Abstract
The bacteriophage Mu strong gyrase site (SGS), required for efficient phage DNA replication, differs from other gyrase sites in the efficiency of gyrase binding coupled with a highly processive supercoiling activity. Genetic studies have implicated the right arm of the SGS as a key structural feature for promoting rapid Mu replication. Here, we show that deletion of the distal portion of the right arm abolishes efficient binding, cleavage, and supercoiling by DNA gyrase in vitro. DNase I footprinting analysis of the intact SGS revealed an adenylyl imidodiphosphate-dependent change in protection in the right arm, indicating that this arm likely forms the T segment that is passed through the cleaved G segment during the supercoiling reaction. Furthermore, in an SGS derivative with an altered right-arm sequence, the left arm showed these changes, suggesting that the selection of a T segment by gyrase is determined primarily by the sequences of the arms. Analysis of the sequences of the SGS and other gyrase sites suggests that the choice of T segment correlates with which arm possesses the more extensive set of phased anisotropic bending signals, with the Mu right arm possessing an unusually extended set of such signals. The implications of these observations for the structure of the gyrase-DNA complex and for the biological function of the Mu SGS are discussed.
The type II topoisomerases facilitate essential cellular processes such as DNA replication and transcription by catalyzing the relaxation, supercoiling, and decatenation of DNA substrates (2, 55). These transactions are achieved by the enzymes catalyzing formation of a transient double-strand break in one DNA molecule—the gate or G segment—and by passing a second DNA duplex—the transported or T segment—through the gap (56). The strand passage event and subsequent reaction cycles are coupled to ATP binding and hydrolysis. DNA gyrase is a type IIA topoisomerase found predominantly in prokaryotes and more recently in plant mitochondria and chloroplasts (54), and it is the only known topoisomerase capable of introducing negative supercoils into DNA.
DNA gyrase contains two kinds of subunits, GyrA and GyrB, with the active form of the enzyme being an A2B2 tetramer (16, 40). The A subunit consists of a 59-kDa N-terminal domain and a 37-kDa C-terminal domain (CTD), the structures of which have each been solved separately (3, 24). The B subunit also consists of two domains, and the 43-kDa N-terminal domain (NTD) of GyrB was the first type II topoisomerase structure to be determined at atomic resolution (57). Functionally, this part of the B subunit binds and hydrolyzes the ATP needed for enzyme activity. The structure of the 47-kDa GyrB C-terminal domain is unknown, although it can be inferred by analogy with the structure of the homologous yeast topoisomerase II enzyme (1, 7).
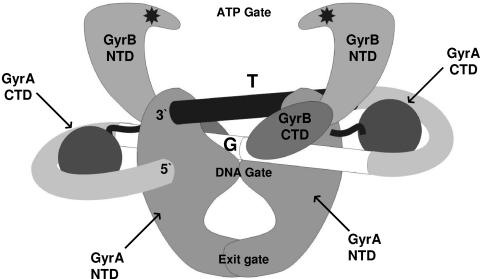
There is a division of labor between the GyrA and GyrB subunits in the gyrase reaction. As illustrated in Fig. 1, the NTD of GyrA forms a dimer linked by two protein bridges, termed the DNA gate and the exit gate (21, 56), such that a central cavity large enough to contain a DNA duplex is formed (24). The DNA gate contains the amino acid residues that bind to and catalyze the double-strand break in the G segment. In the case of gyrase, and indeed all type II topoisomerases, the break is a 4-bp staggered cut yielding 5′ overhangs, with the terminal 5′ end of each strand becoming covalently linked (in Escherichia coli GyrA) to Tyr122 (13). The GyrB N-terminal domains form two lobes of an ATP-operated DNA clamp or ATP gate: this is normally open in the absence of ATP but closes once ATP (or a nonhydrolyzable analogue such as adenylyl imidodiphosphate [ADPNP]) binds. Closure of the ATP gate captures a DNA helix, which becomes the T segment of DNA during the reaction cycle. The binding of ATP (or ADPNP) also induces the structural changes in gyrase that part the G segment, drive the adjacent T segment through the gap into the central cavity of the GyrA dimer, and reseal the G-segment break. Once the break is resealed, the T segment is unable to return directly to the ATP gate; instead, this region of DNA now passes through the GyrA exit gate to leave the complex. The net result is a vectorial or unidirectional passage of the T segment through the enzyme complex.
FIG. 1.
A structural model for the gyrase-DNA complex. The DNA gyrase A and B subunits (GyrA and GyrB) are each composed of an NTD and a CTD. The GyrB NTDs carry the ATP binding sites, starred in the figure, and also define an ATP gate, shown here in the open configuration. The GyrA NTD also forms two protein bridges, the DNA gate and the exit gate, which are here shown closed. The GyrA CTDs are likely linked to the rest of the A subunits by a flexible linker. The possible path of the DNA associated with the enzyme is indicated, with arbitrary 5′ and 3′ designations to delineate the binding site. The continuation of the DNA substrate is not indicated. The core of the DNA site, or G segment, is given as a split white cylinder, the split being the point of 4-bp staggered cleavage. The T segment is shown by the black cylinder, positioned just above the G segment. The mechanistic effects of ATP binding on this structure are expanded upon in the text.
Footprinting analyses with DNase I, exonuclease III, or hydroxyl radicals have established that gyrase protects 120 to 150 bp of DNA from attack, the exact length depending on both the cleavage agent and the DNA sequence used as a substrate (8, 15, 16, 30, 39, 53). The central 25- to 35-bp region of the footprint, corresponding to the G segment, is most heavily protected from degradation. On both sides of the G-segment core, the enzyme protects an additional 50 bp or so of DNA, in which nuclease-hypersensitive sites are present every 10 to 11 bp, suggesting that each arm of the site is wrapped on the gyrase surface. The extent of DNA protection is unique to gyrase; other type II topoisomerases, which are unable to introduce negative supercoils into DNA, yield footprints only around 30 bp in length (17, 37). The GyrA C-terminal domains, a structural component specific to gyrase, form DNA binding regions on the protein and promote the wrapping of DNA around the tetramer (14). This mode of DNA binding ensures that one of the arms itself (or an immediately contiguous region) will be available for capture by GyrB to form the T segment (Fig. 1), and so gyrase defines the precise topological and geometrical arrangement of both T and G segments during supercoiling. This feature, coupled with the vectorial passage of the T segment through the enzyme, ensures the net introduction of negative supercoils into DNA in the gyrase reaction cycle.
The above model of the T segment being significantly wrapped by the enzyme before strand passage and undergoing conformational changes during supercoiling was recently corroborated by atomic force microscopy studies (12), which demonstrated the loss of DNA wrapping by gyrase in the presence of the nucleotide. However, the determinants by which the enzyme selects one arm over the other to form the T segment are currently not well understood. Evidence that the arms of a gyrase site do indeed play differing roles has come primarily from hydroxyl-radical footprinting studies of the complex with the strong pBR322 gyrase site (30). In the absence of ADPNP, the left arm upstream of the G segment (reading the pBR322 sequence 5′ to 3′ in the conventional numbering scheme) was protected against hydroxyl radical attack over a longer distance than the right arm of the site, forming an asymmetric pattern of DNA protection with respect to the core. Addition of ADPNP reduced the extent of protection of the left arm, leading to a more symmetric footprint; in addition, the locations of hypersensitive sites within the left arm shifted by an average of 3 bp. These results showed that a loss of extended protection, coupled with a conformational change, occurs in the pBR322 site left arm once ATP binds, implying that this arm forms the T segment in the supercoiling reaction.
It is also not clear how the sequence itself of a given gyrase site might affect the choice of T segment, or otherwise modulate the activity of the enzyme. A dramatic example of DNA sequence-dependent effects on gyrase function is seen with the bacteriophage Mu strong gyrase site (Mu SGS) (36). The 37-kb Mu genome integrates into host DNA and is replicated by replicative transposition, a pathway that necessitates the ends of the genome be aligned and then synapsed in a defined topological way before DNA replication (5, 31). The Mu SGS at the center of the Mu genome (25) functions as a strong site for gyrase activity in vivo (32) and in vitro (28). We have proposed that the function of host gyrase recruited at the SGS is to actively promote alignment and synapsis of the genome ends by efficiently introducing multiple supercoils into the Mu DNA, thus extruding a Mu chromosomal domain and overcoming the topological constraints of the host nucleoid on the alignment of the prophage ends (34). In support of this model, deletion of the SGS, switching the location of the SGS away from the center of Mu, or inactivation of the host gyrase activity all caused significant delays to the rate of Mu replication (32, 36, 42, 47). That this effect is dependent on the SGS was shown when other gyrase sites, including the major sites from pBR322 or pSC101 (27, 53), were substituted for the SGS: replication of these chimeric phages was significantly impaired compared to wild-type Mu (35). Gyrase sites analogous to the Mu SGS have been found in several Mu-like prophages from bacterial genera such as Neisseria and Haemophilus, but replacement of the Mu SGS by these sites generally had little or no effect in stimulating Mu replication. In fact, the only naturally occurring sequence we have thus far identified as being largely able to substitute for the SGS is a site from the center of a Mu-like prophage in the E. coli O157:H7 Sakai genome (29).
As with the pBR322 site, there is evidence that the arms of the Mu SGS have differing roles in gyrase function. More specifically, genetic data have implicated the SGS right arm as supplying key biological properties of the site (33). (The designation of the right arm of the Mu SGS is based on the published sequence for the Mu genome (25), where the region immediately upstream of the SGS cleavage site is termed the left arm and that immediately downstream is termed the right arm.) A hybrid pBR322-based gyrase site with the cognate right arm replaced by the Mu SGS right arm, for example, was more efficient in supporting Mu replication than the intact pBR322 site; similarly, a site comprising the Mu right arm fused to the core and left arm of the pSC101 site was indistinguishable from wild-type SGS in supporting fast DNA replication. No such effect was apparent when the SGS left arm was used in equivalent hybrid sites. Deletions of either arm of the SGS towards the core of the site also showed differing effects. The majority of the right arm (as well as the SGS core) was needed to support efficient replication, while almost all of the left arm could be deleted before a deleterious effect on replication was observed (33). Separate biochemical studies showed that the SGS differed from other gyrase sites examined in that it was both very efficiently bound by gyrase and conferred efficient supercoiling with an increased degree of processivity (28). Since the biology of the SGS is related to the activity of gyrase at this site, it is reasonable to assume that the SGS right arm is a determinant for these distinctive biochemical properties.
It is becoming apparent that each arm of a given gyrase site plays a differing role in the supercoiling reaction. Furthermore, the right arm of the Mu SGS appears to possess a key function required for efficient Mu replication. The experiments presented here were designed to further our understanding of the structural and functional interactions of the gyrase site arms with the enzyme, with a particular focus on the right arm of the SGS itself. The aim was to shed light on both the criteria by which gyrase selects an arm to form the T segment in the supercoiling reaction and to what extent the right-arm sequence can account for the biological effects of the Mu SGS on Mu replication.
MATERIALS AND METHODS
Enzymes and drugs.
DNA gyrase and topoisomerase I were purified as described previously (23). T4 polynucleotide kinase, Taq DNA polymerase, T4 DNA ligase, and proteinase K were obtained from Invitrogen Life Technologies; AhdI and HincII were from New England Biolabs; and DNase I was from Sigma. Enoxacin (a gift from Pat Higgins) was dissolved in equimolar NaOH and stored at −20°C. Chloroquine phosphate was freshly dissolved in gel running buffer and used immediately.
Sources of gyrase sites and DNA substrates.
Gyrase sites were derived by PCR performed on wild-type Mu DNA, chimeric Mu constructs bearing previously constructed hybrid sites (33), genomic DNA from E. coli strain O157:K7 Sakai (from K. Makino, Osaka University, Osaka), and pSC101 or pBR322 DNA, using oligonucleotides to synthesize 200-bp products centered on the main gyrase cleavage site. Derivatives of the E. coli O157:H7 Sakai (ECs) site (29) were obtained by designing oligonucleotide pairs for PCR on E. coli O157:K7 genomic DNA, such that the first encompassed one intact arm of the ECs site and the other hybridized within the other arm to create an appropriate deletion. The PCR product was then ligated into p1856, and a chimeric Mu prophage was obtained as previously described (35).
Biochemical and biological assays.
All PCR products were purified from 1.5% agarose gels, using the Qiaquick gel extraction kit (QIAGEN). End labeling of DNA fragments was achieved using T4 polynucleotide kinase and [γ32P]ATP (ICN Biomedicals). Labeled DNA was purified by passage through a Sephadex G-50 spin column (Amersham Biosciences). Molecules labeled at a single 5′ end were prepared by radiolabeling the appropriate primer before use in a PCR. For production of 3-kb plasmid substrates, the appropriate PCR products were ligated into HincII-linearized pUC19; closed circular forms of each DNA were purified from cesium gradients, following standard protocols (43). Relaxed forms of each DNA were obtained by subsequent treatment with calf thymus topoisomerase I; these were purified by phenol extraction and ethanol precipitation. Binding, cleavage, and supercoiling assays on linear or circular substrates were performed as previously described (28).
The parent strain for lysogens was E. coli AB1157 recB recC sbcB malF::Mu cts62 (36). Host lysis by prophages bearing the ECs site or a derivative were performed by growing the appropriate lysogen in L broth at 30°C to a density of around 108 cells/ml; these were diluted twofold with L broth and induced by transferring the culture to 42°C. Culture density was subsequently monitored with Klett readings.
Footprinting analysis.
Footprinting reactions comprised 10 mM HEPES (pH 7.5), 24 mM KCl, 5 mM MgCl2, 5 mM dithiothreitol, 15 nM each singly radiolabeled 0.2-kb fragment, and 90 nM DNA gyrase tetramer in a volume of 32 μl. Reaction mixtures were incubated at 37°C for 15 min, and 4 μl of 15 mM ADPNP (prepared in the same buffer) was added; reaction mixtures were then incubated at 37°C for a further 5 min. A solution of DNase I (25 ng/μl) was added, and digestion was continued for 2 min at 37°C: reactions were then immediately quenched by the addition of 10 μl 25% (vol/vol) glycerol and 0.1 M EDTA solution and chilled on ice. Samples were extracted once with an equal volume of phenol and desalted by passage through a Sephadex G-50 spin column. DNA was recovered by ethanol precipitation and resuspended in formamide loading buffer, and bands were resolved on denaturing 8% polyacrylamide gels. These also included reference-sequencing ladders obtained with the Promega fmol sequencing kit and products of the quinolone-induced cleavage reaction as markers. After electrophoresis, gels were dried and exposed to Kodak phosphorimager screens, which were analyzed in a Bio-Rad phosphorimager.
Calculation of DNA flexibility.
DNA flexibility was approximated by summing the stacking energy of a running window of 30 adjacent base steps using an experimentally derived set of base step-specific parameters (38). These parameters correlate better with experimentally determined persistence lengths (52) than any other set tested (A. A. Travers, unpublished observations).
RESULTS
Nomenclature of gyrase sites used in this work.
To compare the regions of different gyrase sites used in these studies, the following scheme was used. The base pairs of each sequence were numbered centered on the middle of the gyrase cleavage site, omitting a base pair of zero, so under this scheme the 4-bp staggered cut made by the enzyme encompassed positions −2, −1, +1, and + 2. The core of each site was defined as the 20-bp region from bp −10 to bp +10. In relation to the core sequence, the left arm comprised the sequence upstream from bp −11 extending to approximately bp −70; similarly, the right arm was the sequence from bp +11 to approximately bp +70. (The sequences of the gyrase sites used in these studies are shown below; see Fig. 7.)
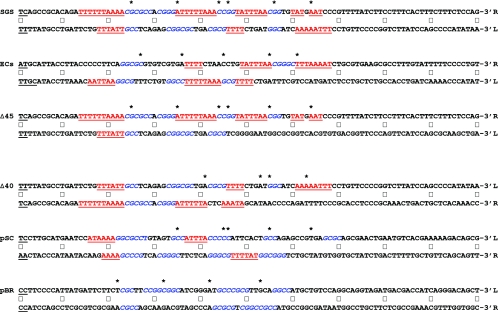
FIG. 7.
Anisotropic flexibility of gyrase sites. The nucleotide sequences of the 200-bp sites are given in full. To facilitate comparison of each arm of the sites directly, the sequence of one of the arms was folded back, an arrangement that places the central 4-bp cleavage site, shown in underlined boldface black type, at the left of the figure. (The ECs cleavage site comprises two overlapping sites, GCAA and AAAT; hence, a total of 6 bp are underlined.) Central dots mark every 10th nucleotide from the cleavage site. The upper line of the each site gives the sequence that runs 5′ to 3′ downstream from the cleavage site, whereas the lower, folded-back sequence is that complementary to the arm upstream from the cleavage site, ensuring that each line of the sequence runs in the conventional 5′-to-3′ direction. A label (L or R) indicates the left or right arm of each site, based on the scheme used throughout this work. Asterisks above the sequences mark the positions of DNase I-hypersensitive sites observed in the absence of ADPNP. GC-rich regions of ≥3 bp in phase with the marked DNase I cleavage sites are shown in blue italics; conversely, AT regions of ≥3 bp in length that are out of phase are in underlined red type. Colorings were done without prejudice, so that the entire length of an A-T or G-C run was included. (In some cases, particularly in the SGS and ECs right arms, some AT runs are so long that these are both in phase and out of phase.) Finally, each sequence is presented such that the presumptive T segments implied by DNase I footprinting are in the upper line and the non-T segments are in the lower line.
The Mu SGS right arm plays a key role in the selection of a preferred gyrase binding and cleavage site.
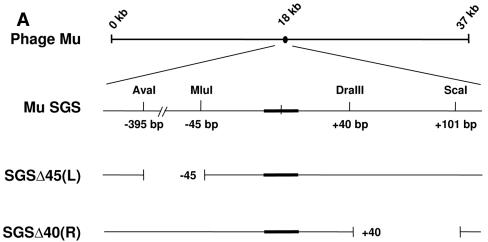
To test the prediction that the unusual biochemical properties of the SGS are a consequence of the right-arm sequence, we examined the binding and cleavage activity of gyrase on DNA fragments containing the SGS or sites with deletions of the right or left arm (Fig. 2A). To make the Mu SGS derivatives, two restriction cuts were made in a central portion of Mu DNA for each construct, removing a fragment of each arm, and the subsequent ends were then blunted and religated. Thus, the remainder of each site was formed from Mu sequence that normally falls outside of the wild-type SGS. The site where the SGS right-arm sequence from bp +40 onwards was replaced by a different sequence was termed SGSΔ40(R) and the site with a similar substitution in the left arm, of sequences from bp −45 outwards, was termed SGSΔ45(L).
FIG. 2.
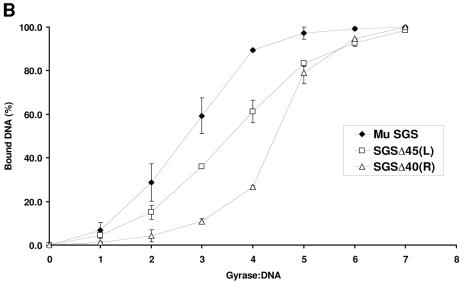
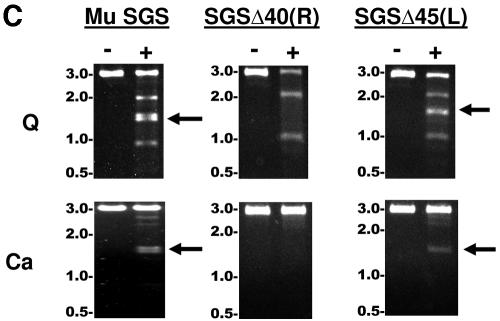
Binding and cleavage by gyrase of Mu SGS and deletion derivatives. (A) The relative position of the SGS in the 37-kb Mu genome is shown by the black oval. An expanded region below shows the core of the SGS as a thickened black line, and the sequence is numbered centered on the point of cleavage, as shown by the small vertical line at the core center. To make the derivative SGS sequences, cuts were made at the naturally occurring restriction sites indicated, the ends blunted where necessary, and the sites were reassembled by blunt-end ligation. (B) DNA fragments 200 bp in length (2.5 nM), with the SGS core falling centrally in each molecule, were incubated with increasing amounts of DNA gyrase at 37°C for 30 min. Bound and unbound molecules were then separated on 5% polyacrylamide gels, and the relative amounts were quantified. Binding curves were plotted as shown, with data points and error bars representing the means ± range of two experiments. (C) Linear recombinant pUC19 plasmids (10 nM), carrying the sites indicated above each gel, were incubated either without (−) or with (+) 40 nM gyrase in the presence of enoxacin (Q) (top) or with Ca2+ (Ca) replacing Mg2+ in the reaction buffer (bottom). After 30 min at 37°C, cleavage was induced by the addition of sodium dodecyl sulfate, followed by proteinase K. Products were resolved on a 1% agarose gel, with sizes of DNA markers (in kilobases) indicated to the left of the gels. The bands at 2 kb and 1 kb produced by enoxacin-dependent DNA cleavage arise from cutting at the pUC19 gyrase site lying just upstream of the bla gene promoter, and the doublet at 1.5 kb, shown by arrows where visible, arises from cleavage at the cloned gyrase sites.
DNA gyrase interacts with a significant length of DNA (>100 bp), and generally does not form stable complexes with shorter (<70-bp) fragments (22, 26). We thus inferred the relative contributions each arm of the Mu SGS makes to the overall affinity for the enzyme using the SGSΔ40(R) and SGSΔ45(L) sites. In these binding studies, the fragments used were obtained by PCR in linear form and were 200 bp long, with the SGS cleavage site centered halfway along the molecule. Figure 2B shows that each site bound gyrase less efficiently than the natural SGS, with the right-arm substitution producing the more deleterious effect.
We next examined the quinolone- and Ca2+-dependent cleavage of DNA by the enzyme. The G-segment break can be trapped by quinolones such as enoxacin (9, 49) or when Mg2+ ions in the reaction mixture are replaced by Ca2+ ions (41). In this work, the different sites were again obtained in linear form by PCR and cloned into pUC19. These recombinant plasmids were then cut with the restriction enzyme AhdI, such that the cloned gyrase sites lie close to the center of the resultant linear 3-kb molecule. Gyrase cleavage at the cloned sites then yielded two bands approximately 1.5 kb in size, while cutting at the strong pUC19 site (28, 48) gave two bands 2 kb and 1 kb in length (although note that these latter bands were not observed when Ca2+ was the cleavage agent). Figure 2C shows that the pUC19+SGSΔ45(L) DNA, like the pUC19+SGS construct, was efficiently cut by gyrase at the cloned sites in the presence of either enoxacin or Ca2+, whereas the pUC19+SGSΔ40(R) construct showed no evidence of cutting at the cloned site under either set of conditions. This result may be explicable, at least in part, by the reduced binding due to the substitution of the right arm sequence from bp +40 onwards (Fig. 2B). Nevertheless, the results of this section emphasize the importance of the SGS right-arm sequence beyond bp +40 in directing gyrase to the DNA and supporting efficient cutting by the enzyme.
The SGS right arm enhances the supercoiling efficiency of gyrase.
To assess the effect the Mu SGS right arm on the supercoiling reaction of gyrase, the pUC19 constructs with various gyrase sites were obtained in relaxed form, and equivalent amounts of substrate were incubated with a range of gyrase concentrations. Figure 3 presents the profiles obtained on agarose-chloroquine gels after the different constructs were incubated with gyrase. With the gel system used here, relaxed DNA molecules become positively supercoiled due to the intercalation of chloroquine and were consequently run with increased mobility. As the gyrase supercoiling reaction (i.e., the introduction of negative supercoils) proceeds, molecules with a moderate amount of negative supercoiling show an initial decrease in mobility, up to a point where their migration is at a minimum. Once the level of negative supercoiling is beyond this point, however, the molecules run once more with increasing mobility, until at extreme levels of negative supercoiling the topoisomers form an unresolved band, almost but not quite with the mobility of the relaxed substrate.
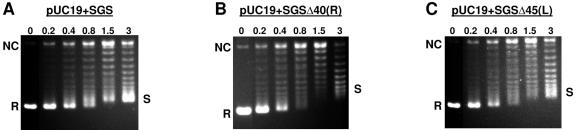
FIG. 3.
Supercoiling of pUC19 plasmids bearing SGS-based gyrase sites. Relaxed pUC19 constructs (10 nM) were incubated at 37°C for 30 min with increasing concentrations of DNA gyrase, with the enzyme concentration (in nanometers) shown above each lane. Samples were resolved on 1% agarose gels with 40-μg/ml chloroquine. R, relaxed substrate; NC, nicked circular DNA; S, negatively supercoiled product.
The pUC19+SGS DNA molecule was a very efficient substrate for DNA gyrase (Fig. 3A) and most of the input material was converted into a highly supercoiled form, at a range of enzyme:DNA ratios. Also, even at levels where the substrate was present in a ≥10-fold molar excess, some fully supercoiled product was still observed: this was visible as a band running just above the relaxed substrate on the gels used here (Fig. 3A; Fig. 4D). This is a hallmark of processive supercoiling by the enzyme (26), where multiple-strand passage events are introduced into the substrate per single binding event. By contrast, the same levels of enzyme were unable to supercoil pUC19+SGSΔ40(R) to the same extent (Fig. 3B). The profile of pUC19+SGSΔ45(L) (Fig. 3C) showed that this site was a more efficient substrate for gyrase than pUC19+SGSΔ40(R), although it was still inferior compared to the wild-type SGS.
FIG. 4.
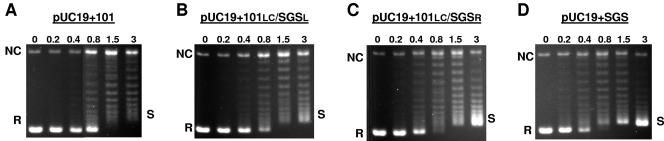
Supercoiling of pUC19 plasmids bearing hybrid gyrase sites. Relaxed pUC19 constructs (10 nM) were incubated at 37°C for 30 min with increasing concentrations of DNA gyrase, with the enzyme concentration (in nanometers) shown above each lane. Samples were resolved on 1% agarose gels containing 40-μg/ml chloroquine. R, relaxed substrate; NC, nicked circular DNA; S, negatively supercoiled product.
Clearly the right arm of the SGS contributes to the efficient use of the site in the supercoiling reaction, but can it also impart efficient supercoiling to other gyrase sites when one arm is replaced by the SGS right arm? This possibility was examined by using some previously constructed hybrid gyrase sites (33) where the right arm of the Mu SGS was fused to the left arm and core of (for example) the major gyrase site from pSC101, yielding a hybrid site designated here as 101LC/SGSR. Similarly, 101LC/SGSL is the designation for the hybrid site made with an equivalent introduction of the SGS left arm. The effects of these sites (when present in pUC19) on the supercoiling activity by gyrase in vitro are shown in Fig. 4. Supercoiling due to the 101LC/SGSR site (Fig. 4C) was visibly improved compared with that due to the pSC101 site alone (Fig. 4A); the reaction profile of the hybrid site approached that of the wild-type SGS (Fig. 4D). In addition, the experiment with the 101LC/SGSL site (Fig. 4B) showed that the left arm of the SGS could improve the ability of the pSC101 site to support gyrase-dependent supercoiling, although this effect was not as dramatic as that seen with the SGS right arm. Taken together, these data establish that the profile and efficiency of gyrase-catalyzed supercoiling can be modified by altering the arm sequence alone of a given gyrase site. In addition, a major determinant of supercoiling efficiency appeared to lie in the Mu SGS right arm distal region starting at bp +40, while the left arm had a less dramatic, but still demonstrable, effect on the enzyme.
The right arm of the gyrase site contained within an E. coli O157 Mu-like prophage confers efficient Mu replication.
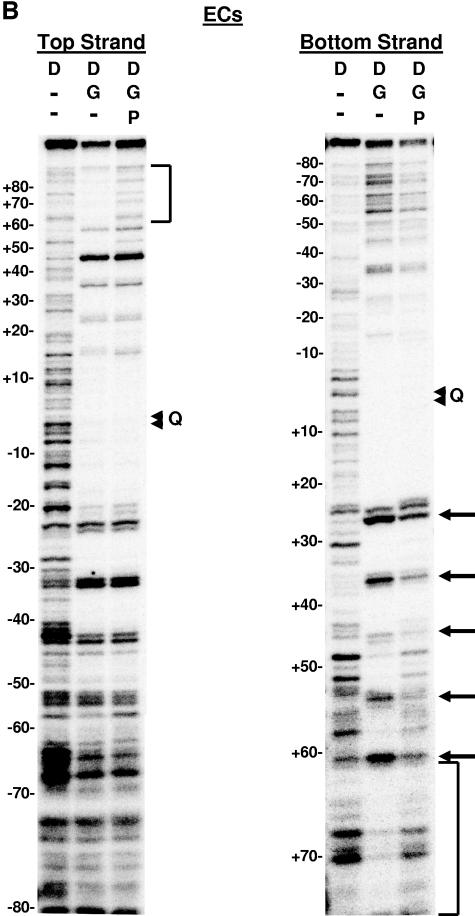
The Mu SGS right-arm sequence starting at bp +40 is further implicated as being important for SGS function, as a virtually identical sequence is found in another gyrase site, ECs, which also supports efficient Mu replication. The ECs site, which originates from the center of the Mu-like prophage resident in the E. coli O157 Sakai genome (11), is the only other naturally occurring site we have found to date that can promote near-normal rates of Mu replication and host lysis when replacing the SGS in a Mu prophage. In addition, the ECs site confers efficient, highly processive supercoiling to the gyrase reaction in vitro (29). The 16-bp stretch of the SGS right arm from bp +42 to bp +57 is also present (with one base change) in an equivalent region of the ECs site right arm, raising the possibility that this region also accounts for the activity of the ECs site in Mu replication.
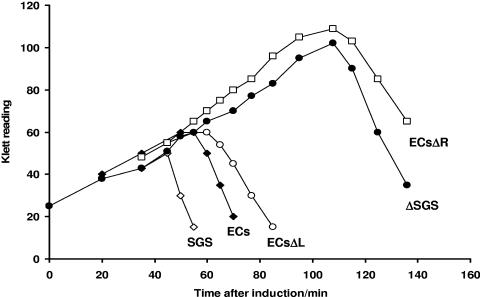
This prediction was tested by performing host lysis assays with chimeric Mu prophages bearing either the intact ECs site or an ECs deletion derivative (Fig. 5). Previous studies showed that prophages with the Mu wild-type SGS or the SGSΔ45(L) sites allowed the same rapid DNA synthesis and lysis after induction; however, the SGSΔ40(R) site resulted in a long delay in lysis and replication equivalent to that observed with a prophage lacking a central gyrase site (33). Substitution of the SGS in a Mu prophage with the wild-type ECs site resulted in only a slight decrease in the kinetics of lysis and DNA replication after induction (29). The data shown in Fig. 5 indicate that replacement of the SGS in a Mu prophage by either the wild-type ECs site or an ECs site with a deletion of the left arm (from bp −43 outwards) allowed very similar kinetics of lysis after induction of the lysogen, while substitution with the ECs carrying a deletion of the right arm (from bp +42 onwards) resulted in the same long delay in lysis observed with the absence of a central gyrase site. This result with ECs derivatives thus mirrors the previous results with Mu SGS derivatives (33) and further underscores the significance of the SGS right-arm distal region in accounting for the effects on Mu replication.
FIG. 5.
Host lysis by chimeric Mu prophage bearing ECs-based sites. The central region of the Mu genome carrying the SGS was replaced by either the wild-type ECs site or a derivative with substituted sequence in the left (ECsΔL) or right (ECsΔR) arms. Host E. coli cells carrying one of these prophages were grown at 30°C, and Mu replication was induced by a temperature shift to 42°C. Wild-type Mu prophage (SGS) or one lacking the entire SGS (ΔSGS) was included as a control. Host lysis was followed by Klett readings of the induced cultures.
The Mu SGS right-arm sequence from bp +40 influences T-segment selection by gyrase.
To determine how the right-arm sequence affects the interaction of the SGS with gyrase in more structural terms and to learn more about the role of the Mu SGS right arm in the context of current models of the gyrase-DNA complex (Fig. 1), we performed footprinting reactions on gyrase bound to the various sites. A previous hydroxyl radical footprinting study on the complex of gyrase with the strong pBR322 site had detected a conformational change and differences in the extent of protection only in the left arm of that site, depending on whether ADPNP was present or not (30). These results implied that the left arm of the pBR322 site forms (or is immediately adjacent to) the T segment. In the present work, we performed analogous footprinting experiments on radiolabeled sites complexed with DNA gyrase, either alone or with ADPNP subsequently added, this time using DNase I as a cleavage agent. Each site examined was obtained by PCR as a 200-bp linear fragment, with the gyrase cleavage site in the center of the sequence. Two sets of footprinting reactions were performed for each site, with DNA labeled at the 5′ end of either the top or bottom strand. In this manner, the top-strand gel for each site resolved most clearly the pattern of DNase I attack on the left-arm sequences from bp −70 to bp −1; similarly, the bottom-strand gel showed the equivalent right-arm data in detail.
The footprints are presented in Fig. 6. The nature of the footprint of gyrase on DNA has been described in detail by many previous studies (8, 15, 16, 30, 39, 53), and we obtained footprints showing many of the same features. Briefly, the core region (G segment) of each site was heavily protected from nuclease attack, compared to the reference latter of bands obtained from treating the DNA sites with DNase I alone. The apparent sizes ranged from 30 to 50 bp, and even for a single site the apparent core size reported by DNase I was somewhat different on each strand. This may reflect a true variation in the size of the core regions themselves or may be due to the particular sequence and structural preferences of DNase I in cleaving DNA. The arm regions extending beyond the core generally exhibited hypersensitive sites with a phased 10-bp spacing, the classic signature of DNA bound on a surface.
FIG. 6.
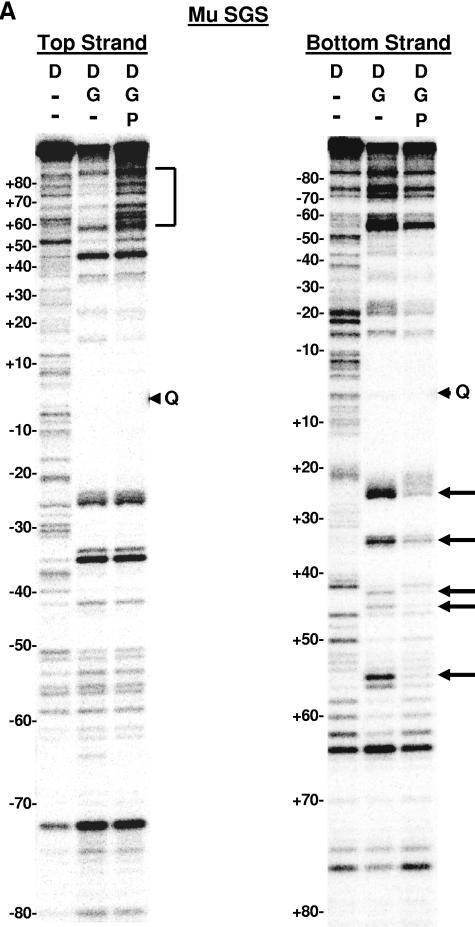
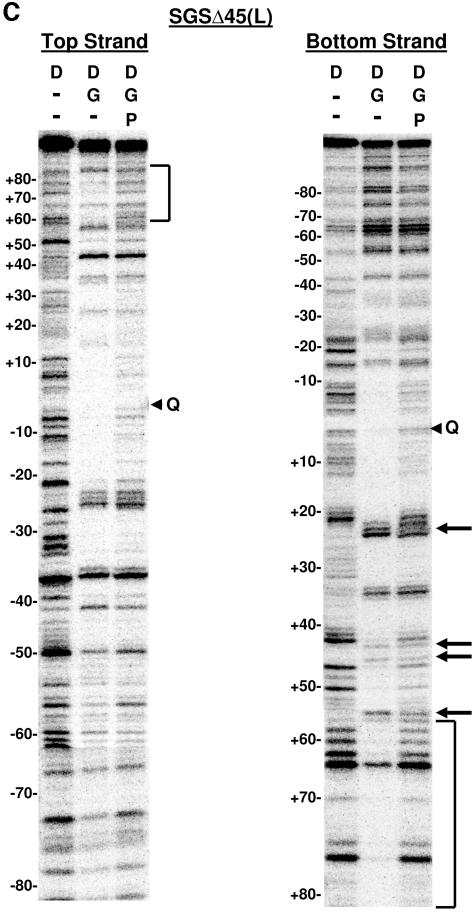
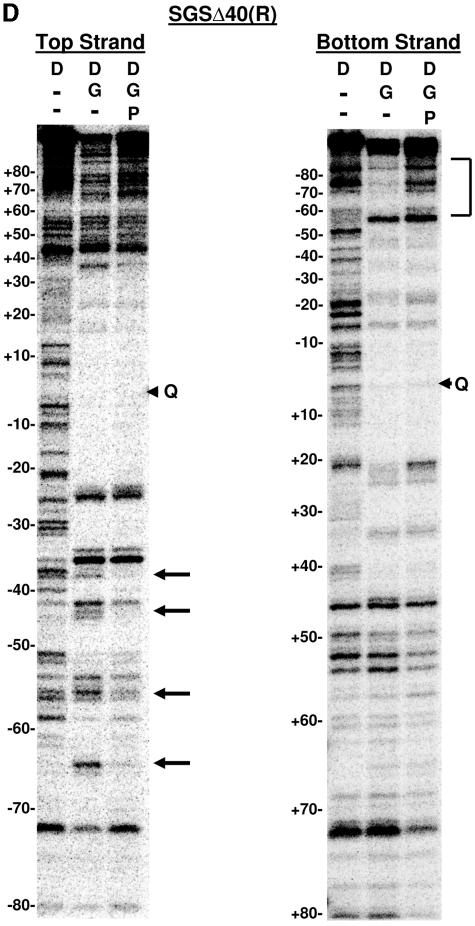
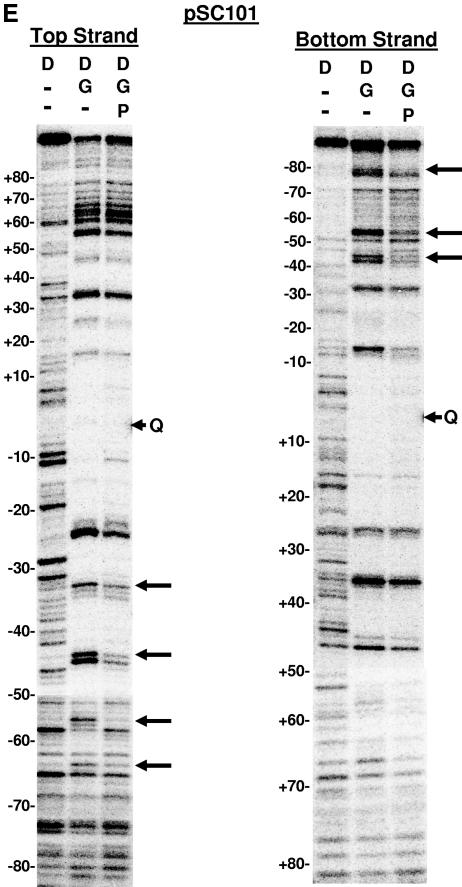
DNase I footprinting on natural and SGS-based gyrase sites. Each site was obtained in the form of a 200-bp linear molecule, radiolabeled at the 5′ end of either the top or bottom strands. Each panel shows the result of a footprinting experiment performed on one of the substrates, as indicated above the gels. The DNA was incubated either alone (D) or in two reactions with DNA gyrase (G) at 37°C for 15 min to allow formation of bound complexes. ADPNP (P) was then added to one enzyme-containing reaction mixture, and all three samples were incubated an additional 5 min. DNase I was then added, and the reactions were quenched after 2 min. DNA samples were purified and separated on 6% denaturing polyacrylamide gels. Arrowheads marked with a Q on the right of each gel show the location of enoxacin-dependent cleavage of each fragment, and the numbers on the left of each panel show the positions of every 10th nucleotide from the center of the gyrase cleavage site, numbered, following the convention used throughout this paper. Vertical brackets highlight regions of additional protection to the DNA arms in the absence of ADPNP that were lost on the addition of nucleotide. Arrows on the right of the gels highlight positions exhibiting increased sensitivity to DNase I in the absence of ADPNP. (A) Mu SGS; (B) ECs; (C) SGSΔ45(L); (D) SGSΔ40(R); (E) pSC101.
With the Mu SGS (Fig. 6A), and perhaps even more clearly with the ECs site (Fig. 6B), the top strand gel revealed a region of DNase I protection in the right arm from approximately bp +60 to bp +80 in the complex without ADPNP, which was no longer apparent once ADPNP was added. In the presence of ADPNP, the pattern of bands in this region on the top-strand gels matched very closely the pattern seen in the lane with DNA alone (albeit with minor variations in cleavage efficiency) (Fig. 6A and B, compare lanes 1 and 3). The fact that the cleavage patterns remain virtually identical suggests the +60 to +80 portion of the right arm was not appreciably bound by gyrase in the presence of ADPNP.
The Mu SGS bottom-strand gel (Fig. 6A) showed that the positions around bp +25, +35, +45, and +56 were more sensitive to DNase I attack in the absence of ADPNP than the reactions with this cofactor present. The ECs footprint (Fig. 6B) also exhibited similar nucleotide-dependent changes to DNase I sensitivity around bp +26, +36, +46, and +55. This 10-bp phasing of hypersensitive sites in each case arises from binding of the DNA on the enzyme surface extending over at least three turns of helix. The same phased hypersensitive sites were also apparent in the region spanning positions +25 to +55 on both top-strand gels, although the addition of ADPNP did not alter the hypersensitivity as much as it did on the bottom-strand gels. There was some protection of the SGS right arm from bp +58 out to bp +80 on the bottom-strand gels in the absence of ADPNP, although this is claim is somewhat hard to justify due to the paucity of bands produced by the DNA alone reaction. This protection was however more apparent on the gels for the ECs site (Fig. 6B) in the equivalent region. Thus, for both the SGS and ECs sites, the extended protection seen on the right arm on the bottom-strand gels in the absence of ADPNP reflected the extended protection to the right arm observed on each top-strand gel.
The patterns of protection to the left arm of both the SGS and ECs sites on the top-strand gels, by contrast, remained essentially unchanged whether ADPNP was present or not. The equivalent regions on the bottom strand again showed a conservation of the pattern of bands (again, with minor variations in efficiency), consistent with the notion that the addition of nucleotide causes no significant alterations to the left arm protection offered by gyrase. The above results show the SGS right arm (as well as the ECs right arm) had major changes in protection induced by nucleotide, while left-arm protection remained relatively constant. By the previous T-segment assignment criteria (30), namely, nucleotide-dependent changes in protection offered by the enzyme and the extension to the protected region on one arm in the absence of ADPNP, these data suggest the SGS right arm preferentially forms the T segment in the supercoiling reaction.
Much the same conclusions can be drawn from SGSΔ45(L) top- and bottom-strand footprints (Fig. 6C). However, the increased protection from bp +60 onwards in the absence of ADPNP was more extensive than in the SGS (compare Fig. 6C, bottom strand, with Fig. 6A), while the modulations to DNase I-hypersensitive sites around bp +25, +35, +45, and +56 were somewhat less prominent than with the wild-type SGS footprint. Also of note, there was a slight loss of protection with this sequence in the core of the footprint once ADPNP was added (Fig. 6C). This is consistent with the T segment held by gyrase in close proximity to the G segment before nucleotide binding (Fig. 1), hence offering additional protection against the nuclease but moving away from the G segment after the strand passage event (12). In summary, the patterns of DNase I footprinting of these three sequences implied that the right arms of each of the ECs, Mu SGS, and SGSΔ45(L) sites form the T segment in the gyrase reaction. In addition, since DNase I cleaves DNA at the minor groove, these patterns imply that the minor grooves with base pairs an integral number of helical turns from the cleavage site, namely, bp +30, +40, +50, etc., point towards the gyrase tetramer.
Significantly, the SGSΔ40(R) fragment (Fig. 6D) did not reproduce these patterns. Instead, the left arm of this deletion site predominantly showed ADPNP-dependent structural changes: specifically, the base pairs around positions −44, −55, and −65 were more sensitive to DNase I without ADPNP, and the region from bp −60 through to −80 was more protected in the absence of nucleotide. The changes to DNase I protection in the right arm of the SGSΔ40(R) site when ADPNP was added were now minimal, certainly much less dramatic than the changes to this arm in either the wild-type SGS or the SGSΔ45(L) sites. (Since the footprinting data report the average features of a large population of molecules, this result is consistent with the notion that gyrase does not necessarily exhibit an absolute preference in selecting an arm to form a T segment.) Taken together, the footprinting data imply that the SGS right arm preferentially forms the T segment in the gyrase reaction but only when the natural sequence from bp +40 onwards is present; without this region of the right arm, sequence gyrase then exhibits a preference for the left arm of the SGS as a T segment.
Two additional naturally occurring gyrase sites, namely, the pBR322 and pSC101 sites, were also subjected to DNase I footprinting. We saw patterns of protection with the pBR322 site by gyrase (data not shown) similar to those reported previously (8, 15, 39), where no clear delineation of a T segment was apparent. With the pSC101 site (Fig. 6E), we observed some minor changes to DNase I hypersensitivity, predominantly with the left arm, although we saw no clear indication of an additional region of protection to the DNA in the absence of ADPNP. It is possible that the pSC101 site may be one where there is no clear preference for a T segment per se. Alternatively, the capture of an extra portion of DNA (as implied by extended DNase I protection in the gyase-DNA complex before ADPNP binding) to form the T segment could be inefficient with this DNA. This possibility is supported by the fact that the pSC101 site, while strongly bound by the enzyme, is inefficient in an in vitro supercoiling reaction (28).
Analysis of isotropic flexibility of the SGS.
To seek an explanation for the patterns of DNase I protection and implied T-segment selection in the gyrase sites studied, we examined the sequences of the sites in detail. A recent extensive analysis of gyrase cleavage sites using both Streptococcus pneumoniae gyrase and topoisomerase IV enzymes has shown that particular bases at certain positions in or near the cleavage site are favored (18). Nevertheless, neither these nor earlier studies using E. coli gyrase (19, 27) revealed specific sequence requirements in the arms of a given site. Instead, we examined how the overall flexibility of the DNA could define in more detail the salient features of an efficient gyrase site and how sequence flexibility might account for the preferences of gyrase in selecting a T segment (as implied by DNase I footprinting).
The flexibility of a DNA sequence can be closely approximated by summing the stacking energies in a given length of sequence (10, 45). We analyzed several gyrase sites in this study, again using 200-bp sequences centered on the point of gyrase cleavage in each case. Our initial analyses of the SGS, ECs, pBR322, and pSC101 sites indicated some common features: namely, a relatively flexible core region, flanked by at least one region of relatively inflexible DNA and at least one, if not both, of the arms consisting of relatively flexible sequences (data not shown). More significantly in the context of this work, however, was that we observed no significant indication that the Mu SGS has a particularly unusual overall flexibility, compared to the patterns observed with the other strong gyrase sites. Nor was the relative overall flexibility of each arm of a gyrase site per se a reliable criterion by which to predict the choice of T segment versus non-T segment (based on the analysis of Fig. 6).
The anisotropic flexibility of a gyrase site is implicated in T-segment selection.
We next extended the analysis of flexibility to consider anisotropic effects. Anisotropic flexibility of a DNA sequence is the ability of the molecule to bend preferentially in a particular direction. A striking example of how the anisotropic flexibility in a DNA sequence influences the structure of a large nucleoprotein complex is the eukaryotic nucleosome. Previous analyses of the sequences that preferentially formed stable nucleosome core particles (6, 44-46) have established that helical phasing of short runs of A and T bases, which create a narrowed minor groove, caused the DNA to wrap around the histone octamer such that the minor grooves of the A/T regions point inwards towards the protein core. This effect was magnified when runs of G and C bases, with a widened minor groove, were also exactly out of phase with the A/T sequences, allowing the G/C minor groove to point outwards away from the histone octamer.
We analyzed the gyrase sequences to determine if similar phasing signals were apparent; the results are summarized in Fig. 7. A very striking pattern was seen in the Mu SGS right arm: base steps such as TA, which are of low stacking energy and high deformability, occurred with an approximately 10-bp phasing over four turns of the DNA helix. These were also interspersed with sequences (usually G-C rich) of high stacking energy and low deformability. The SGS right arm, then, is a sequence amenable to being bent in a particular direction, a feature which would presumably facilitate this arm being wrapped efficiently around the gyrase tetramer (52). Most significantly, three of the G-C sequences correlated well with the strong DNase I cleavages (seen in the absence of ADPNP) at positions +25, +45, and +56 on the top strand, demonstrating that the orientation of the SGS right arm on the surface of the enzyme corresponds to that predicted from a bending of the sequence.
Figure 7 also presents the same analysis of the other gyrase sites used in this study. In the total of eight gyrase site arms from the naturally occurring sites (SGS, ECs, pSC101, and pBR322), anisotropic bending signals were present to a greater or lesser degree. In most cases in the regions spanning bp +20 to +60, the locations of DNase I-hypersensitive sites observed in the absence of ADPNP (Fig. 6) aligned very closely with the sequences likely to point the minor groove away from the enzyme, strongly suggesting that the anisotropic bendability of a DNA sequence is a key parameter in determining the conformation and efficiency of interaction with gyrase.
Additionally, the sequences are presented with the presumptive T segments on the upper line; in all six sites (the four naturally occurring sites and the two Mu SGS derivatives), this arm contained the greater, more continuous length of anisotropic signals than the lower (and, by implication, the non-T-segment) arm. The SGSΔ40(R) substitution had the effect of eliminating a large part of the phased bending signals of the SGS right arm, creating a site not only where the left arm contained the more extensive run of signals by comparison (Fig. 7) but also where the left arm appeared to fulfill the T-segment role (Fig. 6D).
The above analysis suggests to us that gyrase makes the selection of a T segment such that the arm with the greater relative bendability is favored. The SGS right arm contains the longest, most continuous set of bending signals out of all the sequences analyzed. This unusually extended anisotropic signal could well mean that the arm is particularly efficient in facilitating T-segment strand passage by gyrase (and hence efficient supercoiling), accounting in large part for its singular biological and biochemical effects. Detailed analysis of this hypothesis will require the examination of the sequences of additional strong gyrase sites as they become available and examination of the effects of mutationally altering sequences within the SGS right arm.
DISCUSSION
In this paper, we have analyzed the structure and function of the complex of DNA gyrase with various gyrase sites, with particular attention focused on the Mu SGS. We undertook the work presented here with two related goals in mind: to account in molecular terms for the biological function of the SGS and to learn more about how the sequence of a gyrase site influences the selection of an arm to function as a T segment.
SGS and Mu biology.
Our model for the function of the SGS in Mu replication proposes that host gyrase activity at the SGS, located in the center of the Mu genome, results in the extrusion of a Mu-containing domain within the context of the bacterial nucleoid. This in turn brings the prophage ends into close proximity and facilitates synapsis of the genome ends, an obligatory early step in Mu replication and host lysis. We have proposed that the special features of the SGS that allow it to function in this role include highly efficient binding of gyrase and the ability to promote efficient, highly processive supercoiling (28). Genetic analysis showed that the right arm of the SGS was required for rapid Mu DNA replication (33), and now the results of the assays presented here (Fig. 2 to 4) reveal that the right arm of the Mu SGS, from bp +40 onwards, is required for the highly efficient binding, cleavage, and supercoiling by DNA gyrase observed with the SGS. These results thus provide a biochemical rationale for the effect of the SGS right-arm distal region in facilitating efficient Mu replication.
To determine the role of the right arm in the interaction with gyrase, we performed footprinting analyses (Fig. 6) and concluded that the intact SGS right arm likely forms the T segment that is transported through the cleaved G segment during the supercoiling reaction. Interestingly, when the sequence of the SGS right arm beyond bp +40 was replaced by an alternate sequence, the footprinting analysis showed a dramatic change in the patterns of nucleotide-induced protection, strongly suggesting that the left arm of the Mu SGS then fulfilled the T-segment role. This observation implies that the determinants for selection of a T segment by gyrase resides predominantly in the arm sequences, probably in the region at least 40 bp away from the center of the cleavage site.
To help understand the features of the right arm of the SGS that are responsible for its selection as the T segment, an analysis of the sequences of various gyrase sites was undertaken. The analysis indicated that the sites possess a relatively flexible core region, immediately flanked by more-inflexible sequences, followed by at least one arm showing a high degree of flexibility. The overall flexibility of the Mu SGS was not significantly different from the other sites examined, and the relative flexibility of each arm per se was not a clear indicator of which arm of a given gyrase site forms the T segment. Instead, the arm possessing the greater set of phased anisotropic bending signals showed structural characteristics of a T segment in the DNase I analyses. The Mu SGS in particular possesses a more continuous and extended set of such signals than the other gyrase sites analyzed, in that these covered four turns of the DNA helix. Also, substituting this sequence from bp +40 onwards abolished the extent of the phased anisotropic signals and resulted in significant changes to the patterns of DNase I protection, which implied a change in the preference by gyrase for the T segment. These observations lead us to propose that anisotropic flexibility is a key parameter in the selection of a T segment utilized by gyrase in sites that support efficient supercoiling and, in turn, efficient Mu DNA replication.
Implications for the structure of the gyrase-DNA complex and enzyme activity.
Analysis of the isotropic and anisotropic flexibilities of the arms of various gyrase sites also highlights the similarities between the gyrase-DNA complex and the eukaryotic nucleosome. In the latter case, it has been long established that stable nucleosomes are promoted by both isotropically flexible DNA sequences and appropriately phased anisotropic bending signals (51). The most significant difference between the nucleosome and the gyrase-DNA complex, however, is the fact that the latter is a much more dynamic entity. Gyrase functions not just to bind DNA but to catalyze the strand passage event and thus convert topological isomers, predominantly to introduce negative supercoils into DNA. Current models for gyrase supercoiling demand that the T segment undergoes extensive conformational changes during the strand passage event and subsequent supercoiling cycles. Clearly, this would be favored by bendable, flexible sequences. Thus, the sequence-dependent flexibility of a gyrase site is predicted to be important not just for the initial binding of the substrate but also for the ease by which supercoiling is catalyzed at the site in question. The SGS right arm likely fulfils the criteria required by this model by being anisotropically flexible and acting as a T segment in conferring efficient, processive supercoiling by gyrase.
The analysis suggested that one common feature of the efficient gyrase sites was possession of a relatively flexible core region. The two Tyr122 residues of GyrA, the ones which attack the G segment duplex in catalyzing the strand cleavage event, lie too far apart in the crystal structure of the GyrA dimer to readily or simultaneously approach both scissile phosphodiester bonds in the G segment (24). Instead, either the protein, DNA, or both must be distorted if simultaneous cleavage is to occur. Clearly, a relatively flexible core region could facilitate a productive interaction with the enzyme, consistent with the analysis of overall flexibility presented here.
The structure of the CTD of GyrA, which is responsible for the extended interaction with the DNA site and in presenting a T segment to the GyrB clamp, has been solved at atomic resolution (3). The CTD possesses a hexameric β-sheet propeller structure, with four regions of high positive charge on the outer surfaces. These are very likely DNA binding regions, an arrangement which would also be consistent with a recent structural model for the entire GyrA protein (4), which places the GyrA CTD against the N-terminal domain such that these four regions are solvent accessible. Assuming that the G-segment interaction with the tetramer (or more specifically with the GyrA N-terminal domains) initiates complex formation (Fig. 1), each arm would then be free to interact with a separate GyrA CTD. Once wrapped, however, only one of the arms is then presented to the GyrB ATP clamp for strand passage to occur. Although the A2B2 enzyme is a symmetric tetramer, there is an asymmetry in the entire DNA complex in that (for certain sites at least) one arm is predominantly favored as a T segment. The data shown in Fig. 6 and 7 together strongly suggest that the arm with the more extensive set of anisotropic signals will manifest as the preferred T segment. The simplest explanation for this observation is that the arm with the longer bending signals will be more amenable than the other to being wrapped efficiently around one of the available GyrA CTDs and thus being presented to the GyrB clamp to initiate the strand passage event.
In addition, the patterns shown in Fig. 7 imply that the region from around +20 to +60 is likely to be critical in the above regard. Consistent with this, the right-arm sequences that are protected in the presence but not the absence of ADPNP extend from about bp +60 to bp +80 and may correspond to the region that is positioned over the G segment and passed through the cleaved gate. Interestingly, a substitution of the right arm extending from bp +64 to +100 had no deleterious effect on Mu replication (33), which could indicate that the sequence of this protected region is not critical. Rather, any sequence properly positioned by the appropriate wrapping of DNA around the GyrA CTD may suffice.
Finally, it is noteworthy that the SGS right arm, more than all the other arms of sites analyzed, possesses anisotropic signals that remained in phase with the DNase I cleavage sites over four turns of the helix, immediately suggesting that these could promote the wrapping of the DNA around gyrase by favoring simultaneous contact with all four of the GyrA CTD positively charged regions. A notable feature of the SGS right arm is the occurrence of phased TA base steps in this region. These are also characteristic of high-affinity histone octamer binding sites (50, 52), which on nucleosome formation are bent to an extent very similar to that of the DNA wrapped by the GyrA CTD (3, 20). The Mu SGS thus appears to possess those sequence and structural features that enhance the efficiency of interaction with gyrase to a degree where biological consequences (namely, a rapid synapsis of Mu genome ends spaced 40 kb apart) are apparent.
Acknowledgments
We thank Diana Marra-Oram for helpful discussions and insights, P. Higgins for enoxacin, and K. Makino, Osaka University, for a generous gift of E. coli O157:K7 Sakai genomic DNA.
Work in M.L.P.'s laboratory is supported by grant MCB 0090898 from the NSF and work in A.M.'s laboratory is supported by the BBSRC (United Kingdom).
REFERENCES
- 1.Berger, J. M., S. J. Gamblin, S. C. Harrison, and J. C. Wang. 1996. Structure and mechanism of DNA topoisomerase II. Nature 379:225-232. [DOI] [PubMed] [Google Scholar]
- 2.Champoux, J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 3.Corbett, K. D., R. K. Shultzaberger, and J. M. Berger. 2004. The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold. Proc. Natl. Acad. Sci. USA 101:7293-7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costenaro, L., J. G. Grossmann, C. Ebel, and A. Maxwell. 2005. Small-angle X-ray scattering reveals the solution structure of the full-length DNA gyrase a subunit. Structure (Cambridge) 13:287-296. [DOI] [PubMed] [Google Scholar]
- 5.Craigie, R., and K. Mizuuchi. 1986. Role of DNA topology in Mu transposition: mechanism of sensing the relative orientation of two DNA segments. Cell 45:793-800. [DOI] [PubMed] [Google Scholar]
- 6.Drew, H. R., and A. A. Travers. 1985. DNA bending and its relation to nucleosome positioning. J. Mol. Biol. 186:773-790. [DOI] [PubMed] [Google Scholar]
- 7.Fass, D., C. E. Bogden, and J. M. Berger. 1999. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat. Struct. Biol. 6:322-326. [DOI] [PubMed] [Google Scholar]
- 8.Fisher, L. M., K. Mizuuchi, M. H. O'Dea, H. Ohmori, and M. Gellert. 1981. Site-specific interaction of DNA gyrase with DNA. Proc. Natl. Acad. Sci. USA 78:4165-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gellert, M., K. Mizuuchi, M. H. O'Dea, T. Itoh, and J. I. Tomizawa. 1977. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. USA 74:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gotoh, O., and Y. Tagashira. 1981. Locations of frequently opening regions on natural DNAs and their relation to functional loci. Biopolymers 20:1043-1058. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 12.Heddle, J. G., S. Mitelheiser, A. Maxwell, and N. H. Thomson. 2004. Nucleotide binding to DNA gyrase causes loss of DNA wrap. J. Mol. Biol. 337:597-610. [DOI] [PubMed] [Google Scholar]
- 13.Horowitz, D. S., and J. C. Wang. 1987. Mapping the active site tyrosine of Escherichia coli DNA gyrase. J. Biol. Chem. 262:5339-5344. [PubMed] [Google Scholar]
- 14.Kampranis, S. C., and A. Maxwell. 1996. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl. Acad. Sci. USA 93:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirkegaard, K., and J. C. Wang. 1981. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell 23:721-729. [DOI] [PubMed] [Google Scholar]
- 16.Klevan, L., and J. C. Wang. 1980. Deoxyribonucleic acid gyrase-deoxyribonucleic acid complex containing 140 base pairs of deoxyribonucleic acid and an alpha 2 beta 2 protein core. Biochemistry 19:5229-5234. [DOI] [PubMed] [Google Scholar]
- 17.Lee, M. P., M. Sander, and T. Hsieh. 1989. Nuclease protection by Drosophila DNA topoisomerase II. Enzyme/DNA contacts at the strong topoisomerase II cleavage sites. J. Biol. Chem. 264:21779-21787. [PubMed] [Google Scholar]
- 18.Leo, E., K. A. Gould, X. S. Pan, G. Capranico, M. R. Sanderson, M. Palumbo, and L. M. Fisher. 2005. Novel symmetric and asymmetric DNA scission determinants for Streptococcus pneumoniae topoisomerase IV and gyrase are clustered at the DNA breakage site. J. Biol. Chem. 280:14252-14263. [DOI] [PubMed] [Google Scholar]
- 19.Lockshon, D., and D. R. Morris. 1985. Sites of reaction of Escherichia coli DNA gyrase on pBR322 in vivo as revealed by oxolinic acid-induced plasmid linearization. J. Mol. Biol. 181:63-74. [DOI] [PubMed] [Google Scholar]
- 20.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 21.Maxwell, A. 1996. Protein gates in DNA topoisomerase II. Nat. Struct. Biol. 3:109-112. [DOI] [PubMed] [Google Scholar]
- 22.Maxwell, A., and M. Gellert. 1984. The DNA dependence of the ATPase activity of DNA gyrase. J. Biol. Chem. 259:14472-14480. [PubMed] [Google Scholar]
- 23.Maxwell, A., and A. J. Howells. 1999. Overexpression and purification of bacterial DNA gyrase. Methods Mol. Biol. 94:135-144. [DOI] [PubMed] [Google Scholar]
- 24.Morais Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 25.Morgan, G. J., G. F. Hatfull, S. Casjens, and R. W. Hendrix. 2002. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 317:337-359. [DOI] [PubMed] [Google Scholar]
- 26.Morrison, A., N. P. Higgins, and N. R. Cozzarelli. 1980. Interaction between DNA gyrase and its cleavage site on DNA. J. Biol. Chem. 255:2211-2219. [PubMed] [Google Scholar]
- 27.O'Connor, M. B., and M. H. Malamy. 1985. Mapping of DNA gyrase cleavage sites in vivo oxolinic acid induced cleavages in plasmid pBR322. J. Mol. Biol. 181:545-550. [DOI] [PubMed] [Google Scholar]
- 28.Oram, M., A. J. Howells, A. Maxwell, and M. L. Pato. 2003. A biochemical analysis of the interaction of DNA gyrase with the bacteriophage Mu, pSC101 and pBR322 strong gyrase sites: the role of DNA sequence in modulating gyrase supercoiling and biological activity. Mol. Microbiol. 50:333-347. [DOI] [PubMed] [Google Scholar]
- 29.Oram, M., and M. L. Pato. 2004. Mu-like prophage strong gyrase site sequences: analysis of properties required for promoting efficient Mu DNA replication. J. Bacteriol. 186:4575-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orphanides, G., and A. Maxwell. 1994. Evidence for a conformational change in the DNA gyrase-DNA complex from hydroxyl radical footprinting. Nucleic Acids Res. 22:1567-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pathania, S., M. Jayaram, and R. M. Harshey. 2002. Path of DNA within the Mu transpososome. Transposase interactions bridging two Mu ends and the enhancer trap five DNA supercoils. Cell 109:425-436. [DOI] [PubMed] [Google Scholar]
- 32.Pato, M. L. 1994. Central location of the Mu strong gyrase binding site is obligatory for optimal rates of replicative transposition. Proc. Natl. Acad. Sci. USA 91:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pato, M. L., and M. Banerjee. 2000. Genetic analysis of the strong gyrase site (SGS) of bacteriophage Mu: localization of determinants required for promoting Mu replication. Mol. Microbiol. 37:800-810. [DOI] [PubMed] [Google Scholar]
- 34.Pato, M. L., and M. Banerjee. 1996. The Mu strong gyrase-binding site promotes efficient synapsis of the prophage termini. Mol. Microbiol. 22:283-292. [DOI] [PubMed] [Google Scholar]
- 35.Pato, M. L., and M. Banerjee. 1999. Replacement of the bacteriophage Mu strong gyrase site and effect on Mu DNA replication. J. Bacteriol. 181:5783-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pato, M. L., M. M. Howe, and N. P. Higgins. 1990. A DNA gyrase-binding site at the center of the bacteriophage Mu genome is required for efficient replicative transposition. Proc. Natl. Acad. Sci. USA 87:8716-8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng, H., and K. J. Marians. 1995. The interaction of Escherichia coli topoisomerase IV with DNA. J. Biol. Chem. 270:25286-25290. [DOI] [PubMed] [Google Scholar]
- 38.Protozanova, E., P. Yakovchuk, and M. D. Frank-Kamenetskii. 2004. Stacked-unstacked equilibrium at the nick site of DNA. J. Mol. Biol. 342:775-785. [DOI] [PubMed] [Google Scholar]
- 39.Rau, D. C., M. Gellert, F. Thoma, and A. Maxwell. 1987. Structure of the DNA gyrase-DNA complex as revealed by transient electric dichroism. J. Mol. Biol. 193:555-569. [DOI] [PubMed] [Google Scholar]
- 40.Reece, R. J., and A. Maxwell. 1991. DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26:335-375. [DOI] [PubMed] [Google Scholar]
- 41.Reece, R. J., and A. Maxwell. 1989. Tryptic fragments of the Escherichia coli DNA gyrase A protein. J. Biol. Chem. 264:19648-19653. [PubMed] [Google Scholar]
- 42.Resibois, A., M. Pato, P. Higgins, and A. Toussaint. 1984. Replication of bacteriophage Mu and its mini-Mu derivatives. Adv. Exp. Med. Biol. 179:69-76. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Satchwell, S. C., H. R. Drew, and A. A. Travers. 1986. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 191:659-675. [DOI] [PubMed] [Google Scholar]
- 45.Satchwell, S. C., and A. A. Travers. 1989. Asymmetry and polarity of nucleosomes in chicken erythrocyte chromatin. EMBO J. 8:229-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shrader, T. E., and D. M. Crothers. 1989. Artificial nucleosome positioning sequences. Proc. Natl. Acad. Sci. USA 86:7418-7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sokolsky, T. D., and T. A. Baker. 2003. DNA gyrase requirements distinguish the alternate pathways of Mu transposition. Mol. Microbiol. 47:397-409. [DOI] [PubMed] [Google Scholar]
- 48.Stellwagen, N., C. Gelfi, and P. G. Righetti. 2002. The use of gel and capillary electrophoresis to investigate some of the fundamental physical properties of DNA. Electrophoresis 23:167-175. [DOI] [PubMed] [Google Scholar]
- 49.Sugino, A., C. L. Peebles, K. N. Kreuzer, and N. R. Cozzarelli. 1977. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. USA 74:4767-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thastrom, A., L. M. Bingham, and J. Widom. 2004. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J. Mol. Biol. 338:695-709. [DOI] [PubMed] [Google Scholar]
- 51.Travers, A. A. 2004. The structural basis of DNA flexibility. Philos. Trans. A Math Phys. Eng. Sci. 362:1423-1438. [DOI] [PubMed] [Google Scholar]
- 52.Virstedt, J., T. Berge, R. M. Henderson, M. J. Waring, and A. A. Travers. 2004. The influence of DNA stiffness upon nucleosome formation. J. Struct. Biol. 148:66-85. [DOI] [PubMed] [Google Scholar]
- 53.Wahle, E., and A. Kornberg. 1988. The partition locus of plasmid pSC101 is a specific binding site for DNA gyrase. EMBO J. 7:1889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wall, M. K., L. A. Mitchenall, and A. Maxwell. 2004. Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc. Natl. Acad. Sci. USA 101:7821-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, J. C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3:430-440. [DOI] [PubMed] [Google Scholar]
- 56.Wang, J. C. 1998. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 31:107-144. [DOI] [PubMed] [Google Scholar]
- 57.Wigley, D. B., G. J. Davies, E. J. Dodson, A. Maxwell, and G. Dodson. 1991. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351:624-629. [DOI] [PubMed] [Google Scholar]