twice for 15 min in the same buffer containing 150 mM iodacetamide prior to SDS-polyacrylamide gel electrophoresis (SDS-PAGE). The IPG strips were transferred to 10.5 to 14% SDS-PAGE (Criterion Tris-HCl gel; Bio-Rad, Munich, Germany) gels, and electrophoresis was performed at a constant 200 V for 55 min. Large IPG strips (17 cm, pH 5 to 8) were loaded with 700μg of protein, and IEF was conducted for a total of 110,000 V · h. SDS-PAGE was performed at a constant temperature of 20°C with 1 W per gel for approximately 20 h using 10% polyacrylamide gels (25.5 by 20.5 cm). All gels were stained with ruthenium(II)-tris-(bathophenanthroline disulfonate) (RuBPS) as described before (35). Gels were documented with an FX-Scanner (Bio-Rad, Munich, Germany). Analysis and quantification of differential protein spot patterns were performed by using the software Z3 (Compugen, Tel Aviv, Israel). Gel spots were excised and treated using a method described before introducing minor modifications (40). Briefly, the gel pieces were washed with water, dehydrated with acetonitrile (ACN), and digested with trypsin (sequencing grade; Promega). Peptides were extracted and collected in four elution steps (each 15 min, 37°C) using 25 mM NH4HCO3, ACN, 5% formic acid, and again ACN. Extracted peptides were purified using ZipTip C18 microcolumns (Millipore), following the manufacturer's instructions. Proteins were identified by peptide-mass fingerprint (PMF) as well as post-source decay fragmentation data recorded on a Bruker Ultraflex matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer. PMF data were analyzed using an internal MASCOT server at the GBF (version 1.9; Matrix Science) (34) and the NCBI database (restricted to the taxon Pseudomonas aeruginosa). Only peptides with a MASCOT rank of 1 were considered significant and used for the combined peptide score. The criteria used to accept protein identifications based on PMF data included the extent of sequence coverage (minimum of 30%), the number of peptides matched (minimum of 5), and the score of probability (minimum of 70 for the Mowse score). Lower-scoring proteins were either verified manually or rejected.
Biofilm experiments.
Biofilm studies were performed in three-channel flow cells with individual channel dimensions of 1 by 4 by 40 mm supplied with a flow of 3 ml h−1 of AB medium (22) supplemented with 300 μM glucose and where indicated with 50 mM KNO3 (6). The system was assembled and cultures for inoculation were prepared as described before (6, 21). Biofilms were stained with Syto62 (Molecular Probes) to visualize the biofilm matrix. Dead cells in the biofilm were visualized with propidium iodide (Sigma). Microscopy and image acquisition were done as outlined in reference 22 with a Zeiss LSM 510 confocal laser-scanning microscope (CLSM) (Carl Zeiss, Jena, Germany). Quantitative image analysis was done using the COMSTAT software (22). Images were processed using the Imaris software (Bitplane AG, Zurich, Switzerland). For protein analysis, biofilms were grown in silicone tubes (30 cm; inner square, 0.5 cm), in AB medium as described above and supplied with a flow rate of 20 ml h−1.
RESULTS
Proteome analysis of P. aeruginosa under pyruvate fermentation conditions.
Recently, we showed that P. aeruginosa can survive anaerobically for several weeks by pyruvate fermentation in the absence of nitrate, nitrite, or arginine (13). Pyruvate fermentation allows survival but not growth of P. aeruginosa cells during severe anaerobic energy starvation conditions. We used a proteomics approach to identify proteins which are preferentially synthesized during pyruvate fermentation. Long-term survival pyruvate fermentation experiments using cultures grown in LB medium were set up as described previously (13). For this purpose, cells were grown aerobically to an OD578 of 0.3, immediately shifted to anaerobic flasks containing pyruvate at a final concentration of 40 mM, and further incubated for 7 days at 37°C. Viable cell counts of three independent cultures were determined and remained almost constant during the whole experiment (2.8× 108 ± 1.3 × 108 cells per ml of medium at day 0 and 1.6× 108 ± 1.0 × 107 at day 7).
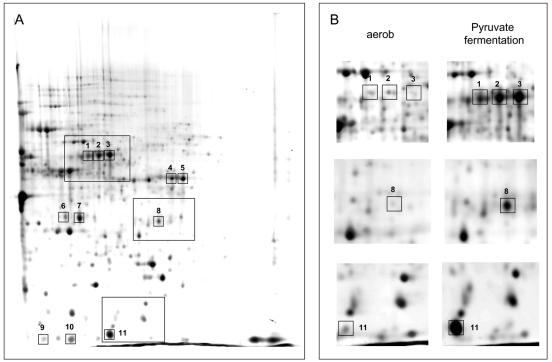
We compared the protein pattern of the aerobically grown culture in phosphate buffered LB medium (OD578 = 0.3) with that of the culture incubated for 7 days under pyruvate fermentation conditions (phosphate-buffered LB medium plus 40 mM pyruvate, Fig. 1A and B). The concentration of 11 proteins was found highly increased at least ninefold under anaerobic pyruvate fermentation conditions (Table 2). MALDI-TOFanalysis identified the 11 separated proteins as representing 6 different proteins. Three proteins involved in arginine fermentation (ArcA, ArcB, and ArcC) were found highly upregulated as well as the outer membrane protein OprL and two proteins, PA3309 and PA4352, which are annotated as conserved hypothetical proteins but which each have universal stress protein motifs (Table 2 and Fig. 1B). None of these proteins was expected to be involved in or contribute to pyruvate fermentation. The results of the proteome analysis raised the question of whether proteins involved in arginine fermentation or the two hypothetical proteins contribute to survival during pyruvate fermentation.
FIG. 1.
(A) Two-dimensional image of crude extracts of P. aeruginosa PAO1 incubated under pyruvate fermentation conditions for 7 days. Outlined areas in the gel indicate zones of the 2D gel that are represented in Fig. 1B. (B) Enlarged 2D gel images showing protein extracts of the P. aeruginosa PAO1 aerobic culture immediately before the shift to anaerobic pyruvate fermentation conditions and the pyruvate fermentation culture shown in Fig. 1A. Numbers of the boxed spots indicate identified proteins that are synthesized at higher levels during pyruvate fermentation; the numbers correlate with the numbers given in Table 2.
TABLE 2.
Identified proteins under pyruvate fermentation in LB mediuma
| Spot no. | PA no. | Protein | Description | Regulation (fold)b |
|---|---|---|---|---|
| 1 | PA5171 | ArcA | Arginine deiminase | 132 |
| 2 | PA5171 | ArcA | Arginine deiminase | 9 |
| 3 | PA5171 | ArcA | Arginine deiminase | Unmatched |
| 4 | PA5172 | ArcB | Catabolic ornithine carbamoyltransferase | 29 |
| 5 | PA5172 | ArcB | Catabolic ornithine carbamoyltransferase | 14 |
| 6 | PA5173 | ArcC | Carbamate kinase | Unmatched |
| 7 | PA5173 | ArcC | Carbamate kinase | 60 |
| 8 | PA4352 | Conserved hypothetical protein | 22 | |
| 9 | PA0973 | OprL | Outer membrane protein | 99 |
| 10 | PA0973 | OprL | Outer membrane protein | Unmatched |
| 11 | PA3309 | Conserved hypothetical protein | 72 |
PA number, protein short name, and description are shown according to the Pseudomonas database (www.pseudomonas.com).
Shown are the ratios of the amount of proteins from a P. aeruginosa PAO1 culture incubated for 7 days under pyruvate fermentation conditions at 37°C in phosphate-buffered LB medium to the amount of protein in a culture grown under aerobic conditions (see Materials and Methods). Proteins were separated by 2D gel electrophoresis and stained with RuBPS (see Materials and Methods for details). Protein ratios of 2D gel images were determined by the Z3 software (Compugen, Tel Aviv, Israel). “Unmatched” describes a protein spot which is detected only in the protein pattern of the pyruvate fermentation culture.
Contribution of arcDABC and PA3309 to anaerobic long-term survival of P. aeruginosa.
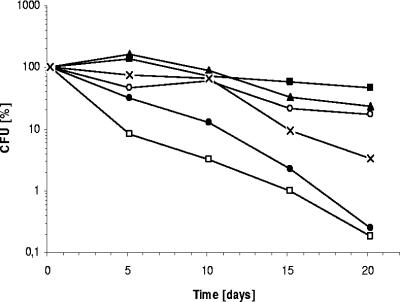
Since we used a complex medium containing arginine, we investigated if arginine fermentation contributes to survival under anaerobic pyruvate fermentation conditions. The ΔarcDABC mutant PAO6251 cannot grow anaerobically by arginine fermentation. Viable cell counts of the ΔarcDABC mutant decreased by a factor of 30 compared to the wild type during 20 days of pyruvate fermentation. In contrast, the viable cell counts of the wild-type control, which was incubated under anaerobic conditions without pyruvate, decreased by a factor of 500 (Fig. 2). These results demonstrated that arginine fermentation contributes to anaerobic long-term survival: however, to a lesser extent than the PA3309 protein.
FIG. 2.
Anaerobic survival of P. aeruginosa wild type (▪), the PAO6251 (ΔarcDABC) mutant (×), the ΔPA4352 mutant (▴), the ΔPA3309 mutant (•), and the complemented ΔPA3309 mutant (○) in the presence of 40 mM pyruvate. CFU of the P. aeruginosa wild type without pyruvate served as a control (□). The P. aeruginosa wild type and mutants were grown aerobically in phosphate-buffered LB medium. At an OD578 of 0.3, cultures were transferred to rubber-stoppered bottles and 40 mM pyruvate was added. Survival under anaerobic conditions without alternative electron acceptors was determined with viable cell counts on agar plates. Graphs represent the results of at least three independent experiments. Standard deviations were 44% until day 5 and below 5% between days 10 and 20 and are omitted for the sake of clarity.
Next, we investigated if the hypothetical protein PA3309 promotes survival during pyruvate fermentation. First, a PA3309 chromosomal knockout mutant strain was constructed as described in Materials and Methods. The PA3309 mutant strain exhibited a clear 400-fold reduction of anaerobic long-term survival over a 20-day period (Fig. 2). We were able to complement the PA3309 mutant by chromosomal integration of pKS13, which contains the cloned PA3309 gene and the putative promoter region (Table 1). Survival rates of the complemented mutant strain (KS29) do not completely reach the wild-type level, but were found significantly increased (100-fold) compared to those of the PA3309 mutant (KS17) and the PA3309 mutant containing the empty mini-CTX2 vector integrated in the attB locus (KS28; data for this mutant not shown). A PA4352 knockout mutant (NB015) without the second Usp-type stress protein showed no reduced survival during pyruvate fermentation (Fig. 2).
Our studies revealed an essential role of PA3309 for anaerobic long-term survival during pyruvate fermentation.
The conserved hypothetical protein PA3309 belongs to the group of anaerobically induced universal stress proteins.
The conserved hypothetical protein PA3309 consists of 151 amino acids with a calculated mass of 16,496 Da and a theoretical pI of 5.31. The Pfam database indicates the presence of a single Usp domain (Pfam accession number PF00582) which originates from the universal stress protein A (UspA) of E. coli. PA3309 shares the single conserved Usp domain, a similar molecular mass, and a moderate amino acid sequence identity of 37% with E. coli UspA. Six Usp-type stress protein paralogues have been identified in E. coli which are produced in response to a variety of different stress conditions, most of them leading to growth arrest (27). We tested if the PA3309 knockout mutant showed similar phenotypes to the E. coli uspA mutant in response to UV stress and stationary-phase survival. However, no similar behavior was observed (data not shown). Despite the observed phenotype of the PA3309 knockout mutant during pyruvate fermentation, we did not observe a defect in anaerobic denitrifying growth using different media supplemented with 50 mM nitrate. However, PA3309 contributes to survival of severe anaerobic energy stress conditions. We incubated wild-type P. aeruginosa and the PA3309 mutant anaerobically in LB medium without nitrate, nitrite, or pyruvate. Within 20 days, cell numbers of the wild-type strain decreased by a factor of 9.0 × 102, while cell numbers of the mutant strain decreased by a factor of 9.7 × 104.
PA3309 is induced upon a shift to anaerobic conditions by the oxygen regulator Anr.
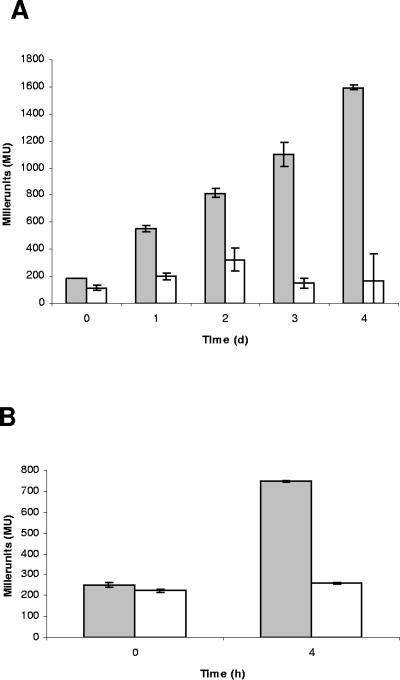
We studied the regulatory behavior of the PA3309 promoter towards various environmental stimuli and corresponding metabolic conditions with a chromosomal transcriptional promoter-lacZ reporter gene fusion. We monitored β-galactosidase activities of the PPA3309-lacZ reporter gene fusion (KS06) during pyruvate fermentation for the first 4 days. The β-galactosidase activities increased linearly up to 1,594 ± 20 Miller units during the first 4 days (Fig. 3A), indicating strong anaerobic induction. Anr is a global transcriptional regulatory protein of the Crp-Fnr family which activates gene expression in P. aeruginosa in response to oxygen limitation (16, 39). We detected a putative Anr box 86 bp upstream of the translational start codon in the presumed promoter region of PA3309 using tools of the PRODORIC database (30). We also checked dependence of the PA3309 promoter on Anr. No significant increase was detected when the PPA3309-lacZ reporter gene fusion was monitored in the anr mutant strain PAO6261 (Fig. 3A). We also measured a strong increase in β-galactosidase activity of the PPA3309-lacZ reporter gene fusion in wild-type P. aeruginosa upon a shift to anaerobic conditions in the absence of pyruvate. Again, under these conditions the anr mutant failed to induce the PA3309 promoter activity (Fig. 3B). Clearly, Anr induces anaerobic expression of PA3309 independent of the presence of pyruvate.
FIG. 3.
(A) Diagram showing the expression pattern of the P. aeruginosa wild type containing the PPA3309-lacZ reporter gene fusion (strain KS06; gray bars) and the anr mutant (PAO6261) containing the PPA3309-lacZ reporter gene fusion (KS08, white bars) during the first 4 days of a pyruvate fermentation experiment. Cells were grown in phosphate buffered LB medium at 37°C (for details see Materials and Methods). Time point 0 is the aerobic culture shortly before the transfer to an anaerobic flask. (B) Diagram showing the expression pattern of the P. aeruginosa wild type containing the PPA3309-lacZ reporter gene fusion (strain KS06; gray bars) and the anr mutant (PAO6261) containing the PPA3309-lacZ reporter gene fusion (KS08; white bars) in a shift experiment. Cells were grown at 37°C in LB medium plus 50 mM KNO3 up to an OD578 of 0.4 (time point 0) and transferred to a rubber-stoppered flask for a subsequent 4-h incubation (time point 4). β-Galactosidase activities were determined at the indicated time points. Experiments were repeated three times.
PA3309 is induced in stationary phase.
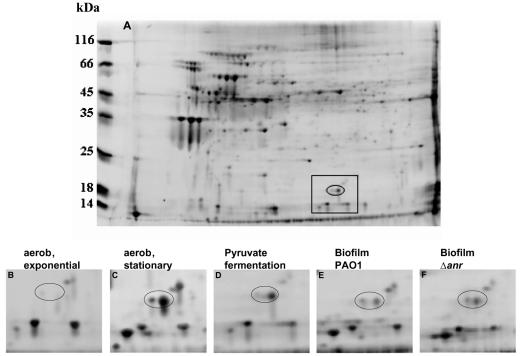
Further examination of the PPA3309-lacZ behavior during various anaerobic growth phases revealed a 5.3-fold induction during anaerobic exponential growth compared to aerobic exponential growth. Interestingly, the PA3309 promoter was further induced 1.8-fold in the anaerobic stationary phase (Table 3). Next, we investigated the behavior of the PA3309 promoter during the aerobic stationary phase. To our surprise, a 5.4-fold induction was observed. Gene expression of PA3309 in the aerobic stationary phase was further confirmed by 2D gel electrophoresis (Fig. 4B and C). In contrast to pyruvate fermentation, where we identified only one protein spot representing PA3309 (Fig. 4D), two spots representing PA3309 were identified in the protein pattern of aerobic stationary-phase cells (Fig. 4C). This might indicate a phosphorylation of PA3309 as reported for E. coli UspA (15). The Anr regulator was the candidate responsible for the aerobic induction of the PPA3309-lacZ reporter gene fusion upon entry into the stationary phase, since the strong respiration of a high-cell-density culture leads to oxygen limitation. However, expression of the PPA3309-lacZ reporter gene fusion in the anr mutant still increased 4.2-fold upon entry in the aerobic stationary phase (Table 3). Therefore, promoter activity in the aerobic stationary phase is independent of Anr. Expression of the uspA, uspC, uspD, and uspE genes in E. coli upon entry in the stationary phase requires stringent control via the nucleotide guanosine 3′, 5′-bisdiphosphate (ppGpp) and the RelA protein (14, 18, 26). In P. aeruginosa, the RelA protein (PA0934) synthesizes ppGpp in response to amino acid starvation conditions or in the stationary phase (12, 42). Deletion of the P. aeruginosa relA gene abolishes the production of ppGpp and also decreases production of RpoS (12, 42). Expression of the P. aeruginosa PA3309 promoter in a P. aeruginosa relA mutant remained unchanged compared to that under wild-type conditions (Table 3). Furthermore, we did not detect increased PA3309 promoter activity when wild-type cells faced carbon starvation during growth in 1/20 LB medium (data not shown). Therefore, the stringent response system does not contribute to PA3309 regulation under the tested conditions.
TABLE 3.
Expression of the PPA3309-lacZ reporter gene fusion in P. aeruginosa wild type and mutants
| P. aeruginosa strain | β-Galactosidase activity (Miller units) or fold inductiona
|
|||||
|---|---|---|---|---|---|---|
| Aerobic
|
Fold inductiond | Anaerobic
|
Fold inductione | |||
| Exponential phaseb | Stationary phasec | Exponential phaseb | Stationary phasec | |||
| Wild type | 214.4 ± 9.6 | 1,149.2 ± 70.4 | 5.4 | 1,339.1 ± 171.7 | 2,377.5 ± 270.8 | 1.8 |
| anr mutant | 218.1 ± 8.0 | 914.1 ± 23.5 | 4.2 | NDf | ND | ND |
| rpoS mutant | 284.3 ± 164.1 | 1,160.5 ± 500.1 | 4.1 | 956.2 ± 117.6 | 2,262.4 ± 0 | 2.4 |
| gacA mutant | 229.7 ± 11.3 | 985.4 ± 47.0 | 4.3 | 1,161.2 ± 118.8 | 2,524.9 ± 154.4 | 2.2 |
| rhlR mutant | 248.5 ± 4.0 | 1,342.2 ± 87.5 | 5.4 | 1,314.8 ± 56.3 | 2747.9 ± 818.2 | 2.1 |
| lasR mutant | 217.1 ± 2.9 | 1,139.8 ± 79.4 | 5.3 | 1,172.6 ± 44.8 | 2,106.6 ± 65.5 | 1.8 |
| relA mutant | 455.5 ± 20.8 | 1,577.5 ± 70.0 | 3.5 | ND | ND | ND |
Shown are the β-galactosidase activities of the PPA3309-lacZ reporter gene fusion in wild-type PAO1 and various mutants under aerobic and anaerobic conditions as described in Materials and Methods.
Exponential growth phase at an OD578 of 0.5 or 0.3 (aerobic and anaerobic culture, respectively).
Stationary phase represents 16 h after entry into the stationary phase.
Fold induction from aerobic exponential phase to aerobic stationary phase.
Fold induction from anaerobic exponential phase to anaerobic stationary phase.
ND, not determined.
FIG. 4.
(A) 2D gel image of crude extracts of P. aeruginosa PAO1 incubated under pyruvate fermentation conditions for 7 days. The boxed area contains the one or two protein spots marked by an ellipse, which were identified to be PA3309 and represented in detail in panels B to F. Enlarged 2D gel images showing the area outlined in panel A of protein extracts of P. aeruginosa PAO1 aerobic culture (B) growing exponentially at an OD578 of 0.4; (C) 16 h after entering stationary phase; (D) after 7 days under pyruvate fermentation; (E) of a 6-day-old biofilm (P. aeruginosa PAO1), and (F) a protein extract of the anr mutant PAO6261 grown as a biofilm for 6 days. Growth conditions of the biofilm experiments are outlined in Materials and Methods.
The stationary-phase sigma factor RpoS activates gene expression upon entry into the stationary phase. Anr-dependent genes like azu encoding the blue copper protein azurin have been reported to be RpoS dependent (45). However, an rpoS mutant showed no influence on the PA3309 promoter activity under aerobic or anaerobic stationary-phase conditions (Table 3).
In Mycobacterium smegmatis, a two-component regulator similar to the Mycobacterium tuberculosis DevR protein was shown to induce expression of three genes encoding Usp-type proteins (3, 32). DevR shares domain organization and 37% identity on the amino acid sequence level to the P. aeruginosa GacA regulator of the global GacA/GacS system (global antibiotics and cyanide control). This system regulates the expression of multiple phenotypes in pseudomonads. Again, no change in PA3309 promoter activity was found in a gacA mutant strain (Table 3). Similar observations were made using lasR and rhlR mutants carrying defects in both quorum-sensing systems of P. aeruginosa (Table 3).
So far, we have not checked if PA3309 expression in the stationary phase is RecA/FtsK dependent, as shown for E. coli uspA (10). However, since the PA3309 deletion mutant is not sensitive to UV exposure (data not shown), a RecA/FtsK-dependent regulation seems unlikely.
Expression of PA3309 in biofilms.
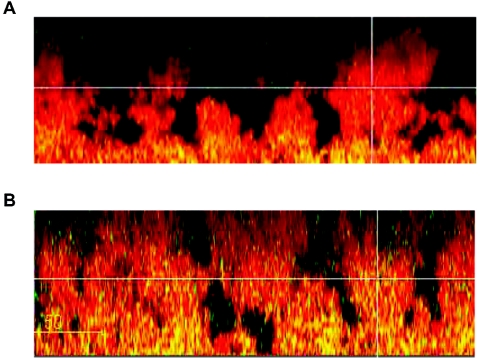
We checked the spatial distribution of PA3309 promoter induction within biofilms, its dependence on the Anr regulatory protein, and the effect of nitrate. To visualize the promoter activity of PA3309 in biofilms, a PPA3309-gfp reporter gene fusion was constructed and transferred to the P. aeruginosa chromosome (see Materials and Methods). Biofilms were grown for 6 days in flow cells and inspected by CLSM. As depicted in Fig. 5A, PA3309 promoter activity visualized by GFP fluorescence, represented as yellow areas in the red-colored biofilm matrix, was only detectable in the deeper layers of the biofilm. Quantitative analysis of the biofilms using COMSTAT revealed a twofold increase of GFP fluorescence when biofilms grew in the presence of nitrate (Fig. 5B). Since GFP requires small amounts of oxygen for activity, the deeper layers of the inspected biofilms were not strictly anaerobic but oxygen limited. In control experiments, biofilms were stained with propidium iodide to visualize dead cells. Only a small portion of the biofilm cells (1 to 2%) were not alive (data not shown).
FIG. 5.
Vertical sections showing spatial structures of 6-day-old P. aeruginosa KS15 expressing a PPA3309-gfp transcriptional fusion. Biofilms were grown in AB minimal medium containing 300 μM glucose without (A) and supplemented with (B) 50 mM KNO3. Scale bars represent 50 μm. The biofilms were stained with Syto62 (Molecular Probes) to visualize the biofilm matrix. Promoter activity is visualized by GFP fluorescence, represented as yellow areas in the red-colored biofilm matrix. Using the COMSTAT software (22), GFP was calculated to be expressed in 13.9% of the total biomass in the absence of nitrate and 29.8% when medium contained 50 mM nitrate.
To examine if the expression of PA3309 in biofilms is Anr dependent, a proteome approach was applied. Biofilms of P. aeruginosa PAO1 and the anr mutant strain PAO6261 were grown for 6 days under aerobic conditions, since the anr mutant is unable to grow and, consequently, cannot form biofilms under strict anaerobic conditions. Figure 4E and F show the partial enlargements of a 2D gel image containing two PA3309 protein spots. Both strains contain similar amounts of PA3309. Therefore, we conclude that under these microaerobic conditions induction of the PA3309 promoter is independent of Anr, comparable to the Anr-independent induction in the aerobic stationary phase.
DISCUSSION
During pyruvate fermentation, the survival of P. aeruginosa depends on PA3309, a protein with the signature domain of universal stress proteins (Usp). The PA3309 protein contains a single Usp domain and shares 37% amino acid sequence identity with UspA of E. coli. Universal stress proteins have been identified in bacteria, archaea, plants, and fungi. Six different Usp proteins (UspA, UspC, UspD, UspE, UspF, and UspG) are present in E. coli (27). Although much is known about the regulation, the function of Usp-type proteins still remains unknown (27). The E. coli proteins are produced in response to a large number of different stresses, including DNA damage, stationary phase, and growth arrest. In some cases, Usp-type proteins have been linked to the resistance to DNA-damaging agents and respiratory uncouplers (18). However, a P. aeruginosa PA3309 deletion mutant shows no comparable phenotype to an E. coli uspA mutant strain, regarding UV resistance and survival in the stationary phase (data not shown). The phenotype of the PA3309 mutant seems to be restricted to anaerobic conditions. Besides the essential role of PA3309 during pyruvate fermentation, PA3309 also contributes to survival during long-term anaerobic energy starvation in the absence of pyruvate. During these stress conditions, cell numbers of the wild type decrease dramatically by a factor of 9.0 × 102 while cell numbers of the PA3309 mutant decrease by a factor of 9.7 × 104. Consistent with the anaerobic phenotypes, regulation of the PA3309 gene was found to be dependent on the oxygen-sensing regulatory protein Anr. The β-galactosidase activities of a PPA3309-lacZ reporter gene fusion remained unchanged in an anr mutant strain upon a shift to anaerobic conditions, while they increased four- to eightfold (Fig. 3) in wild-type P. aeruginosa. As shown in Fig. 3, this anaerobic induction of the PA3309 promoter is independent of the presence of pyruvate. The regulatory behavior is consistent with the observed anaerobic phenotype of the PA3309 mutant. However, we also observed production of the PA3309 protein in the aerobic stationary phase and monitored a fivefold-increased β-galactosidase activity of the PPA3309-lacZ reporter gene fusion. We showed that this induction in the stationary phase is independent of Anr. It is also independent of other regulators which induce promoter activity in the stationary phase, during starvation, or at high cell densities, such as RpoS, RelA, and GacA and the quorum-sensing regulators RhlR and LasR. We also did not find a phenotype of the PA3309 mutant during the aerobic stationary phase in LB medium with or without supplemented pyruvate as reported for the E. coli uspA mutant. These experiments clearly indicate a different role and regulation of the Usp-type protein PA3309 in P. aeruginosa compared to the Usp proteins in E. coli. Stationary-phase expression of the P. aeruginosa PA3309 promoter is independent of RelA, which induces expression of usp genes in E. coli, and GacA, which shows similarities to DevR of M. smegmatis.
To identify the role of PA3309 during the aerobic stationary phase, we are currently investigating the PA3309 promoter in more detail and aim to identify the regulator involved.
We also investigated the spatial distribution of PA3309 promoter induction within biofilms. Biofilms are dense bacterial communities attached to a surface and surrounded by an exopolysaccharide matrix. Differences in density and architecture determine the access to nutrients and oxygen within the biofilm. Oxygen limitation can start within the first 30 μm below the surface of an aerobically grown P. aeruginosa biofilm (48). Proteome studies of P. aeruginosa biofilms indicate that a large portion of a mature biofilm population is under oxygen limitation (38). Moreover, P. aeruginosa forms even more robust biofilms when grown anaerobically in the presence of nitrate (50). Since the CF airway mucus is anaerobic (47), anaerobic biofilms might mirror a persistent infection situation in the CF lung. Oxygen limitation contributes to antibiotic resistance of the oxygen-restricted layers of aerobically grown biofilms (4). Previously, increased expression of PA3309 in aerobic and anaerobic biofilms was described (50). Our biofilm experiments clearly showed an induction of the PA3309 promoter in the deeper layers of biofilms and an increased promoter activity when biofilms were grown in the presence of nitrate. However, 2D gel electrophoresis revealed that PA3309 production in biofilms is independent of the oxygen-sensing regulator Anr. Currently, we are investigating if PA3309 also contributes to survival of cells in deeper layers of a biofilm.
2D gel analysis of P. aeruginosa cells during pyruvate fermentation revealed production of a second Usp-type stress protein, PA4352, as well as proteins involved in arginine fermentation. While a PA4352 mutant had no phenotype during pyruvate fermentation, a mutant with a defect in arginine fermentation, ΔarcDABC, showed decreased survival. Since we used a complex medium and shifted the culture to anaerobic conditions during the early exponential phase, minor amounts of arginine could support survival. We determined arginine levels in LB medium to be approximately 1.6 mM (data not shown), but these amounts do not allow survival in the absence of pyruvate (see the wild-type control in Fig. 2). However, the mutant with a defect in arginine fermentation also grows poorly, even in aerobiosis (17). This suggests that the arginine deiminase operon in general supports growth and survival of P. aeruginosa, including pyruvate fermentation.
This is the first report of an Usp-type stress protein in P. aeruginosa which contributes to anaerobic survival and pyruvate fermentation and which is produced in response to anaerobiosis in an Anr-dependent manner. Usp-type stress proteins recently gained attention from investigations using Mycobacteria. Data from proteome and transcriptome analysis showed that Usp-type proteins of Mycobacteria were produced upon oxygen limitation and phagocytosis (33). The stationary-phase regulator DevR, which is required for oxygen starvation, was identified to control usp gene expression in Mycobacterium smegmatis (3, 32). Oxygen limitation has been shown to induce a transition from active growth to a nonreplicative persistent stage important for Mycobacterium tuberculosis latency and infection (33). Our results indicate that an Usp-type protein in P. aeruginosa also contributes to anaerobic survival and may play a role in survival in anaerobic mucus plaques in CF lungs.
Acknowledgments
We thank Dieter Jahn for critically reading the manuscript. We are indebted to Dieter Haas (Universite de Lausanne, Switzerland) for providing the P. aeruginosa gacA, arcDABC, and anr mutants. We thank E. P. Greenberg (University of Iowa, Ames) for supplying the P. aeruginosa rpoS mutant and Miguel Camara (University of Nottingham,Nottingham, Great Britain) for providing the plasmids pSB219.9A and pSB224.10A for construction of the lasR and rhlR knockout mutants in P. aeruginosa. We thank H. P. Schweizer (University of Colorado, Boulder) for providing the pEX18Ap, mini-CTX-lacZ, pFLP2, and mini-CTX2 plasmids.
This investigation was funded by funds from the Deutsche Forschungsgemeinschaft, the German Research Centre for Biotechnology, and the Fonds der Chemischen Industrie. K.S. was supported by the DFG-European Graduate College 653.
REFERENCES
- 1.Beatson, S. A., C. B. Whitchurch, A. B. T. Semmler, and J. S. Mattick. 2002. Quorum sensing is not required for twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 184:3598-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becher, A., and H. P. Schweizer. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. BioTechniques 29:948-950, 952. [DOI] [PubMed] [Google Scholar]
- 3.Boon, C., and T. Dick. 2002. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J. Bacteriol. 184:6760-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borriello, G., E. Werner, F. Roe, A. M. Kim, G. D. Ehrlich, and P. S. Stewart. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 48:2659-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson, C. A., and J. L. Ingraham. 1983. Comparison of denitrification by Pseudomonas stutzeri, Pseudomonas aeruginosa, and Paracoccus denitrificans. Appl. Environ. Microbiol. 45:1247-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, Jr., A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 8.Davies, K. J., D. Lloyd, and L. Boddy. 1989. The effect of oxygen on denitrification in Paracoccus denitrificans and Pseudomonas aeruginosa. J. Gen. Microbiol. 135:2445-2451. [DOI] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 10.Diez, A., N. Gustavsson, and T. Nystrom. 2000. The universal stress protein A of Escherichia coli is required for resistance to DNA damaging agents and is regulated by a RecA/FtsK-dependent regulatory pathway. Mol. Microbiol. 36:1494-1503. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, N. W., and B. W. Holloway. 1971. Pleiotrophy of p-fluorophenylalanine-resistant and antibiotic hypersensitive mutants of Pseudomonas aeruginosa. Genet. Res. 18:185-197. [DOI] [PubMed] [Google Scholar]
- 12.Erickson, D. L., J. L. Lines, E. C. Pesci, V. Venturi, and D. G. Storey. 2004. Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster. Infect. Immun. 72:5638-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eschbach, M., K. Schreiber, K. Trunk, J. Buer, D. Jahn, and M. Schobert. 2004. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 186:4596-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farewell, A., A. A. Diez, C. C. DiRusso, and T. Nyström. 1996. Role of the Escherichia coli FadR regulator in stasis survival and growth phase-dependent expression of the uspA, fad, and fab genes. J. Bacteriol. 178:6443-6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freestone, P., T. Nystrom, M. Trinei, and V. Norris. 1997. The universal stress protein, UspA, of Escherichia coli is phosphorylated in response to stasis. J. Mol. Biol. 274:318-324. [DOI] [PubMed] [Google Scholar]
- 16.Galimand, M., M. Gamper, A. Zimmermann, and D. Haas. 1991. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J. Bacteriol. 173:1598-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamper, M., A. Zimmermann, and D. Haas. 1991. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J. Bacteriol. 173:4742-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gustavsson, N., A. Diez, and T. Nystrom. 2002. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 43:107-117. [DOI] [PubMed] [Google Scholar]
- 19.Hanna, S. L., N. E. Sherman, M. T. Kinter, and J. B. Goldberg. 2000. Comparison of proteins expressed by Pseudomonas aeruginosa strains representing initial and chronic isolates from a cystic fibrosis patient: an analysis by 2-D gel electrophoresis and capillary column liquid chromatography-tandem mass spectrometry. Microbiology 146:2495-2508. [DOI] [PubMed] [Google Scholar]
- 20.Hassett, D. J., J. Cuppoletti, B. Trapnell, S. V. Lymar, J. J. Rowe, S. Sun Yoon, G. M. Hilliard, K. Parvatiyar, M. C. Kamani, D. J. Wozniak, S. H. Hwang, T. R. McDermott, and U. A. Ochsner. 2002. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Deliv. Rev. 54:1425-1443. [DOI] [PubMed] [Google Scholar]
- 21.Heydorn, A., B. K. Ersboll, M. Hentzer, M. R. Parsek, M. Givskov, and S. Molin. 2000. Experimental reproducibility in flow-chamber biofilms. Microbiology 146:2409-2415. [DOI] [PubMed] [Google Scholar]
- 22.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 23.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 24.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 25.Hoiby, N., H. Krogh Johansen, C. Moser, Z. Song, O. Ciofu, and A. Kharazmi. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect. 3:23-35. [DOI] [PubMed] [Google Scholar]
- 26.Kvint, K., C. Hosbond, A. Farewell, O. Nybroe, and T. Nystrom. 2000. Emergency derepression: stringency allows RNA polymerase to override negative control by an active repressor. Mol. Microbiol. 35:435-443. [DOI] [PubMed] [Google Scholar]
- 27.Kvint, K., L. Nachin, A. Diez, and T. Nystrom. 2003. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6:140-145. [DOI] [PubMed] [Google Scholar]
- 28.Lam, J., R. Chan, K. Lam, and J. W. Costerton. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28:546-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. M. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Munch, R., K. Hiller, H. Barg, D. Heldt, S. Linz, E. Wingender, and D. Jahn. 2003. PRODORIC: prokaryotic database of gene regulation. Nucleic Acids Res. 31:266-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nystrom, T., C. Larsson, and L. Gustafsson. 1996. Bacterial defense against aging: role of the Escherichia coli ArcA regulator in gene expression, readjusted energy flux and survival during stasis. EMBO J. 15:3219-3228. [PMC free article] [PubMed] [Google Scholar]
- 32.O'Toole, R., M. J. Smeulders, M. C. Blokpoel, E. J. Kay, K. Lougheed, and H. D. Williams. 2003. A two-component regulator of universal stress protein expression and adaptation to oxygen starvation in Mycobacterium smegmatis. J. Bacteriol. 185:1543-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Toole, R., and H. D. Williams. 2003. Universal stress proteins and Mycobacterium tuberculosis. Res. Microbiol. 154:387-392. [DOI] [PubMed] [Google Scholar]
- 34.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 35.Rabilloud, T., J. M. Strub, S. Luche, A. van Dorsselaer, and J. Lunardi. 2001. A comparison between Sypro Ruby and ruthenium II tris (bathophenanthroline disulfonate) as fluorescent stains for protein detection in gels. Proteomics 1:699-704. [DOI] [PubMed] [Google Scholar]
- 36.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 37.Rompf, A., C. Hungerer, T. Hoffmann, M. Lindenmeyer, U. Romling, U. Gross, M. O. Doss, H. Arai, Y. Igarashi, and D. Jahn. 1998. Regulation of Pseudomonas aeruginosa hemF and hemN by the dual action of the redox response regulators Anr and Dnr. Mol. Microbiol. 29:985-997. [DOI] [PubMed] [Google Scholar]
- 38.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawers, R. G. 1991. Identification and molecular characterization of a transcriptional regulator from Pseudomonas aeruginosa PAO1 exhibiting structural and functional similarity to the FNR protein of Escherichia coli. Mol. Microbiol. 5:1469-1481. [DOI] [PubMed] [Google Scholar]
- 40.Shevchenko, A., I. Chernushevich, M. Wilm, and M. Mann. 2000. De novo peptide sequencing by nanoelectrospray tandem mass spectrometry using triple quadrupole and quadrupole/time-of-flight instruments. Methods Mol. Biol. 146:1-16. [DOI] [PubMed] [Google Scholar]
- 41.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 42.van Delden, C., R. Comte, and M. Bally. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 183:5376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Delden, C., and B. H. Iglewski. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vander Wauven, C., A. Piérard, M. Kley-Raymann, and D. Haas. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vijgenboom, E., J. E. Busch, and G. W. Canters. 1997. In vivo studies disprove an obligatory role of azurin in denitrification in Pseudomonas aeruginosa and show that azu expression is under control of rpoS and ANR. Microbiology 143:2853-2863. [DOI] [PubMed] [Google Scholar]
- 46.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Doring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, K. D., P. S. Stewart, F. Xia, C.-T. Huang, and G. A. McFeters. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye, R. W., D. Haas, J.-O. Ka, V. Krishnapillai, A. Zimmermann, C. Baird, and J. M. Tiedje. 1995. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of Fnr. J. Bacteriol. 177:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593-603. [DOI] [PubMed] [Google Scholar]
- 51.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]