Abstract
Corynebacterium glutamicum contains genes for 13 two-component signal transduction systems. In order to test for their essentiality and involvement in the adaptive response to phosphate (Pi) starvation, a set of 12 deletion mutants was constructed. One of the mutants was specifically impaired in its ability to grow under Pi limitation, and therefore the genes lacking in this strain were named phoS (encoding the sensor kinase) and phoR (encoding the response regulator). DNA microarray analyses with the C. glutamicum wild type and the ΔphoRS mutant supported a role for the PhoRS system in the adaptation to Pi starvation. In contrast to the wild type, the ΔphoRS mutant did not induce the known Pi starvation-inducible (psi) genes within 1 hour after a shift from Pi excess to Pi limitation, except for the pstSCAB operon, which was still partially induced. This indicates an activator function for PhoR and the existence of at least one additional regulator of the pst operon. Primer extension analysis of selected psi genes (pstS, ugpA, phoR, ushA, and nucH) confirmed the microarray data and provided evidence for positive autoregulation of the phoRS genes.
Phosphorus (P) is an essential nutrient for all cells and required, e.g., for the biosynthesis of nucleotides, DNA, and RNA and in addition for the functional regulation of protein activity by phosphorylation. The common phosphorus source is inorganic phosphate (Pi), and cells have developed mechanisms for the acquisition, assimilation, and storage of phosphate. Under phosphate starvation, many bacteria induce the synthesis of proteins that enable them to use the limiting phosphate resources more efficiently and to make alternative phosphorus sources accessible. The corresponding genes are collectively named Pi starvation-inducible genes, or psi genes. The phosphate starvation response, in particular its regulation, has been most carefully studied in Escherichia coli (33) and Bacillus subtilis (11). In both species, two-component signal transduction systems consisting of a histidine kinase and a response regulator play a prominent role.
In E. coli, induction of the Pi starvation genes is dependent on the PhoR-PhoB two-component system. Under Pi limitation, the histidine kinase PhoR phosphorylates the response regulator PhoB, and PhoB∼P in turn activates transcription of at least 31 genes, which form the Pho regulon (33). The genes include the phoBR operon; the pstSCAB-phoU operon, encoding an ABC transporter for high-affinity Pi uptake and a regulatory protein; the ugpBAECQ operon, encoding an sn-glycerol 3-phosphate ABC uptake system and glycerophosphoryl diester phosphodiesterase; the phoA-psiF operon, encoding alkaline phosphatase and a protein of unknown function; phoE, encoding an anion-specific porin; phoH, encoding an ATP-binding protein of unknown function; and the phnCDEFGHIJKLMNOP operon, encoding proteins involved in the uptake of phosphonates and their degradation via the C-P lyase pathway. Thus, when Pi is scarce, E. coli takes up Pi by an ATP-driven high-affinity transport system, mobilizes Pi outside the cytoplasm by phosphatases and esterases, and forms proteins for the uptake and degradation of organophosphates and phosphonates.
In B. subtilis, the response to Pi starvation is more complex, since it involves the induction both of a specific Pho regulon and of the σB-dependent general stress response. The Pho regulon is controlled by a two-component system composed of the histidine kinase PhoR and the response regulator PhoP. It includes the phoPR operon; phoA and phoB for two alkaline phosphatases; phoD for an alkaline phosphatase/phosphodiesterase with a putative role in cell wall teichoic-acid turnover; the pstSACB1B2 operon; the tuaABCDEFGH operon, encoding proteins which are responsible for the synthesis of teichuronic acid, which replaces the teichoic acid in the cell walls of phosphate-starved cells; glpQ, encoding glycerophosphoryl diester phosphodiesterase; and ydhF, encoding a lipoprotein (2, 11). The tagAB and tagDEF operons, which encode protein involved in teichoic acid biosynthesis, are repressed by PhoP under Pi starvation (20). Besides PhoR-PhoP, the ResD-ResE two-component system, the response regulator Spo0A, and the AbrB regulator are also involved in the control of Pho regulon genes (11).
Corynebacterium glutamicum is a gram-positive bacterium belonging to the order Actinomycetales and has gained considerable interest because of its use in the large-scale biotechnological production of l-glutamate and l-lysine (6). Phosphorus constitutes 1.5% to 2.1% of the cell dry weight of C. glutamicum (18), part of which is present as polyphosphate (16, 24). In a previous study, we analyzed the Pi starvation response of C. glutamicum by using whole-genome DNA microarrays (12). The comparison of the mRNA profiles before and at different times after a shift from Pi excess to Pi starvation led to the identification of a group of genes that are specifically required to cope with a limited Pi supply. This group includes the pstSCAB operon, encoding an ABC transporter for high-affinity Pi uptake; the ugpAEBC operon, encoding an ABC transporter for the uptake of glycerol 3-phosphate; glpQ1, encoding a glycerophosphoryl diester phosphodiesterase; ushA, encoding a secreted enzyme with UDP sugar hydrolase and 5′-nucleotidase activity (27); nucH, encoding a putative secreted nuclease which possibly serves a role in liberating Pi from extracellular nucleic acids; NCgl2959/Cg3393, which may encode a cell wall-associated phosphatase (34); phoH1, encoding an ATPase of unknown function; and the pctABCD operon, encoding an ABC transport system which might be involved in the uptake of a yet-unknown phosphorus-containing compound (12).
In parallel to the elucidation of the Pi starvation stimulon of C. glutamicum, we initiated studies aimed at the identification of the regulators controlling the Pi starvation response. Evidence derived from other bacteria showed that two-component regulatory systems are prime candidates for this function. Here we present evidence that one of the 13 two-component systems of C. glutamicum, named PhoS-PhoR, is involved in the adaptation of C. glutamicum to limiting Pi concentrations.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains, plasmids, and oligonucleotides used or constructed for this work are listed in Tables S1, S2, and S3, respectively (see the supplemental material). For the construction of plasmids, E. coli DH5α was used as a host and routinely grown aerobically at 37°C on a rotary shaker (120 rpm) in LB medium (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl) or on LB agar plates (LB medium with 1.5% [wt/vol] agar). C. glutamicum ATCC 13032 (1) and its derivatives were cultivated aerobically on a rotary shaker (120 rpm) at 30°C either in LB medium, in brain heart infusion medium supplemented with 0.5 M sucrose, in CGIII medium (21), or in CGXII minimal medium (15) containing 30 mg/liter protocatechuic acid as an iron chelator and 40 g/liter glucose as a carbon and energy source. CGXII medium contains 13 mM phosphate in the forms of KH2PO4 and K2HPO4 as its sole phosphorus source. In order to obtain phosphate-limited conditions (0.065 mM), the concentration of these two compounds was reduced from 1 g/liter to 0.005 g/liter. If appropriate, kanamycin was added to a final concentration of 25 μg/ml (C. glutamicum) or 50 μg/ml (E. coli).
For an analysis of the influence of different phosphate concentrations on the growth of C. glutamicum strains, they were first grown overnight in CGIII medium. After being washed, the cells were cultured for 24 h in CGXII medium under Pi-limiting conditions (0.065 mM) and then inoculated into CGXII medium containing either 0.065 mM phosphate (Pi limitation) or 13 mM phosphate (Pi excess). Growth was followed by a measurement of the optical density at 600 nm (OD600).
For an analysis of the response of C. glutamicum to a shift from Pi excess to Pi limitation by DNA microarray or primer extension analysis, cells of the wild type or the ΔphoRS mutant were first precultured in CGIII medium and then grown for 24 h in CGXII medium under Pi-sufficient conditions and finally inoculated into the same medium to an initial OD600 of 0.6. Exponentially growing cells from this Pi-sufficient culture (OD600, 4 to 5) were harvested and washed. RNA was prepared from one aliquot, whereas the other aliquot was used to inoculate cultures with a medium containing a limiting Pi concentration (0.065 mM). RNA was prepared 10, 30, 60, or 90 min after the Pi downshift.
Construction of C. glutamicum deletion mutants.
Deletion mutants of C. glutamicum lacking citAB, cgtSR1, cgtRS2, cgtRS3, cgtRS5, cgtRS6, cgtSR7, cgtSR8, cgtRS9, cgtSR10, or cgtSR11 were constructed via a two-step homologous-recombination procedure involving crossover PCR (19) and the suicide vector pK19mobsacB (28) as described previously (23). The deletions were verified by PCR using primers annealing outside the regions involved in recombination (e.g., oligonucleotides cgtRS3-out-fw and cgtRS3-out-rv). In the cases of the ΔphoRS (=ΔcgtRS3), ΔcitAB, and ΔcgtRS9 mutants, Southern blot analysis was also performed (23) and confirmed the deletions (data not shown).
Construction of plasmid pEKEx2-phoRS.
The phoRS coding sequence was amplified from the chromosomal DNA of the C. glutamicum wild type by PCR using the oligonucleotides phoRS-SbfI-fw and phoRS-KpnI-rv (Table S3 in the supplemental material) and the Expand High Fidelity PCR system (Roche Diagnostics, Mannheim, Germany). The primer phoRS-SbfI-fw introduces an SbfI restriction site 40 bp upstream of the phoR start codon. The primer phoRS-KpnI-rv introduces a KpnI restriction site after the stop codon of phoS and adds eight codons (WSHPQFEK) before the stop codon, leading to a PhoS protein with a carboxy-terminal StrepTag-II (29). The resulting PCR product (2,263 bp) was cleaved with SbfI and KpnI, gel purified using the QIAEX kit (QIAGEN, Hilden, Germany), and cloned into the E. coli-C. glutamicum shuttle vector pEKEx2 (7) cut with the same enzymes, resulting in plasmid pEKEx2-phoRS. DNA sequence analysis confirmed that the cloned phoRS genes were identical to the published wild-type sequence (13).
Preparation of total RNA for DNA microarray and primer extension experiments.
RNA from C. glutamicum was prepared from 20-ml culture aliquots using the RNeasy kit (QIAGEN, Hilden, Germany) with on-column DNase I treatment as described previously (22). Isolated RNA samples were checked for purity by denaturing formaldehyde agarose gel electrophoresis, quantified by UV spectrophotometry, and stored at −70°C until use.
DNA microarray analysis.
Whole-genome DNA microarrays for C. glutamicum were generated as described previously (12, 17). Identical amounts (20 to 25 μg) of total RNA were used for random hexamer-primed synthesis of fluorescently labeled cDNA by reverse transcription with Superscript II (Invitrogen, Karlsruhe, Germany) and the fluorescent nucleotide analogues FluoroLink Cy3-dUTP (green) and Cy5-dUTP (red) (Amersham Pharmacia, Little Chalfont, United Kingdom) as described previously (17). The labeled cDNA probes were purified and concentrated by using Microcon YM-30 filter units (Millipore, Bedford, Mass.). Subsequently, the mixed Cy3- and Cy5-labeled cDNAs containing 1.2 μg poly(A)/ml (Sigma, Munich, Germany) as a competitor, 30 mM HEPES, and 0.3% sodium dodecyl sulfate in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) were hybridized to whole-genome arrays in a humid chamber for 5 to 16 h at 65°C. After hybridization, the arrays were washed in a mixture of 1× SSC and 0.03% sodium dodecyl sulfate and finally in 0.05× SSC. Immediately after stringent washing, the fluorescence intensities at 635 and 532 nm were determined using a GenePix 4000 laser scanner (Axon, Inc., Union City, Calif.), and the images were processed by using GenePix 3.0 software. Data were normalized to the average ratio for C. glutamicum genomic DNA. The normalized ratio of median fluorescence was taken to reflect the relative RNA abundance for spots with green or red fluorescent signals that were at least threefold greater than the median fluorescence background signal. For statistical analysis of the gene expression data, P values for the independent replicate experiments were calculated based on the Student t test by using log-transformed fluorescence ratios for individual genes on the one hand and for genomic DNA on the other hand. In a first series of experiments (performed in triplicate), the mRNA levels of the ΔphoRS mutant 10, 30, and 60 min after a shift from Pi excess (13 mM) to Pi limitation (0.065 mM) were compared to the mRNA levels before the shift (zero time). In a second series of experiments, the mRNA levels of the wild type were compared with the mRNA levels of the ΔphoRS mutant before and 60 min after a shift from Pi excess to Pi limitation.
Primer extension analysis.
Nonradioactive primer extension analysis of the genes pstS, ugpA, phoR, ushA, and nucH was performed as described previously (8, 9) using IRD800-labeled oligonucleotides (Table S3 in the supplemental material) and 20 μg isolated RNA as a template. The lengths of the primer extension products were determined by running the four lanes of a DNA-sequencing reaction mixture set up using the same oligonucleotide as that usedfor reverse transcription alongside the primer extension products. The templates for DNA sequencing were obtained by PCR using the oligonucleotide pairspstS_promreg_fw/pstS_promreg_rv, ugpA_promreg_ fw/ugpA_promreg_rv,phoR_promreg_fw/phoR_promreg_rv, ushA1/ushA2, and nucH1/nucH2 (for sequences, see Table S3 in the supplemental material).
RESULTS
In silico and deletion analyses of C. glutamicum two-component systems.
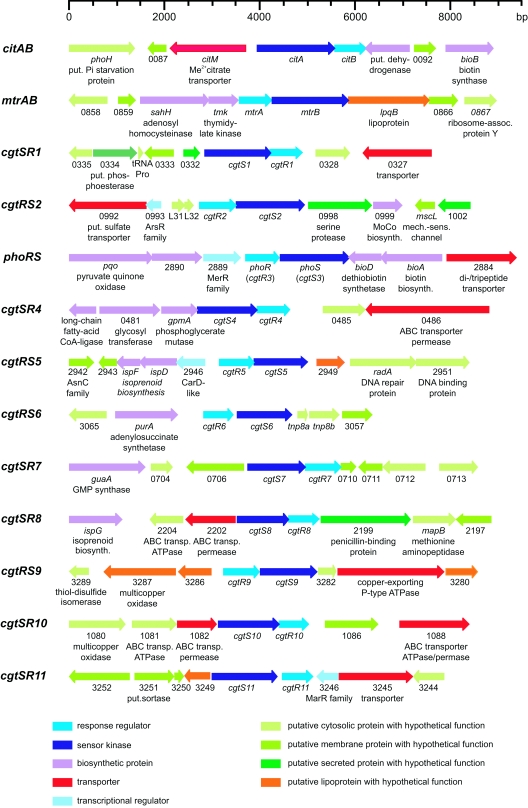
During the annotation of the C. glutamicum genome sequence (13), genes encoding 13 sensor kinases and 13 response regulators were identified. In Fig. 1, maps of the corresponding genomic loci and the designations given to the corresponding genes are shown. As is evident from this figure, each sensor kinase gene is paired with a response regulator gene, suggesting that the corresponding proteins form the cognate partners of a typical two-component signal transduction system. No orphan sensor kinase or response regulator genes are present in the genome. Bioinformatic analysis indicated that each of the 13 sensor kinases possesses at least one transmembrane helix, indicating that all are integral membrane proteins (Table S4 in the supplemental material). All sensor kinases contained the two domains that are characteristic for these proteins, i.e., the histidine kinase acceptor domain containing the phosphorylated histidine residue and the ATP-binding domain. Analysis of the response regulators predicted that all contain the characteristic amino-terminal receiver domain and a carboxy-terminal DNA-binding domain. Thus, all two-component systems of C. glutamicum appear to have roles in gene regulation.
FIG. 1.
Maps of the 13 C. glutamicum genome loci containing genes for two-component signal transduction systems. The different colors indicate different functions of the encoded proteins or different localizations of proteins with unknown functions. The numbers are derived from GenBank accession no. BX927147 (13). put., putative; Me, metal; assoc., associated; biosynth., biosynthesis; mech.-sens., mechano-sensitive; CoA, coenzyme A; transp., transporter.
Except for the CitAB system, which belongs to a family of two-component systems controlling the uptake and metabolism of citrate or dicarboxylates (5, 10, 14, 26), obvious predictions of the stimuli recognized by the sensor kinases and of the target genes regulated by the response regulators were not possible. In a recent analysis of the MtrAB system, we provided experimental evidence that this system is involved in the regulation of genes involved in cell wall metabolism and osmoregulation (22).
For functional analysis of the two-component systems, deletion mutants, each lacking one pair of sensor kinase/response regulator genes, were constructed using a previously established method (23). In these deletion mutants, only the first six codons of the promoter-proximal gene and the last 12 codons of the promoter-distal gene are still present, whereas the intervening region is replaced by a 21-bp sequence tag. For 12 two-component systems, the corresponding deletion strains could be obtained, showing that these genes are not essential (Table S1 in the supplemental material). In the case of the cgtSR4 genes, however, no deletion mutants were obtained, suggesting that cgtS4 and/or cgtR4 is essential.
Screening of two-component deletion mutants for growth under Pi limitation.
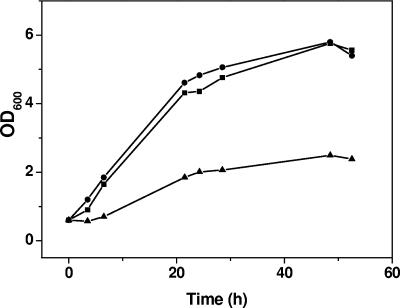
In order to determine whether one of the C. glutamicum two-component systems is involved in the phosphate starvation response, the set of 12 deletion mutants was tested for growth in glucose minimal medium containing different Pi concentrations. One deletion strain (ΔcgtRS3 mutant) showed a growth defect under Pi-limiting conditions (0.065 mM) but not under Pi excess (13 mM), indicating that the cgtRS3 genes might have a specific function in the Pi starvation response. When the ΔcgtRS3 mutant was transformed with a plasmid containing the cgtRS3 genes (pEXEx2-phoRS), the growth defect under Pi limitation was abolished and the complemented strain grew almost like the wild type (Fig. 2). This experiment confirmed that the growth phenotype of the mutant was caused by the deletion of the cgtRS3 genes. In this work the corresponding genes were renamed phoR for phosphate response regulator and phoS for phosphate sensor kinase and the mutant was renamed the ΔphoRS mutant. The coding regions of phoR and phoS are separated by 8 bp, suggesting cotranscription of the two genes.
FIG. 2.
Growth of the C. glutamicum 13032/pEKEx2 (▪), ΔphoRS/pEKEx2 (▴), and ΔphoRS/pEKEx2-phoRS (•) strains in CGXII minimum medium with 40 g/liter glucose and 0.065 mM phosphate. Cells were precultured in CGXII medium with 0.065 mM phosphate for 24 h.
The sensor kinase encoded by PhoS is composed of 485 amino acids (52,365 Da). It contains two putative transmembrane helices extending from residues 44 to 64 and 184 to 200, enclosing an extracytoplasmic domain of 120 amino acid residues. The carboxy-terminal cytoplasmic portion of PhoS contains a HAMP domain (3) extending from residues 185 to 255, a histidine kinase A acceptor domain extending from residues 266 to 330, and a DNA gyrase B/Hsp90-like ATPase domain extending from residues 373 to 485. The histidine residue which presumably is phosphorylated is located at position 276. The response regulator encoded by PhoR is composed of 235 amino acids (26,350 Da) and contains two conserved domains, i.e., an amino-terminal receiver domain extending from residues 9 to 128 and a carboxy-terminal DNA binding domain extending from residues 156 to 230. The aspartate residue which presumably is phosphorylated by PhoS is located at position 59.
Gene expression changes in the ΔphoRS mutant after a shift from Pi-sufficient to Pi-limiting conditions.
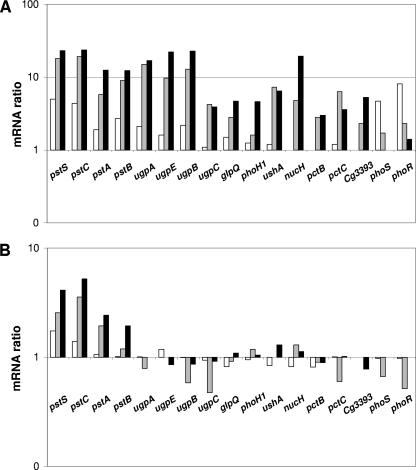
The growth defect under Pi-limiting conditions indicated that the ΔphoRS mutant is impaired in its ability to induce the phosphate starvation genes (see the introduction). Therefore, the changes in gene expression occurring 10, 30, and 60 min after a shift from Pi excess to Pi starvation were analyzed as described previously for the wild type (12) and as outlined in Materials and Methods. Overall, 34 and 47 genes of the C. glutamicum ΔphoRS mutant showed a ≥2-fold-increased and a ≥2-fold-decreased mRNA level, respectively, at least at one time point after the shift (Table 1). Except for the pst genes, none of the prominent phosphate starvation-inducible genes, e.g., the ugpAEBC operon, glpQ1, phoH1, ushA, or nucH, were induced either 10, 30, or 60 min after the shift (Fig. 3B). This is in marked contrast to the situation in the wild type (Fig. 3A) and supports a critical role for the PhoRS two-component system in the phosphate starvation response of C. glutamicum. In the case of the pstSCAB operon, a twofold- to fivefold-increased mRNA level was observed within 1 hour after the shift. Since this induction level is lower than in the wild type, the data suggest also that the pst operon is controlled by the PhoRS system.
TABLE 1.
Genes showing ≥2-fold-altered mRNA ratios in the C. glutamicum ΔphoRS mutant 10, 30, or 60 min after a shift from Pi excess to Pi limitation
| C. glutamicum locus tag no. | NCBI no. | Annotationc | Gene | mRNA ratio (after shift/ before shift to Pi-limiting conditions)a
|
||
|---|---|---|---|---|---|---|
| 10 min | 30 min | 60 min | ||||
| cg0160 | NCgl0123 | Hypothetical protein | 0.43b | 0.34 | 0.65 | |
| cg0229 | NCgl0181 | Glutamine 2-oxoglutarate aminotransferase, large subunit | gltB | 1.28 | 1.95 | 2.59b |
| cg0230 | NCgl0182 | Glutamine 2-oxoglutarate aminotransferase, small subunit | gltD | 6.41b | 5.83b | 6.24b |
| cg0310 | NCgl0251 | Catalase | katA | 3.49b | 3.24b | 1.55b |
| cg0464 | NCgl0375 | Copper-exporting P-type ATPase | ctpA | 10.32b | 21.51 | 5.16b |
| cg0467 | NCgl0378 | Put. hemin ABC transporter, periplasmic binding protein | hmuT | 0.42b | 0.40 | 0.63b |
| cg0468 | NCgl0379 | Put. hemin ABC transporter, permease | hmuU | 0.33 | 0.83b | |
| cg0469 | NCgl0380 | Put. hemin ABC transporter, ATPase | hmuV | 0.41b | 0.50 | 0.69 |
| cg0470 | NCgl0381 | Conserved secreted protein | 0.16b | 0.22b | 0.58 | |
| cg0500 | NCgl0405 | Transcriptional regulator, LysR family | 2.07b | 2.74 | 2.21b | |
| cg0569 | NCgl0465 | Cation-transporting P-type ATPase | 8.60b | 4.77 | 2.65b | |
| cg0589 | NCgl0482 | Put. Fe3+ siderophore ABC transporter, ATPase | 0.06b | 0.10b | 0.44 | |
| cg0590 | NCgl0483 | Put. Fe3+ siderophore ABC transporter, permease | 0.22b | 0.12 | 0.51 | |
| cg0591 | NCgl0484 | Put. Fe3+ siderophore ABC transporter, permease | 0.15b | 0.21 | 0.26 | |
| cg0759 | NCgl0628 | Methylcitrate dehydratase | prpD2 | 0.33b | 0.47b | 0.55b |
| cg0760 | NCgl0629 | Methylisocitrate lyase | prpB2 | 0.37b | 0.53b | 0.49b |
| cg0762 | NCgl0630 | Methylcitrate synthase | prpC2 | 0.22b | 0.39 | 0.40 |
| cg0767 | NCgl0635 | Put. siderophore-interacting protein | 0.16b | 0.18 | 0.62 | |
| cg0768 | NCgl0636 | Put. Fe3+ siderophore ABC transporter, ATPase | 0.25b | 0.09 | 0.36 | |
| cg0769 | NCgl0637 | Put. Fe3+ siderophore ABC transporter, permease | 0.16b | 0.09 | 0.23 | |
| cg0771 | NCgl0639 | Put. Fe3+ siderophore ABC transporter, periplasmic binding protein | 0.14b | 0.10 | 0.29 | |
| cg0812 | NCgl0678 | Acetyl/propionyl-CoA carboxylase, beta subunit | dtsR1 | 0.26b | 0.24b | 0.48b |
| cg0921 | NCgl0773 | Put. siderophore-interacting protein | 0.13b | 0.12b | 0.51b | |
| cg0922 | NCgl0774 | Put. Fe3+ siderophore ABC transporter, periplasmic binding protein | 0.11b | 0.12b | 0.27b | |
| cg0924 | NCgl0776 | Put. Fe3+ siderophore ABC transporter, periplasmic- binding protein | 0.07b | 0.10b | 0.18b | |
| cg0926 | NCgl0777 | Put. Fe3+ siderophore ABC transporter, permease | 1.24 | 0.17 | ||
| cg0927 | NCgl0778 | Put. Fe3+ siderophore ABC transporter, permease | 0.24b | 0.28b | 0.38b | |
| cg0928 | NCgl0779 | Put. Fe3+ siderophore ABC transporter, ATPase | 0.65b | 0.12 | 0.82 | |
| cg0957 | NCgl0802 | Fatty acid synthase | fas-1B | 0.46 | 0.16 | 0.27b |
| cg1120 | NCgl0943 | Transcriptional regulator, AraC-family | ripA | 0.21b | 0.20 | 0.42b |
| cg1121 | NCgl0944 | Conserved membrane protein | 0.49b | 0.92 | 1.26 | |
| cg1343 | NCgl1141 | Nitrate reductase, beta subunit | narH | 4.40b | 1.91 | 1.08b |
| cg1344 | NCgl1142 | Nitrate reductase, alpha subunit | narG | 2.26b | 1.87 | 1.13 |
| cg1405 | NCgl1200 | Put. siderophore-interacting protein | 0.34b | 0.17 | 0.46 | |
| cg1412 | NCgl1205 | Sugar ABC transporter, permease | 0.48b | 0.79 | 0.91 | |
| cg1413 | NCgl1206 | Sugar ABC transporter, periplasmic binding protein | 0.40b | 0.78 | 0.54 | |
| cg1418 | NCgl1209 | Put. Fe3+ siderophore periplasmic binding protein | 0.22b | 0.25 | 0.50b | |
| cg1419 | NCgl1210 | Transporter, bile acid:Na+ symporter family (TC 2.A.28) | 0.39b | 0.35 | 0.48 | |
| cg1447 | NCgl1232 | Put. Co/Zn/Cd exporter, cation diffusion facilitator family (TC 2.A.4) | 7.16 | 6.08 | 7.18b | |
| cg1695 | NCgl1444 | Put. DNA binding protein | 3.17b | 2.18 | 2.05b | |
| cg1737 | NCgl1482 | Aconitase | acn | 1.87 | 3.69b | 2.25b |
| cg1785 | NCgl1521 | Ammonia permease | amt | 1.77 | 2.07b | 4.46 |
| cg1930 | NCgl1646 | Put. secreted trypsin-like serine protease | 0.14b | 0.21b | 0.51b | |
| cg2118 | NCgl1859 | Transcriptional regulator of sugar metabolism, DeoR family | 4.08b | 0.93 | 0.69b | |
| cg2120 | NCgl1861 | Enzyme II of fructose phosphotransferase system | ptsF | 3.64b | 0.66 | 0.41b |
| cg2181 | NCgl1915 | Oligopeptide ABC transporter, periplasmic binding protein | oppA | 0.68b | 0.53b | 0.31b |
| cg2183 | NCgl1917 | Oligopeptide ABC transporter, permease | oppC | 0.71 | 0.58 | 0.44b |
| cg2234 | NCgl1959 | Put. Fe3+ siderophore binding protein | 0.26b | 0.24 | 0.55b | |
| cg2283 | NCgl2001 | Conserved hypothetical protein | 0.33b | 0.36 | 0.62 | |
| cg2422 | NCgl2127 | Lipoate-protein ligase | lipB | 1.99b | 2.17 | 1.46b |
| cg2423 | NCgl2128 | Lipoic acid synthase | lipA | 1.65b | 2.12b | 1.26b |
| cg2726 | NCgl2393 | Put. membrane protein | 15.60b | 3.46 | 4.46b | |
| cg2743 | NCgl2409 | Fatty acid synthase | fas-IA | 0.42b | 0.39 | 0.46b |
| cg2777 | NCgl2434 | Put. membrane protein | 0.47b | 0.37 | 0.55b | |
| cg2782 | NCgl2439 | Ferritin | ftn | 18.82b | 17.89 | 12.14b |
| cg2843 | NCgl2483 | Phosphate ABC transporter, ATPase | pstB | 1.02b | 1.19 | 1.94b |
| cg2844 | NCgl2484 | Phosphate ABC transporter, permease | pstA | 1.06b | 1.93 | 2.43b |
| cg2845 | NCgl2485 | Phosphate ABC transporter, permease | pstC | 1.39b | 3.55 | 5.21b |
| cg2846 | NCgl2486 | Phosphate ABC transporter, binding protein | pstS | 1.74b | 2.55 | 4.10b |
| cg2925 | NCgl2553 | Enzyme II of sucrose phosphotransferase system | ptsS | 3.83b | 0.93b | 0.56b |
| cg3116 | NCgl2717 | Phosphoadenosine phosphosulfate reductase | cysH | 2.53b | 0.95 | |
| cg3118 | NCgl2718 | Sulfite reductase (hemoprotein) | cysI | 3.17b | 1.41 | 0.79 |
| cg3132 | NCgl2731 | Put. membrane protein | 2.53b | 1.00 | 1.13b | |
| cg3195 | NCgl2787 | Flavin-containing monooxygenase | 0.40b | 0.53 | 0.88 | |
| cg3227 | NCgl2817 | Put. l-lactate dehydrogenase (quinone) | lldD | 18.31b | 11.82 | 1.32 |
| cg3281 | NCgl2859 | Cation-transporting P-type ATPase | 3.77b | 2.03 | 1.47b | |
| cg3282 | NCgl2860 | Put. secreted lipoprotein | 14.95b | 4.27 | 4.54 | |
| cg3286 | NCgl2864 | Put. secreted lipoprotein | 3.54b | 2.44 | 2.67b | |
| cg3287 | NCgl2865 | Put. secreted multicopper oxidase | 4.13b | 2.81 | 1.57 | |
| cg3303 | NCgl2877 | Transcriptional regulator, PadR-like family | 7.37b | 4.16 | 4.18b | |
| cg3327 | NCgl2897 | Starvation-induced DNA protection protein | dps | 3.76b | 3.51 | 2.44b |
| cg3335 | NCgl2904 | Malic enzyme | mez | 0.46b | 0.45 | 0.38b |
| cg3386 | NCgl2952 | Put. maleylacetate reductase | tcbF | 0.41b | 0.37 | 0.49 |
| cg3387 | NCgl2953 | Put. myo-inositol:H+ symporter (TC 2.A.1.1.26) | 0.32b | 0.32 | 0.50 | |
| cg3390 | NCgl2956 | Put. sugar phosphate isomerase/epimerase | 0.38b | 0.58 | 1.15b | |
| cg3391 | NCgl2957 | Myo-inositol 2-dehydrogenase | idhA1 | 0.41b | 0.43 | 0.38 |
| cg3404 | NCgl2970 | Put. Fe3+ siderophore periplasmic binding protein | 0.06b | 0.07b | 0.18 | |
The mRNA ratios are averages from at least two of three experiments. Only values for open reading frames whose mRNA ratio was altered at least twofold either 10, 30, or 60 min after the Pi downshift are shown.
P < 0.05 as determined by a t test.
Put., putative; CoA, coenzyme A.
FIG. 3.
Relative mRNA levels of phosphate starvation-inducible genes in the C. glutamicum wild type (A) and the ΔphoRS strain (B). The ratios represent the mRNA levels 10 min (white bars), 30 min (gray bars), and 60 min (black bars) after the onset of Pi starvation versus the mRNA level immediately before the onset of Pi starvation. The experiment was performed in triplicate, and average mRNA ratios were calculated. The criterion used for selection of RNA ratios was a signal-to-noise ratio of ≥3 for either Cy3 or Cy5 fluorescence. The data for the wild type (A) were taken from our previous studies (12).
In the hierarchical cluster analysis of genes showing altered expression in the wild type after a shift from Pi excess to Pi limitation, five subgroups were identified (12). Whereas subgroup 1 harbored genes characteristic of phosphate starvation, subgroups 2 to 5 contained genes of which most are apparently not directly related to phosphate metabolism, i.e., genes putatively involved in copper metabolism (subgroup 2, with transiently increased expression after the shift), in protocatechuate degradation (subgroup 3, with transiently increased expression after the shift), in protein synthesis (subgroup 4, with continuously decreasing expression after the shift), or in iron uptake (subgroup 5, with transiently decreased expression after the shift). The changed expression of these genes is presumably due to the exchange of the culture medium in the course of the experiment (subgroups 2, 3, and 5) or to the reduced growth rate of the cells after the shift to Pi limitation (subgroup 4). Whereas the mRNA levels of the majority of the genes belonging to subgroup 1 were unaltered after the shift in the ΔphoRS strain, many of the genes belonging to the other subgroups, in particular subgroups 2 and 5, showed alterations in expression profiles similar to those in the wild type. This result supports the assumption that the phoRS deletion specifically affects the expression of the phosphate starvation-inducible genes.
In a second set of DNA microarray experiments, the mRNA levels of the wild type and the ΔphoRS mutant were directly compared before and 60 min after a shift from Pi excess (13 mM) to Pi limitation (0.065 mM). As expected from the previous experiment, the majority of phosphate starvation-inducible genes (pstS, pstA, pstB, ugpA, ugpE, ugpB, ugpC, glpQ1, phoH1, ushA, nucH, pctB, pctC, and cg3393) showed higher mRNA levels in the wild type than in the ΔphoRS mutant after the shift (data not shown).
Primer extension analysis.
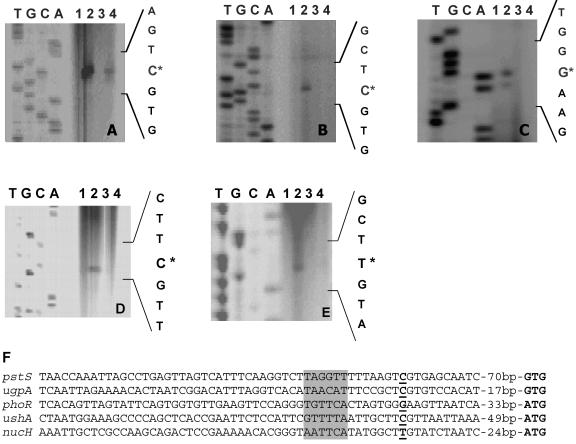
In order to verify the DNA microarray data and to determine the transcriptional start sites, primer extension analyses were performed for prominent psi genes, i.e., the pstSCAB operon, the upgAEBC operon, the phoRS operon, ushA, and nucH (Fig. 4). In these experiments, RNA of the wild type and of the ΔphoRS mutant grown either under Pi excess (13 mM) or under Pi limitation (0.065 mM) for 10 min (phoRS), 60 min (pst, ugp), or 90 min (ushA, nucH) were used. The growth conditions and the preparation of RNA were identical to those in DNA microarray experiments. In the case of the wild type, primer extension products were observed for all five tested genes with RNA from cells grown under Pi limitation but not with RNA from cells grown under Pi excess, confirming that the expression of these genes is induced under Pi limitation. In the case of the ΔphoRS mutant, primer extension products for ugpA, phoR, ushA, and nucH were observed neither with RNA from cells grown under Pi excess nor with RNA from cells grown under Pi limitation. Again, this confirms the results of the microarray data, which indicate that induction of these genes under Pi starvation is dependent on the PhoRS two-component system. In the case of pstS, a primer extension product was obtained with RNA from ΔphoRS cells grown under Pi limitation but not with RNA from ΔphoRS cells grown under Pi excess. The signal intensity of the pstS primer extension product was lower in the ΔphoRS mutant than in the wild type (Fig. 4A), which is in agreement with the DNA microarray data, where the induction ratio was lower in the ΔphoRS strain than in the wild type. The primer extension data thus support the assumption that induction of the pstSCAB operon by Pi starvation is dependent on PhoRS and one or several additional regulators which are not yet known. The fact that no primer extension product was obtained for phoR with RNA from ΔphoRS cells grown under Pi limitation suggests a positive autoregulation of the phoRS genes.
FIG. 4.
Comparison of mRNA levels and determination of the transcriptional start sites of the C. glutamicum genes pstS (A), ugpA (B), phoR (C), ushA (D), and nucH (E) by primer extension analysis. The reverse transcriptase reactions were performed with the oligonucleotides pstS_prext2, ugpA_prext2, phoR_prext1b, ushA80prext, and nucH90prext for these four genes, respectively, and 20 μg of total RNA was isolated from the following strains: the wild type grown under phosphate excess (lane 1); the wild type 60 min (A and B), 10 min (C), or 90 min (D and E) after a shift from 13 mM Pi to 0.065 mM Pi (lane 2); the ΔphoRS mutant grown under Pi excess (lane 3); and the ΔphoRS mutant 60 min (A and B), 10 min (C), or 90 min (D and E) after a shift from 13 mM Pi to 0.065 mM Pi (lane 4). The transcriptional start sites are indicated by asterisks. The corresponding sequencing reactions were generated by using the same IRD-800-labeled oligonucleotide as in the primer extension reactions as well as PCR products which cover the region of the respective transcriptional start site as the template DNA. In panel F, the promoter regions derived from the primer extension studies were aligned. The transcriptional start points are shown in bold and underlined, the proposed start codons are indicated in bold, and putative −10 regions are shaded in gray.
The transcriptional start points identified by the primer extension experiments were located 81 bp upstream of the GTG start codon of pstS, 28 bp upstream of the proposed GTG start codon of ugpA, 44 bp upstream of the ATG start codon of phoR, 60 bp upstream of the ATG start codon of ushA, and 35 bp upstream of the ATG start codon of nucH. Analysis of the promoter sequences (Fig. 4F) revealed motifs with similarity to the proposed −10 region of C. glutamicum (25) but no obvious PhoR binding site.
DISCUSSION
In many bacteria whose phosphate starvation response has been studied in some detail, e.g., E. coli, B. subtilis, Synechocystis spp. (32), or Streptomyces spp. (30, 31), two-component signal transduction systems play a central regulatory role in this process. In the work presented here, phenotypic and genetic evidence that this is also the case in C. glutamicum was obtained. Screening of 12 C. glutamicum deletion mutants, each lacking one specific two-component system, revealed that one of them, the ΔphoRS strain, is specifically impaired in its ability to grow under Pi limitation. Complementation showed that the phoRS deletion is responsible for the growth impairment of this strain. Independent evidence for an involvement of the phoRS genes in the Pi starvation response was obtained previously during the elucidation of the phosphate starvation stimulon of C. glutamicum by DNA microarrays (12). In these studies, the mRNA levels of phoS and phoR were strongly increased (fivefold and eightfold, respectively) 10 min after a shift from Pi excess to Pi limitation but reached preshift levels within 60 to 90 min. Of the other 24 genes encoding proteins of two-component systems, only cgtR9 also showed a >4-fold-altered expression after the shift. Its mRNA level was sevenfold increased 30 min after the shift and remained increased up to 120 min after the shift (12). Preliminary results suggest that the CgtSR9 two-component system is involved in copper metabolism, since the ΔcgtSR9 mutant has a lower copper resistance than the wild type (M. Brocker and M. Bott, unpublished data). It therefore seems likely that the CgtSR9 two-component system is responsible for the induction of the genes involved in copper metabolism (subcluster 2) that were identified in the hierarchical cluster analysis (12). Consequently, PhoRS is the only two-component system of C. glutamicum whose synthesis is rapidly and transiently induced in response to a shift to Pi limitation.
Transcriptome comparisons with DNA microarrays of the ΔphoRS mutant before and after a shift from Pi excess to Pi limitation showed that in contrast to what occurs in the wild type, none of the characteristic psi genes except pstSCAB (e.g., ugpAEBC, glpQ1, phoH, ushA, or nucH) was induced within 60 min after the shift (Fig. 3). The results of the microarrays regarding the expression pattern of the pst, ugp, and phoRS genes were confirmed independently by primer extension analyses (Fig. 4), which also provided evidence for positive autoregulation of the phoRS genes. The induction of the pstSCAB genes in the ΔphoRS mutant suggests the presence of one or more additional regulators which are not yet known. A situation where more than one regulator is involved in the phosphate starvation response has also been found in B. subtilis (4).
Bioinformatic analyses revealed that the PhoS and PhoR proteins of C. glutamicum are highly conserved within the suborder Corynebacterineae. The sensor kinase PhoS orthologs from Corynebacterium efficiens (locus tag CE2493), Corynebacterium diphtheriae (DIP1935), Mycobacterium avium subsp. paratuberculosis (MAP0592), and Mycobacterium tuberculosis (Rv0758) possess 73%, 51%, 44%, and 44% sequence identity, respectively. The response regulator PhoR orthologs from C. efficiens (CE494), C. diphtheriae (DIP1936), Mycobacterium avium subsp. paratuberculosis (MAP0591), and Mycobacterium tuberculosis (Rv0757) possess 91%, 81%, 65%, and 65% sequence identity, respectively. However, an involvement of theseorthologs in phosphate regulation has not been shown. The results provided here for C. glutamicum certainly support such a function.
In summary, our results show that 12 of the 13 two-component systems of C. glutamicum are not essential, and they provide clear evidence for an involvement of the C. glutamicum PhoRS two-component system in the adaptation to phosphate-limiting conditions. Our future studies will focus on the identification of the direct target genes and the binding site of the response regulator PhoR.
Supplementary Material
Acknowledgments
This work was supported by the German Ministry of Education and Research (BMBF) within the framework of genome research on prokaryotic organisms (GenoMik) through grant 031U113D/031U213D.
We thank Hermann Sahm for continuous support.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abe, S., K. Takayama, and S. Kinoshita. 1967. Taxonomical studies on glutamic acid producing bacteria. J. Gen. Appl. Microbiol. 13:279-301. [Google Scholar]
- 2.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 4.Birkey, S. M., W. Liu, X. H. Zhang, M. F. Duggan, and F. M. Hulett. 1998. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol. Microbiol. 30:943-953. [DOI] [PubMed] [Google Scholar]
- 5.Bott, M., M. Meyer, and P. Dimroth. 1995. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol. Microbiol. 18:533-546. [DOI] [PubMed] [Google Scholar]
- 6.Eggeling, L., and M. Bott (ed.). 2005. Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, Fla.
- 7.Eikmanns, B. J., E. Kleinertz, W. Liebl, and H. Sahm. 1991. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene 102:93-98. [DOI] [PubMed] [Google Scholar]
- 8.Engels, S., C. Ludwig, J.-E. Schweitzer, C. Mack, M. Bott, and S. Schaffer. 2005. The transcriptional activator ClgR controls transcription of genes involved in proteolysis and DNA repair in Corynebacterium glutamicum. Mol. Microbiol. 57:576-591. [DOI] [PubMed] [Google Scholar]
- 9.Engels, S., J. E. Schweitzer, C. Ludwig, M. Bott, and S. Schaffer. 2004. clpC and clpP1P2 gene expression in Corynebacterium glutamicum is controlled by a regulatory network involving the transcriptional regulators ClgR and HspR as well as the ECF sigma factor σH. Mol. Microbiol. 52:285-302. [DOI] [PubMed] [Google Scholar]
- 10.Gerharz, T., S. Reinelt, S. Kaspar, L. Scapozza, and M. Bott. 2003. Identification of basic amino acid residues important for citrate binding by the periplasmic receptor domain of the sensor kinase CitA. Biochemistry 42:5917-5924. [DOI] [PubMed] [Google Scholar]
- 11.Hulett, F. M. 2002. The Pho regulon, p. 193-201. In J. A. Sonenshein and R. M. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 12.Ishige, T., M. Krause, M. Bott, V. F. Wendisch, and H. Sahm. 2003. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 185:4519-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Krämer, B. Linke, A. C. McHardy, F. Meyer, B. Möckel, W. Pfefferle, A. Pühler, D. A. Rey, C. Rückert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegrabe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 14.Kaspar, S., R. Perozzo, S. Reinelt, M. Meyer, K. Pfister, L. Scapozza, and M. Bott. 1999. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol. Microbiol. 33:858-872. [DOI] [PubMed] [Google Scholar]
- 15.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambert, C., D. Weuster-Botz, R. Weichenhain, E. W. Kreutz, A. A. De Graaf, and S. M. Schoberth. 2002. Monitoring of inorganic polyphosphate dynamics in Corynebacterium glutamicum using a novel oxygen sparger for real time P-31 in vivo NMR. Acta Biotechnol. 22:245-260. [Google Scholar]
- 17.Lange, C., D. Rittmann, V. F. Wendisch, M. Bott, and H. Sahm. 2003. Global expression profiling and physiological characterization of Corynebacterium glutamicum grown in the presence of l-valine. Appl. Environ. Microbiol. 69:2521-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liebl, W. 2005. Corynebacterium taxonomy, p. 9-34. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, Fla.
- 19.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, W., S. Eder, and F. M. Hulett. 1998. Analysis of Bacillus subtilis tagAB and tagDEF expression during phosphate starvation identifies a repressor role for PhoP∼P. J. Bacteriol. 180:753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menkel, E., G. Thierbach, L. Eggeling, and H. Sahm. 1989. Influence of increased aspartate availability on lysine formation by a recombinant strain of Corynebacterium glutamicum and utilization of fumarate. Appl. Environ. Microbiol. 55:684-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Möker, N., M. Brocker, S. Schaffer, R. Krämer, S. Morbach, and M. Bott. 2004. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 54:420-438. [DOI] [PubMed] [Google Scholar]
- 23.Niebisch, A., and M. Bott. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175:282-294. [DOI] [PubMed] [Google Scholar]
- 24.Pallerla, S. R., S. Knebel, T. Polen, P. Klauth, J. Hollender, V. F. Wendisch, and S. M. Schoberth. 2005. Formation of volutin granules in Corynebacterium glutamicum. FEMS Microbiol. Lett. 243:133-140. [DOI] [PubMed] [Google Scholar]
- 25.Patek, M., J. Nesvera, A. Guyonvarch, O. Reyes, and G. Leblon. 2003. Promoters of Corynebacterium glutamicum. J. Biotechnol. 104:311-323. [DOI] [PubMed] [Google Scholar]
- 26.Reinelt, S., E. Hofmann, T. Gerharz, M. Bott, and D. R. Madden. 2003. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J. Biol. Chem. 278:39189-39196. [DOI] [PubMed] [Google Scholar]
- 27.Rittmann, D., U. Sorger-Herrmann, and V. F. Wendisch. 2005. Phosphate starvation-inducible gene ushA encodes a 5′ nucleotidase required for growth of Corynebacterium glutamicum on media with nucleotides as the phosphorus source. Appl. Environ. Microbiol. 71:4339-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 29.Skerra, A., and T. G. Schmidt. 2000. Use of the Strep-Tag and streptavidin for detection and purification of recombinant proteins. Methods Enzymol. 326:271-304. [DOI] [PubMed] [Google Scholar]
- 30.Sola-Landa, A., R. S. Moura, and J. F. Martin. 2003. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. USA 100:6133-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sola-Landa, A., A. Rodriguez-Garcia, E. Franco-Dominguez, and J. F. Martin. 2005. Binding of PhoP to promoters of phosphate-regulated genes in Streptomyces coelicolor: identification of PHO boxes. Mol. Microbiol. 56:1373-1385. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, S., A. Ferjani, I. Suzuki, and N. Murata. 2004. The SphS-SphR two component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in Synechocystis. J. Biol. Chem. 279:13234-13240. [DOI] [PubMed] [Google Scholar]
- 33.Wanner, B. L. 1996. Phosphorus assimilation and control of the phosphate regulon, p. 1357-1381. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 34.Wendisch, V. F., and M. Bott. 2005. Phosphorus metabolism, p. 377-396. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, Fla.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.