Abstract
The proteins of the trithorax and Polycomb groups maintain the differential expression pattern of homeotic genes established by the early embryonic patterning system during development. These proteins generate stable and heritable chromatin structures by acting via particular chromosomal memory elements. We established a transgenic assay system showing that the Polycomb group response elements bxd and Mcp confer epigenetic inheritance throughout development. With previously published data for the Fab7 cellular memory module, we confirmed the cellular memory function of Polycomb group response elements. In Drosophila melanogaster, several of these memory elements are located in the large intergenic regulatory regions of the homeotic bithorax complex. Using a transgene assay, we showed that transcription through a memory element correlated with the relief of silencing imposed by the Polycomb group proteins and established an epigenetically heritable active chromatin mode. A memory element remodeled by the process of transcription was able to maintain active expression of a reporter gene throughout development. Thus, transcription appears to reset and change epigenetic marks at chromosomal memory elements regulated by the Polycomb and trithorax proteins. Interestingly, in the bithorax complex of D. melanogaster, the segment-specific expression of noncoding intergenic transcripts during embryogenesis seems to fulfill this switching role for memory elements regulating the homeotic genes.
Developmental decisions resulting in differential gene expression patterns need to be maintained over many rounds of cell divisions. The proteins of the Polycomb group (PcG) and trithorax group (trxG) lock determined gene expression states by generating higher-order chromatin structures which are stable and heritable. The members of the PcG protein family form silencing complexes and maintain the repressed state, whereas the conversely acting trxG proteins promote transcription and maintain the active and open chromatin mode (for a recent review, see reference 13).
Well-known targets for this type of regulation, termed cellular memory, are the homeotic gene clusters whose products determine axial cellular identities in multicellular organisms. In Drosophila melanogaster, the bithorax complex (BX-C) contains the three homeotic genes Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B). Parasegment-specific cell identity of parts of the thorax and abdomen is achieved by differential and tightly controlled expression of each homeotic gene along the anterior-posterior axis of the developing fly (21, 35).
The bithorax complex is organized into nine large cis regulatory units (abx/bx, bxd/pbx, and iab-2 to iab-8), which are responsible for the correct parasegment-specific expression patterns of the three homeotic genes. Embryonically acting maternal and segmentation genes set the parasegment-specific expression pattern of homeotic genes. In the cis regulatory sequences, chromosomal elements such as bithoraxoid (bxd), Miscadastral pigmentation (Mcp), and Frontabdominal-7 (Fab7) were identified as Polycomb group response elements (PREs). They are subsequently necessary to maintain the early-repressed expression state of the corresponding homeotic genes in appropriate segmental domains (5, 11, 15, 18, 28). However, at these chromosomal loci, colocalization of both PcG and trxG proteins was revealed (30, 33, 36). Chromosomal memory elements thus seem to contain targets for Polycomb group (PRE) as well as trithorax group (TRE) proteins, suggesting that these elements are responsible for supporting both transcriptionally silenced and activated states.
By using a transgenic assay, we have previously shown that DNA encompassing the Fab7 PRE fulfills this role. In order to mimic the endogenous situation of a homeotic gene, activated transiently by a segmentation gene, a GAL4-inducible system was used (4, 41). The Fab7 PRE was linked to a UAS-lacZ gene and mini-white as a transformation marker. In the absence of GAL4, the Fab7 element acts as a silencer, repressing both reporter genes. However, upon GAL4 activation during embryogenesis, the UAS-lacZ reporter becomes expressed homogenously. Although the GAL4 pulse was given only during embryogenesis, expression of the mini-white gene in the eyes was still observed in adult flies. Since expression of the mini-white gene is required only at late pupal stages, this result demonstrated that a GAL4-activated Fab7 maintains open chromatin conformations and shows expression of the adjacent reporter gene even after removal of the GAL4 transactivator (8, 9). Consequently, Fab7 can promote either a silenced or activated chromatin state throughout development. The combinatorial function of PREs and trithorax group response elements was termed the cellular memory module (CMM) (7).
At present, little is known about the molecular mechanism underlying the CMM function. Silencing appears to be the default state of the element, as PcG proteins are generally recruited to PREs in a parasegment position-independent manner. Transgenes containing PREs in general substantially silence nearby reporter genes as well as transformation markers such as the white gene. However, the mechanism by which the sequence specificity is achieved still remains unclear. If silencing is a default state, an important question is how CMMs can lose their repressive function and become heritably activated in a segment-specific manner. There is accumulating evidence that the transcriptional state of the target gene triggers the activity mode of the CMM: tethering of PcG proteins either by GAL4 or the LexA DNA-binding domain has revealed that Polycomb silencing depends on the time window in which the repressing fusion proteins are expressed (27, 31).
Thus, it becomes important to know how distantly located CMMs can sense the activity of the genes they are supposed to control. In the bithorax complex, the Fab7 CMM is located 43 kb downstream of the Abd-B promoter. Other apparent memory elements such as Mcp are even further away from their target transcription unit. Indeed, it has always been a major enigma why the bithorax complex regulatory elements are scattered over such an extended chromosomal territory and why such large intergenic control regions exist. Another surprising result is the finding that these intergenic regulatory regions appear to be transcribed during certain times of development.
It is now more than a decade since noncoding RNAs were identified in these regions. These transcripts first appear during the cellular blastoderm stage. Lipshitz et al. (22) reported three RNAs expressed in the bxd unit. Furthermore, the regulatory region between the abd-A and Abd-B genes expresses noncoding RNAs in a parasegment-specific manner (1, 12, 34). Although the function of these presumably nontranslated RNAs is currently not understood, it was suggested that they might be involved in the regulation of homeotic gene expression (12, 17).
In this work we show that the CMM function, originally identified for the Fab7 element, also applies to other chromosomal elements containing characterized bithorax complex PREs such as Mcp and bxd. In our transgene assay, we observed that the activation of CMM was accompanied by transcription through the element. Indeed, removal of sequences containing the promoter of the transcripts prevented activation of a CMM. Thus, transcription through a CMM appears to be necessary to remodel chromatin so as to produce a stable and heritable active memory state. We suggest that this is the functional role of the intergenic transcripts, as they are expressed in the appropriate spatial and temporal manner to switch the corresponding CMMs in a parasegment-specific fashion.
MATERIALS AND METHODS
DNA vectors and plasmid subclones.
To construct the plasmids for P-element transformation, the bxd, pBX3384, and the Mcp region, pBX5989 (both plasmids are described in reference 36) as well as the Fab7 1,800-bp boundary/PRE and 870-bp PRE fragment from the p5F24 15,2 clone (8) were amplified by PCR. New NotI restriction sites were introduced at both ends. The resulting sequences mapped to positions 218244 to 220528, 110063 to 114533, 82554 to 84355, and 82554 to 83433, numbered according to the bithorax complex sequence (24), respectively. The PCR fragments are cloned via the NotI site into the pUZ vector (8, 23).
Fly strains and handling.
Embryos of strain w1118 were used as a host for generating the transgenic lines. The hsp70-GAL47-1 driver (4) on the second chromosome or a modified version of it was used. The modified hsp70-GAL4 driver was constructed by inserting the GAL4 driver on the CyO chromosome upon expression of the transposase Δ2-3 (P-jumper; w[*]; ry[506] Sb[1] P{ry[+] 2-3} 99B/TM6B; 2536 from the Bloomington Stock Center) and screening for progeny in which the mini-white marker segregated together with CyO. Afterwards, the white marker gene was mutagenized in an ethyl methanesulfonate screen.
Transgenic flies carrying the Fab7 subfragments.
pGR1 flies carry the 1.8-kb Fab7 fragment in the proximal-distal orientation, and pGR2 flies carry it in the distal-proximal orientation. pGR3 flies have an insertion of the 870-bp Fab7 PRE construct in the proximal-distal orientation, and pGR4 flies have an insertion of the 870-bp Fab7 PRE construct in the distal-proximal orientation, as shown in Fig. 1E. Established fly lines carrying the hsp70-GAL4 driver and homozygous for the Fab7 construct were kept at 28°C or 25°C to ensure repression of the two reporters. The Fab71 fly strain (14) was kindly provided by F. Karch. The Oregon R and Canton S strains were used for in situ hybridization experiments in wild-type flies. Embryos were collected on apple juice agar plates at 25°C. All heat shock experiments were performed at 37°C in a water bath for 1 h as described by Cavalli and Paro (7). LacZ expression in embryos was examined as published previously (40).
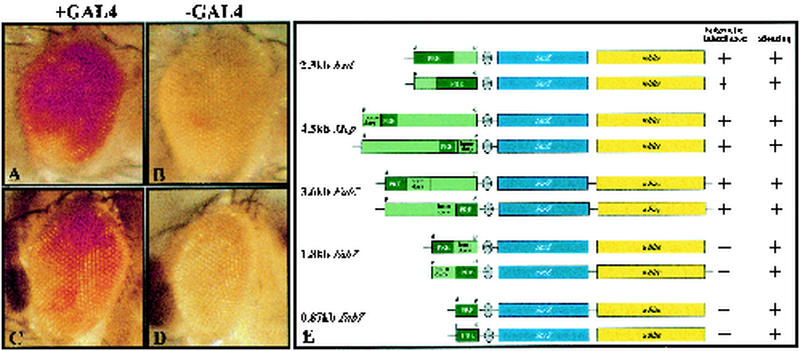
FIG. 1.
CMM function of bxd and Mcp. GAL4 was expressed during embryogenesis (A and C) by transferring embryos to a 37°C water bath for 1 h. Afterwards, heat-shocked lines were raised at 18°C until adulthood, like their control siblings (B and D), which were kept at 18°C constantly. Activated flies showed derepression of the mini-white gene. (E) The bxd, Mcp, and Fab7 subfragments were cloned into the pUZ transformation vector in both orientations (d, distal; p, proximal). From these constructs, transgenic flies were generated. The bxd and Mcp strains inherit the activated state of the two reporter genes, whereas flies carrying the Fab7 subfragments do not, as illustrated. The boundary and the PRE region depicted in the Mcp element are based on information published by Karch et al. (18) and Strutt et al. (36). The separation between boundary and PRE is arbitrary and not based on functional analysis. Constructs which displayed pairing-sensitive silencing or showed CMM function are marked with +; otherwise, a − is indicated.
In situ and slot blot hybridizations.
In situ hybridization was performed as described previously (37) with the following modifications. RNAs were digoxigenin labeled with the Roche digoxigenin RNA labeling mix (no. 1 277 073) and purified with the QIAquick PCR purification kit (no. 28106) from Qiagen. Hybridization solution contained 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 50% formamide, 0.1% Tween 20, and 5 mg of Torula yeast RNA (Sigma) per ml. Hybridization was performed at 65°C overnight. Antidigoxigenin-AP Fab fragments (no. 1 093 274) were used according to the Roche protocol. For staining, a nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) stock solution (no. 1 681 451) from Roche was used. RNA was blotted on a GeneScreen Plus hybridization transfer membrane (no. NEF 976). The same 3.6-kb distal-proximal Fab7 RNA probe as used for in situ hybridization was used for detection according to the Northern blotting protocol taken from the Roche digoxigenin system user's guide for filter hybridization.
RESULTS
CMMs in the bithorax complex.
Fab7, initially identified in the bithorax complex as a PRE, is able to recruit members of the PcG to establish a gene silencing complex. However, Fab7 is also necessary to maintain transcriptional activity of Abd-B, meaning that it is able to convey either an open or closed chromatin conformation through several rounds of cell divisions (8). An interesting question for us was whether this is a general function of chromosomal elements containing PREs. For this reason, we tested two other well-known bithorax complex elements, bxd and Mcp, for their ability to confer cellular memory.
The bxd element is located upstream of the Ubx gene, and bxd mutations alter expression levels of the UBX protein in parasegments 5 and 6 and to a lesser extent in the subsequent more posterior parasegments (17). The Mcp element is located between iab4 and iab5. The Mcp1 deletion (19) ectopically expresses Abd-B in parasegment 9 (10), giving rise to a gain-of-function phenotype, which means that the abdominal segment A4 is transformed to the identity of A5. Both bxd and Mcp were genetically identified as PREs (5, 6, 18, 28, 29), and chromatin immunoprecipitation analysis also shows strong Polycomb protein binding (36) at both cis-acting sequences.
To determine whether bxd and Mcp elements are able to promote cellular memory, transgenic fly lines carrying constructs were established as depicted in Fig. 1E. All seven independent transgenic bxd lines isolated that had the PRE inserted in the proximal-distal orientation expressed the reporters in a variegated manner. Six out of 11 bxd lines carrying the opposite PRE orientation showed pairing-sensitive silencing. In the case of the Mcp transgenes, for both orientations five independent lines were analyzed, and for each orientation, two lines showed pairing-sensitive silencing. Homogenous derepression of both reporters was obtained when mutants of the PcG were crossed to these lines (data not shown), verifying the PcG dependence of the elements tested.
To address the question of whether bxd and Mcp have a CMM function, GAL4 was expressed to activate lacZ via the inducible enhancer UAS (4). Upon GAL4 induction during embryogenesis, lacZ expression was homogenously expressed in the whole embryo in all bxd and Mcp lines tested, which showed pairing-sensitive silencing under repressed conditions. When heat-shocked bxd embryos were transferred back to 18°C and allowed to develop to adulthood, more than 90% of the offspring displayed partial or homogenous mini-white derepression in the eyes (Fig. 1A and B). The other bxd lines examined showed a similar or weaker activation of the mini-white gene. In addition, switching of the CMM into an active mode was independent of the orientation of the PRE to the enhancer.
Similarly, the Mcp lines tested were also able to inherit an active epigenetic chromatin state through mitosis. Upon GAL4 induction during embryogenesis, in several different lines derepression of the mini-white gene was observed in the eyes (Fig. 1C and D) after completion of development. Notably, the switch of the Mcp CMM from a silenced to activated chromatin state resulted in a variegated rather than homogenous pattern; 33% of the GAL4-pulsed Mcp-4.1 line displayed prominent derepression in the eyes, and the rest showed partial expression of the mini-white gene. More than 60% of the nontreated control siblings showed only small patches. The remaining 40% had slightly increased expression.
The silencing ability of Mcp was strongly temperature dependent. As observed with other phenotypes which are PcG dependent, silencing was reduced at lower temperatures and could be restored by propagating one generation at 28°C. Together, these results suggest that PREs are flanked by TREs and function in general as CMMs, thus acting as switchable chromosomal elements which “freeze” epigenetically induced transcriptional states and inherit either the silenced or activated mode during mitosis.
Proximal portion of Fab7 is important for activation and epigenetic maintenance.
The Fab7 element was shown to be responsible for the spatial regulation of the Abd-B gene in parasegments 11 and 12 (14). Genetic as well as transgenic analysis revealed that Fab7 expresses two major functions which were separable into two regions: a minimal fragment sufficient for parasegment-specific enhancer blocking, called the Fab7 boundary, and an adjacent PcG-dependent silencer, the iab7 PRE (15, 26). Despite this detailed knowledge of the organization of the Fab7 element, it remains unexplained how its state controls active chromatin that can be inherited through mitosis in a parasegment-specific manner. For a better understanding of how CMMs function, we used our transgenic assay system to analyze either the boundary and PRE sequences or just the PRE sequences separately (Fig. 1E).
Many of the homozygous lines obtained from the constructs with shortened versions of the original Fab7 fragment displayed a variegated eye color, which is subject to pairing-sensitive silencing. To test if these trimmed Fab7 elements could also be switched to a heritable open chromatin state, a hsp70-GAL4 driver was stably crossed into the chromosome. Embryos were heat shocked and analyzed for lacZ expression, and the adults were analyzed for mini-white expression. Reminiscent of the strong repression in the eye, the lacZ pattern in GAL4-activated embryos was variegated and of lower frequency than in lines carrying the full-length Fab7. In several lines, the majority showed variegated staining rather than homogenous staining. Also, lacZ expression in non-heat-shocked embryos was almost completely repressed, indicating a stronger silencing compared to lines with the full-length construct, where background with patchy expression was observed.
When we analyzed the adults derived from heat-shocked embryos carrying either of the Fab7 subfragments, we found that none of the lines displayed a red eye phenotype due to mini-white expression (two to four lines of each construct were tested). GAL4-pulsed flies were indistinguishable from control flies kept continuously at 18°C. Therefore, neither the PRE alone nor the boundary combined with the PRE was sufficient to convey epigenetic inheritance of the active CMM state. The common feature of all these transgenic flies is the lack of the proximal part of the full-length Fab7. This particular sequence therefore appears to be necessary for CMM activity to generate the activated CMM state.
Activated CMMs are transcribed.
All of the CMMs that we tested are located in intergenic regions of the bithorax complex. Noncoding transcripts were mapped to some of these regions. These are expressed at a time when the transition between the initiation of homeotic gene expression by the maternal and segmentation genes and the maintenance phase by the PcG/trxG control occurs. We wondered whether there might be a correlation between the switch of a CMM to activity and the appearance of a transcript. First, we tested our transgenic CMM lines for transcription of the chromosomal elements bxd, Mcp, and Fab7. If these CMM-containing sequences are indeed transcribed upon GAL4 activation, we would be able to correlate these RNAs with a novel biological function.
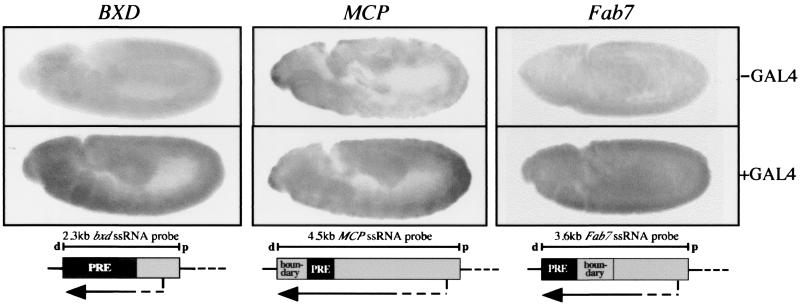
To test this hypothesis, we performed RNA in situ hybridization of fly lines carrying either the Fab7, MCP, or BXD transgene. The embryos were heat shocked to induce a GAL4 pulse and treated as described previously. Each transgenic memory element was probed with a strand-specific, digoxigenin-labeled RNA corresponding to the respective full-length CMM. Indeed, GAL4 pulse-activated embryos revealed transcription through the CMM compared to nonactivated control embryos. Figure 2 shows induced transcripts in transgenic lines capable of switching their CMM status. The transcription of these RNAs varied from a patchy to homogenous expression pattern. The memory element of each tested line was transcribed in its proximal-distal orientation independent of the CMM insertion on the transgene. Non-heat-shocked control siblings occasionally displayed these transcripts in a weaker manner, similar to lacZ background expression. Thus, there was a new transcript dependent on the GAL4 pulse given to these embryos.
FIG. 2.
CMMs are transcribed upon ubiquitous GAL4 expression during embryogenesis: in situ hybridization of embryos carrying the bxd, Mcp, or Fab7 construct. The strand-specific probes detected transcripts in the proximal-distal (d, distal; p, proximal) orientation relative to the bithorax complex. Transcripts were mostly expressed in a homogenous manner. In some lines, silencing was not completely removed, and a variegated expression pattern was observed (see Mcp line). In the absence of GAL4, minimal background expression was observed. ssRNA, single-stranded RNA.
The Fab7 transcript was also shown to be dependent on the initial GAL4 pulse by slot blot analysis. The expression of Fab7 was increased by a factor of 2 to 3 (Fig. 3). A rather surprising observation was that Fab7 CMM RNA was detected only after enrichment for polyadenylated molecules in slot blots and not in total RNA extracts. Extensive analysis of these RNAs in more detail by Northern blot and primer extension analyses did not yield a convincing result regarding the location of the promoter. Since we detected the Fab7 RNA by in situ hybridizations or slot blot analysis only in polyadenylated RNA-enriched samples, we reasoned that these must be very rare transcripts.
FIG. 3.
CMM of heat shock (hs)-induced FLW-1/Fab71 is transcribed two- to threefold more compared with non-heat-shocked controls. A digoxigenin-labeled 3.6-kb Fab7 RNA probe (distal-proximal [d-p] orientation) was hybridized to a slot blot of 5 μg of polyadenylated RNA of various lines. The endogenous Fab7 locus was transcribed weakly in the wild type (wt). Additional controls included 10 ng of in vitro-transcribed Fab7 RNA.
Transgenic lines carrying the boundary/PRE or the PRE subfragment only were not able to convey an epigenetic open chromatin state, as shown. If transcription through these sequences is essential for activating a CMM, a GAL4 pulse should not lead to expression of such RNAs in the truncated transgenic lines. Indeed, upon GAL4 expression, we were not able to detect RNAs derived from the transgenic CMM by in situ hybridization (data not shown). This argues again for the requirement of the proximal portion of the Fab7 CMM for activation. More importantly, it also supports the idea that transcription is involved in switching a silent CMM into an activated state.
The in situ hybridization data and the slot blot analysis revealed an increase in CMM-dependent transcripts in our transgenic assay system upon GAL4 pulsing. GAL4 must activate a cryptic promoter which results in the synthesis of a transcript through the CMM. As these transcripts appear at the same time point at which CMMs are switched and are not detectable in fly lines lacking the proximal portion of the Fab-7 CMM, we reasoned that such transcription may be required to switch a CMM.
CMMs are transcribed in a parasegment-specific manner.
If these noncoding RNAs play a role in regulating parasegment gene expression, one would expect that they would be transcribed in a corresponding temporal and spatial pattern. To study these expression patterns, we performed in situ hybridization experiments with strand-specific RNA probes designed for the detection of transcripts containing the bxd, Mcp, or Fab7 region (Fig. 4).
FIG. 4.
Transcription pattern of cis regulatory intergenic sequences in the bithorax complex. RNA in situ hybridization was performed with 1- to 12-h embryos. cis regulatory sequences are marked by red bars. Homeotic genes according to their most common splicing form are illustrated in black, and the in situ hybridization probes are marked in blue. The upper panels show embryos of the blastula stage, whereas the lower panels depict embryos around germ band extension. All the transcripts depicted are in distal-to-proximal orientation, detected by strand-specific probes. Transcripts remained through embryogenesis or appeared at later stages, as shown for probe D 1E1. The anterior expression domains of CMMs and intergenic cis regulatory sequences are transcribed in a linear pattern along the distal-proximal axis of the bithorax complex. Probe D 1E1 detects transcripts in the most posterior parasegment 14, although only after germ band retraction. Fab7 hybridized to transcripts in parasegments 12 to 14; Mcp to parasegments 10 to 14; iab4 to parasegments 8 to 14; and the bxd PRE to parasegments 6 to 12.
Hybridizing wild-type embryos with the bxd probe revealed an expression pattern between 50 and 25% of egg length during the early blastula stage. With continuing development, this single stripe subdivided into seven distinct stripes which could be linked to parasegments 6 to 12 (Fig. 4A). This also reflects the expression domains of the Ubx transcript, which is triggered by the bxd unit.
Using the Mcp RNA probe for hybridization, three prominent stripes were observed during the blastula stage (Fig. 4C). These were maintained throughout embryonic development in parasegments 10 to 14. Since parasegment 9 transforms into the identity of parasegment 10 by ectopic Abd-B expression in Mcp deletions, the staining pattern of the transcripts detected with the Mcp probe coincided with the Abd-B transcripts. Here again, the most anterior marked parasegment stained was the one which was expected to promote Abd-B transcription in parasegment 10.
In the case of Fab7 in situ hybridization experiments, RNA was detected in a single stripe at the posterior end of the blastula, and later it was found subdivided in parasegments 12 to 14 (Fig. 4D). Similar to bxd and Mcp, Fab7 was also transcribed in regions where it is expected to be in an open chromatin formation to promote Abd-B expression. Using a probe detecting transcripts in the iab4 region, we obtained the same results as Cumberledge et al. (12), who also found transcription in parasegments 8 to 14 (Fig. 4B). In addition, they identified two noncoding, polyadenylated RNAs which originated from the same 6.8-kb primary transcript in the iab4 regulatory region. Finally, a probe designed distal to the Abd-B 5′ end detected transcription in the most posterior parasegment (Fig. 4E).
Thus, the colinear alignment of these regulators in the bithorax complex cluster is also mirrored in a parasegment-specific expression pattern of noncoding RNAs which are transcribed in the same orientation as the homeotic genes. Interestingly, in all chromosomal regions addressed, the lowest transcription levels were detected in parasegments which are actually controlled by the CMM. In the following, more posterior parasegments, the amount of RNA was increased.
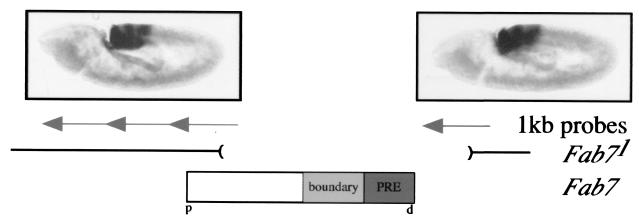
Next, we wondered whether noncoding RNAs were also present when the CMM was deleted. The Fab71 deletion (14) excises a 4.0-kb fragment encompassing the Fab7 boundary and PRE. This results in transformation of abdominal segment 6 (A6) into a copy of A7 in adult flies. In situ hybridization probes detecting RNAs on either the proximal or distal site of the deletion revealed transcripts similar to the wild type (Fig. 5), suggesting that the promoter generating this transcript localizes more distally (39). This experiment also showed that Fab7 did not contain cis-acting elements necessary for activation of the intergenic transcript. Deletion of the cis-acting PRE in the Fab71 mutant, however, removed PcG silencing, thereby circumventing the need to remodel silenced chromatin by the intergenic transcript. Taken together, these results correlate the temporal and spatial expression of these noncoding RNAs through regulatory chromosomal elements with their corresponding parasegments.
FIG. 5.
Upstream and downstream region of the Fab71 deletion are transcribed. Fab71 embryos were in situ hybridized to 1-kb probes flanking the Fab71 deletion. Distal (d) as well as proximal (p) probes generated the same staining pattern observed for the 3.6-kb Fab7 element. The probes hybridized in parasegments 12 to 14.
DISCUSSION
With this work, we show that besides the Fab7 element, two other PRE-containing elements of the bithorax complex, bxd and Mcp, depict CMM functions. Thus, these elements are necessary not only to maintain the repressed state of homeotic genes through their bound PcG silencing proteins but also to maintain active expression states. Indeed, the concept of a CMM appears to be used generally for other developmental regulators whose expression state is required to be heritably maintained. A CMM was also identified at the hedgehog gene used for imaginal disk patterning (24a). CMMs sense decisions made by the patterning system and “freeze” either the silenced or activated expression state of the target gene. Here we show that transcription through the CMM is associated with the mitotically heritable activated mode of a CMM.
We showed that a larval GAL4 pulse, unlike an embryonic pulse, was not able to switch the Fab7 CMM to a mitotically heritable state (7). While a larval GAL4 pulse would indeed relieve silencing and activate the reporter lacZ transiently, after removal of GAL4, expression of lacZ would return to basal levels. This suggested that, during embryogenesis, specific events occur, imposing a mitotically stable activated CMM state. During the process of trimming the original 3.6-kb Fab7 CMM to what we considered to be the core element composed of the boundary and PREs, we realized that shortened constructs lacked CMM functions while retaining the silencing function (Fig. 1). Surprisingly, we found that the full-size Fab7 fragment produced a transcript upon GAL4 activation, while the shorter, nonactivatable fragments lacked this transcript.
We propose that the Fab7 element contains a cryptic promoter which is located in the proximal part of the CMM. This is also supported by the analysis of transgenes carrying the Fab7 fragment in the inverse orientation. These had an activatable CMM function and produced a transcript from the proximal part of the fragment. Thus, the fortuitous combination in our transgenes of a cryptic promoter activatable by GAL4 led us to uncover a transcript through the CMM which was able to generate a mitotically stable activated CMM state.
The identification of embryonic transcripts encoded by the CMMs on our transgene constructs suggested a possible mechanism for activation of memory elements in the bithorax complex. At this point, it was intriguing to observe that in the bithorax complex, many intergenic transcripts pass through known CMMs. Indeed, we found that these transcripts were expressed exactly at the time when the corresponding CMMs need to be parasegment specifically activated. The specific spatial distribution observed further supports the view of a nonrandom functional role of these RNAs.
Noncoding intergenic transcripts have already been identified in the bithorax complex (17, 22, 34). However, their functional role remained unclear. In those cases for which cDNAs were available, no obvious coding capacity was observed, suggesting a role different from that of an mRNA (12, 20). Interestingly, even when a presumptive CMM was located within a transcription unit (the abx/bx element in the Ubx transcription unit), an early, short-lived transcript was identified (16). Thus, the transcription process generating the intergenic bithorax complex RNAs has all the hallmarks necessary to act as a CMM switching mechanism.
It could be argued that the occurrence of these early intergenic transcripts is a by-product of a relatively open embryonic chromatin state allowing spurious transcription. However, other genes located in or near the bithorax complex do not become activated at this time of development (24), arguing against a global deregulation by “loose” chromatin structures. Additionally, the intergenic transcripts are under the same strict temporal and spatial regulation as the homeotic protein encoding RNA transcripts. Interestingly, in parallel work by Hogga and Karch and by Bender and Fitzgerald (3, 16), it was found that ectopic transcription in the endogenous bithorax complex interferes with the establishment of PcG silencing. Their work identified particular mutant fly lines with P element or transgene insertions in the bithorax complex producing long transcripts. They demonstrate that ectopic transcription through iab cis regulatory elements of the bithorax complex results in homeotic phenotypes that are reminiscent of spatially deregulated CMM functions.
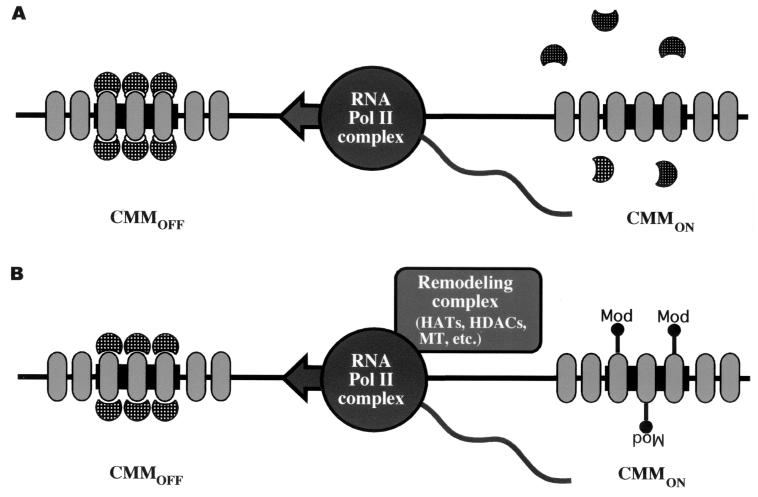
What molecular mechanisms are associated with the embryonic CMM switch? We believe that it is the machinery generating the transcripts which is crucial for the switch. It could be envisaged that passage of an RNA polymerase II complex through a CMM silenced by default displaces the corresponding repressive PcG protein, thus interfering with the establishment of PcG silencing complexes at CMMs or actively removing silencing complexes set by default (Fig. 6). As an activated CMM is mitotically heritable, an additional epigenetic mark might then be placed during the transcription process. Accordingly, the RNA polymerase II complex might “piggyback” enzymatic remodeling complexes which set epigenetic marks by histone modifications (38). Indeed, hyperacetylation of H4 was shown to be heritable in the case of the Fab7 CMM (8).
FIG. 6.
Transcriptional remodeling of CMMs. Initially, gap and segmentation genes set up the expression pattern of the homeotic genes. CMMs are subsequently required to maintain the preset homeotic gene expression patterns throughout development by distinguishing transcriptionally active and silenced chromatin. Transcription through a CMM switches the element from the silenced to activated state. We can envisage two possible, not mutually exclusive, functions to achieve this heritable switch. In model A, the RNA polymerase (Pol) II complex passes through CMMs in intergenic regions, displacing the repressive complexes. As a consequence, chromatin becomes accessible for further epigenetic modifications. In model B, RNA polymerase II “piggybacks” remodeling complexes which epigenetically mark histones. These modifications are stably sustained throughout development and inherit an open chromatin conformation (ovals, nucleosomes; half-circles, silencing complexes; Mod, epigenetic modification of histones like histone acetyltransferase [HAT], histone deacetylase [HDAC], or methyltransferase [MT]).
Another system in which transcription regulation at gene clusters is controlled by chromatin states is the β-globin locus. Its chromatin structures and genetic arrangements are comparable to those of the bithorax complex. The β-globin locus contains five genes clustered in a linear arrangement. However, at each time point during development, only one gene is transcribed. Interestingly, mice carrying a transgenic human β-globin locus display intergenic transcripts in the same direction as β-globin gene transcription. They are found in the vicinity of active genes, can be up to 30 kb long and were implicated in chromatin remodeling and maintenance of active domains (2, 32). In transgenes, the enhancer and 5′ hypersensitive site HS2 of the β-globin locus control region were found to be transcribed. The transcript originated within the enhancer and continued to the linked gene irrespective of the position and orientation of the enhancer (20). Due to this finding that non-mRNA transcription starts in transgenes, it was suggested that intergenic transcription is a general characteristic of active loci.
The finding of functional intergenic transcripts in the bithorax complex could shed new insight on another puzzle related to homeobox gene (HOX) clusters, the colinearity phenomenon (21, 25). The arrangement of the genes in the HOX clusters in the 5′-3′ orientation reflects their sequential expression pattern in the posterior-anterior axis of the developing organism. This colinearity was maintained during evolution, suggesting an underlying functional relevance. The finding that regulatory modules are interconnected by transcripts would argue for a need to maintain the sequential order along the chromosome. The polarity of the transcription process would switch CMMs in an ordered and sequential manner. This could be the underlying force keeping the ordered arrangement of regulatory elements and their target genes together. While in D. melanogaster, the regulatory intergenic transcript might have become broken up, in other HOX complexes, long-range transcripts expressed in the 5′-3′ orientation might fulfill this sequential opening in one process. Thus, it will be interesting to observe whether intergenic transcripts with related CMM switching functions exist in HOX clusters of other organisms.
Acknowledgments
Gerhard Rank and Matthias Prestel contributed equally to this work.
We thank Francois Karch for sending fly lines, Marc Hild for technical advice, Gabi Schwarz for help with in situ hybridizations, and Leonie Ringrose for helpful comments on the manuscript. We are grateful to Francois Karch and Welcome Bender for sharing and allowing us to cite unpublished results.
R.P. was supported by grants from the Deutsche Forschungsgemeinschaft and the Fond der Chemischen Industrie.
REFERENCES
- 1.Akam, M., and A. Martinez-Arias. 1985. The distribution of Ultrabithorax transcripts in Drosophila embryos. EMBO J. 4:1689-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashe, H. L., J. Monks, M. Wijgerde, P. Fraser, and N. J. Proudfoot. 1997. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 11:2494-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, W., and D. P. Fitzgerald. Transcription activates repressed domains in the Drosophila bithorax complex. Development, in press. [DOI] [PubMed]
- 4.Brand, A. H., A. S. Manoukian, and N. Perrimon. 1994. Ectopic expression in Drosophila. Methods Cell Biol. 44:635-654. [DOI] [PubMed] [Google Scholar]
- 5.Busturia, A., and M. Bienz. 1993. Silencers in Abdominal-B, a homeotic Drosophila gene. EMBO J. 12:1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busturia, A., A. Lloyd, F. Bejarano, M. Zavortink, H. Xin, and S. Sakonju. 2001. The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development 128:2163-2173. [DOI] [PubMed] [Google Scholar]
- 7.Cavalli, G., and R. Paro. 1998. Chromo domain proteins: linking chromatin structure to epigenetic regulation. Curr. Opin. Cell Biol. 10:354-360. [DOI] [PubMed] [Google Scholar]
- 8.Cavalli, G., and R. Paro. 1998. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93:505-518. [DOI] [PubMed] [Google Scholar]
- 9.Cavalli, G., and R. Paro. 1999. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science 286:955-958. [DOI] [PubMed] [Google Scholar]
- 10.Celniker, S. E., S. Sharma, D. J. Keelan, and E. B. Lewis. 1990. The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 9:4277-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan, C. S., L. Rastelli, and V. Pirrotta. 1994. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 13:2553-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cumberledge, S., A. Zaratzian, and S. Sakonju. 1990. Characterization of two RNAs transcribed from the cis-regulatory region of the abd-A domain within the Drosophila bithorax complex. Proc. Natl. Acad. Sci. USA 87:3259-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francis, N. J., and R. E. Kingston. 2001. Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell. Biol. 2:409-421. [DOI] [PubMed] [Google Scholar]
- 14.Gyurkovics, H., J. Gausz, J. Kummer, and F. Karch. 1990. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 9:2579-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagstrom, K., M. Müller, and P. Schedl. 1996. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 10:3202-3215. [DOI] [PubMed] [Google Scholar]
- 16.Hogga, I., and F. Karch. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with Polycomb mediated silencing. Development, in press. [DOI] [PubMed]
- 17.Hogness, D. S., H. D. Lipshitz, P. A. Beachy, D. A. Peattie, R. B. Saint, M. Goldschmidt-Clermont, P. J. Harte, E. R. Gavis, and S. L. Helfand. 1985. Regulation and products of the Ultrabithorax domain of the bithorax complex. Cold Spring Harbor Symp. Quant. Biol. 50:181-195. [DOI] [PubMed] [Google Scholar]
- 18.Karch, F., M. Galloni, L. Sipos, J. Gausz, H. Gyurkovics, and P. Schedl. 1994. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 22:3138-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karch, F., B. Weiffenbach, M. Peifer, W. Bender, I. Duncan, S. Celniker, M. Crosbi, and E. B. Lewis. 1985. The abdominal region of the bithorax complex. Cell 43:81-96. [DOI] [PubMed] [Google Scholar]
- 20.Kong, S., D. Bohl, C. Li, and D. Tuan. 1997. Transcription of the HS2 enhancer toward a cis-linked gene is independent of the orientation, position, and distance of the enhancer relative to the gene. Mol. Cell. Biol. 17:3955-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, E. B. 1978. A gene complex controlling segmentation in Drosophila. Nature 276:565-570. [DOI] [PubMed] [Google Scholar]
- 22.Lipshitz, H. D., D. A. Peattie, and H. S. Hogness. 1987. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes Dev. 1:307-322. [DOI] [PubMed] [Google Scholar]
- 23.Lyko, F., J. D. Brenton, M. A. Surani, and R. Paro. 1997. An imprinting element from the mouse H19 locus functions as a silencer in Drosophila. Nat. Genet. 16:171-173. [DOI] [PubMed] [Google Scholar]
- 24.Martin, C. H., C. A. Mayeda, C. A. Davis, C. L. Ericsson, J. D. Knafels, D. R. Mathog, S. E. Celniker, E. B. Lewis, and M. J. Palazzolo. 1995. Complete sequence of the bithorax complex of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 92:8398-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Maurange, C., and R. Paro. A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development. Genes Dev., in press. [DOI] [PMC free article] [PubMed]
- 25.McGinnis, W., and R. Krumlauf. 1992. Homeobox genes and axial patterning. Cell 68:283-302. [DOI] [PubMed] [Google Scholar]
- 26.Mihaly, J., I. Hogga, J. Gausz, H. Gyurkovics, and F. Karch. 1997. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124:1809-1820. [DOI] [PubMed] [Google Scholar]
- 27.Muller, J. 1995. Transcriptional silencing by the Polycomb protein in Drosophila embryos. EMBO J. 14:1209-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller, J., and M. Bienz. 1991. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. EMBO J. 10:3147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller, M., K. Hagstrom, H. Gyurkovics, V. Pirrotta, and P. Schedl. 1999. The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 153:1333-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orlando, V., E. Jane, P. V. Chinwalla, P. Harte, and R. Paro. 1998. Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 17:5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poux, S., R. Melfi, and V. Pirrotta. 2001. Establishment of Polycomb silencing requires a transient interaction between Polycomb and ESC. Genes Dev. 15:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ristaldi, M. S., D. Drabek, J. Gribnau, D. Poddie, N. Yannoutsous, A. Cao, F. Grosveld, and A. M. Imam. 2001. The role of the −50 region of the human gamma-globin gene in switching. EMBO J. 20:5242-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rozovskaia, T., S. Tillib, S. Smith, Y. Sedkov, O. Rozenblatt-Rosen, S. Petruk, T. Yano, T. Nakamura, L. Ben-Simchon, J. Gildea, C. M. Croce, A. Shearn, E. Canaani, and A. Mazo. 1999. Trithorax and ASH1 interact directly and associate with the trithorax group-responsive bxd region of the Ultrabithorax promoter. Mol. Cell. Biol. 19:6441-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez-Herrero, E., and M. Akam. 1989. Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development 107:321-329. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez-Herrero, E., I. Vernós, R. Marco, and G. Morata. 1985. Genetic organisation of Drosophila bithorax complex. Nature 313:108-113. [DOI] [PubMed] [Google Scholar]
- 36.Strutt, H., G. Cavalli, and R. Paro. 1997. Colocalization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 16:3621-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tautz, D., and C. Pfeifle. 1989. A non-radioactive method for the localisation of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98:81-85. [DOI] [PubMed] [Google Scholar]
- 38.Wittschieben, B. O., G. Otero, T. de Bizemont, J. Fellows, H. Erdjument-Bromage, R. Ohba, Y. Li, C. D. Allis, P. Tempst, and J. Q. Svejstrup. 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4:123-128. [DOI] [PubMed] [Google Scholar]
- 39.Zhou, J., H. Ashe, C. Burks, and M. Levine. 1999. Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Development 126:3057-3065. [DOI] [PubMed] [Google Scholar]
- 40.Zink, B., Y. Engström, W. J. Gehring, and R. Paro. 1991. Direct interaction of the Polycomb protein with Antennapedia regulatory sequences in polytene chromosomes of Drosophila melanogaster. EMBO J. 10:153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zink, D., and R. Paro. 1995. Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J. 14:5660-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]