Abstract
An open reading frame (draSO) encoding a putative sulfite oxidase (SO) was identified in the sequence of chromosome II of Deinococcus radiodurans; the predicted gene product showed significant amino acid sequence homology to several bacterial and eukaryotic SOs, such as the biochemically and structurally characterized enzyme from Arabidopsis thaliana. Cloning of the Deinococcus SO gene was performed by PCR amplification from the bacterial genomic DNA, and heterologous gene expression of a histidine-tagged polypeptide was obtained in a molybdopterin-overproducing strain of Escherichia coli. The recombinant protein was purified to homogeneity by nickel chelating affinity chromatography, and its main kinetic and chemical physical parameters were determined. Northern blot and enzyme activity analyses indicated that draSO gene expression is constitutive in D. radiodurans and that there is no increase upon exposure to thiosulfate and/or molybdenum(II).
Sulfite oxidases (SOs) (E.C.1.8.3.1) are molybdenum-coordinating proteins belonging to the broad class of enzymes that comprise three separate families, the bacterial-eukaryotic xanthine oxidase family, the eukaryotic sulfite oxidase family, and the bacterial Me2SO reductase family, all of which are associated with metabolic maintenance of the redox balance in prokaryotic and eukaryotic cells (17, 23). They catalyze the two-electron oxidation of sulfite to sulfate (SO32− + H2O → SO42− + 2H+ + 2 e−) with oxygen and/or heme-coordinated iron ions as the final electron acceptor (16, 20).
Electron transport is guaranteed by a molybdenum center, called the molybdopterin cofactor (MoCo), consisting of a pterin cofactor with molybdenum coordinated by a cysteine ligand from the enzyme and two oxo groups (16, 23). The homodimeric chicken and human sulfite oxidases (4, 13, 14, 22, 31) carry molybdenum as the MoCo and can use oxygen as the electron acceptor with extremely low reactivity, since they possess a heme b binding domain that allows anaerobic oxidation of sulfite (23, 32). Among bacteria, the sulfite oxidase from Starkeya novella (formerly Thiobacillus novellus) consists of a subunit containing one MoCo assembled with a heme-type cytochrome c552 subunit that is separately expressed, which forms an heterodimeric active SO (19, 36, 37).
The SO from Arabidopsis thaliana (At-SO) is homodimeric and homologous to animal sulfite oxidases (18, 34), but it contains only the MoCo domain without any heme-binding domain or alternatively bound heme, as indicated by its spectral properties (10) and by the lack of activity with cytochrome c, which requires heme as a mediator for electron transfer from the MoCo (9).
In all organisms these enzymes also play a key role in cell detoxification by sulfite, a strong nucleophilic and reducing compound, and catalyze oxidation to the more innocuous form sulfate (20). All living cells are exposed to exogenous sulfite generated in the environment from the pollutant sulfur dioxide, which is produced by some geological and industrial processes in addition to other processes; it can also accumulate intracellularly as a product of metabolic reactions involving organic sulfur compounds, such as sulfur-containing amino acids (6, 11, 20). In organisms that are capable of heterotrophic, mixotrophic, chemolithoautotrophic, and phototrophic growth, elemental sulfur oxidation is one of the most energy-yielding reactions as elemental sulfur is an intermediate in sulfur-oxidizing metabolism (5, 12, 21, 35). In fact, elemental sulfur is the electron donor in aerobic and facultative aerobic archaea; it is oxidized via sulfite and thiosulfate in a pathway involving sulfite:acceptor oxidases and has recently been proven to be coupled to the aerobic respiratory chain, indicating that there is a connection between oxygen reduction and sulfur oxidation (24).
Inspection of the genome sequence (38) of the polyextremophile Deinococcus radiodurans (2) strain R1 revealed that in chromosome II there is an open reading frame encoding a putative sulfite oxidase (DraSO) that exhibits significant levels of similarity to corresponding proteins from members of the Bacteria and Eukarya. Here we describe cloning and heterologous expression of the draSO gene and biochemical characterization of its gene product. This is the first Fe/heme-independent SO identified in bacteria, and in contrast to all animal and bacterial SOs characterized previously and like A. thaliana SO (10), DraSO lacks the heme domain and exhibits barely detectable activity when cytochrome c is used as an electron transfer mediator.
MATERIALS AND METHODS
Strains, plasmids, enzymes, and chemicals.
Escherichia coli TOP10F′ (15) was used for plasmid propagation; strains BL21-CodonPlus(DE3)-RIL (Stratagene, La Jolla, Calif.) and TP1000 (ΔmobAB) (29), kindly provided by Giuseppe Perugino (IBP, CNR, Naples, Italy) and Tracy Palmer (Department of Molecular Microbiology, John Innes Centre, Norwich, United Kingdom), were the hosts used for gene expression. All strains were routinely cultured in Luria-Bertani medium with ampicillin (100 μg/ml).
D. radiodurans DSM 20539 was supplied by Deutsche Sammlung von Mikroorganismen (Braunschweig, Germany) and was grown aerobically at 30°C in medium containing 0.5% yeast extract, 0.2% tryptone, 0.2% glucose and either 0.5% (wt/vol) sodium thiosulfate or sodium molybdate or both. All nutrients and agar were purchased from Difco.
Plasmid pQE80-At-sox, carrying the SO gene from A. thaliana, was a gift from Ralf R. Mendel (Botanical Institute, Technical University of Braunschweig).
Isolation of the draSO gene from D. radiodurans.
The gene encoding the D. radiodurans sulfite oxidase (DraSO) was amplified by PCR under the conditions described by Saiki (33); the 5′ primer (5′-ATTCACTAGTGATCATATGCTGATTACCCGAC-3′) contained the ATG start codon and an NdeI site (indicated by boldface type and underlining, respectively). The 3′ primer, 5′-GAATTCGATTCTCGAGTTATGACTTCTCCTTG-3′, contained a recognition site for the XhoI endonuclease (underlined) for suitable insertion into the pET28c expression vector. The amplicon was ligated into the T cloning site of plasmid pGEM T Easy (Promega) and was sequenced by the Primm DNA Sequencing Lab at TIGEM-IGB Institute of CNR, Naples, Italy.
A homology comparison and multiple alignment of the DraSO protein with other molybdenum-coordinating sulfite-oxidizing enzymes were performed using the BLAST and ClustalW programs, respectively, available on the Internet.
Total RNA extraction and Northern analysis.
D. radiodurans cultures were grown in the medium described above; aliquots (50 ml) of cells were harvested when the cultures reached the mid-exponential phase, on average at an optical density at 600 nm of about 0.8. Total RNA was extracted by the method described for Bacillus subtilis total RNA by Yoshida et al. (40). For Northern blot analysis the different RNAs extracted (14 μg) were electrophoretically separated together with RNA molecular weight standards (MBI Fermentas) and were blotted onto a Hybond-XP nylon membrane (Amersham Corp.). Hybridization at 66°C with the α-32P-labeled draSO probe (the PCR-amplified coding sequence) and autoradiography were performed by using standard procedures.
Gene expression and protein purification.
The draSO coding sequence obtained by PCR and cloned in pGEM T Easy was digested with NdeI and XhoI and inserted into the expression vector pET28c (Novagen) between the NdeI and XhoI sites. Another expression vector, pTrcHis-draSO, was obtained by excising the 5′ six-histidine-tagged coding sequence from pET-draSO with the NcoI/XhoI restriction enzymes and inserting the DNA fragment into the NcoI/SalI pTRc99A plasmid (Pharmacia Biotech).
E. coli BL21-CodonPlus(DE3)-RIL competent cells were transformed with pET-draSO, whereas E. coli strain TP1000 was used as the host for gene expression from the pTrcHis-draSO plasmid. In all cases recombinant clones were grown at 22°C in Luria-Bertani medium until the A600 was approximately 1.0. Then 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and the cells were incubated at 22°C for 16 h. Expression and purification of A. thaliana recombinant SO in E. coli TP1000 transformed with pQE80-At-sox were performed as described by Eilers et al. (10).
Lysis for preparation of protein extracts from both D. radiodurans and recombinant E. coli was performed by ultrasonic treatment as described previously for preparation of recombinant thioredoxin from Bacillus acidocaldarius by Pedone et al. (30), with 10 mM Tris HCl (pH 8.0) containing 0.7 mM phenylmethylsulfonyl fluoride as the lysis buffer, and the cytosol fractions were cleared by centrifugation at 13,000 × g. After addition of sodium phosphate (pH 8.0), imidazole, and sodium chloride to final concentrations of 50 mM, 10 mM, and 0.3 M, respectively, the extract was loaded onto a His-Select HF nickel affinity gel (Sigma) column (0.5 by 3.0 cm) connected to a fast protein liquid chromatography system (Amersham Pharmacia Biotech) with a flow rate of 1.0 ml/min; after washing with 30 ml of 10 mM imidazole buffer, fractionation was obtained with a linear gradient of 0.0 to 0.5 M imidazole buffered at pH 7.0. Specific fractions were dialyzed against 10 mM Tris-HCl (pH 8.0), concentrated fivefold by ultrafiltration on Centricon (10-kDa cutoff; Millipore), and stored at −20°C in 20% glycerol.
Protein determination and enzyme assays.
The protein concentration was determined as described by Bradford (3), using the Bio-Rad protein staining assay and bovine serum albumin as the standard.
The UV-visible absorption of the protein (1.0 mg/ml) was measured using a Cary 100 double-beam spectrophotometer (Varian); analysis of the reduced DraSO was performed after extensive deaeration of protein samples and buffers, as well as addition of 400 μM sulfite, immediately before the spectra were recorded.
SO was routinely assayed at 55°C in 50 mM Tris-HCl (pH 8.0) containing 0.1 mM EDTA. The reaction was monitored with a Cary double-beam spectrophotometer (Varian) by monitoring the reduction of ferricyanide (ε = 1,020 M−1 cm−1) at 420 nm. One unit of SO activity was defined as conversion of 1 μmol of sulfite per min (10). Cytochrome c reductase activity was determined as described previously (7).
Steady-state kinetic measurements were determined aerobically at 55°C and pH 8.0 with a 1.0-cm-light-path cuvette in 1.0 ml (final volume), using a saturating concentration of ferricyanide (400 μM) and various concentrations of sulfite between 0.050 and 2.0 mM (saturating concentration).
The dependence on pH was investigated using 50 mM potassium phosphate buffer (pH 5.0 to 8.0), 25 mM Tris-HCl buffer (pH 7.0 to 9.0), and glycine/NaOH buffer (pH 9.0 to 11.0). The data were obtained by performing the enzyme assay at 55°C. The dependence of enzyme activity on temperature was determined in the temperature range from 25 to 65°C in 50 mM Tris-HCl (pH 8.0).
The thermal stability of DraSO (150 μg/ml in 10 mM Tris-HCl [pH 8.0]) was studied over the temperature range from 40 to 65°C by assaying aliquots incubated for different times at 55°C with the ferricyanide assay.
Electrophoretic analysis.
Electrophoresis was performed with a Bio-Rad Mini Protean II cell unit at room temperature. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed as described by Laemmli (25) on 10% polyacrylamide gels, and proteins were revealed by staining with Coomassie brilliant blue R250 (Bio-Rad). Nondenaturing electrophoresis was performed on continuous 7.5% polyacrylamide gels at an alkaline pH in glycine/NaOH buffer (pH 10.0). Proteins were revealed by staining with Coomassie brilliant blue R250 (Bio-Rad) and by the sulfite oxidase activity visualized as described by Cohen and Fridovich (8), using a modification of the procedure of Lyric and Suzuki (27). For analytical isoelectric focusing, samples were applied to a 5% polyacrylamide gel containing 2% Bio-Lyte 3/10 ampholytes (Bio-Rad) and were focused using a Mini IEF cell (Bio-Rad) together with Bio-Rad IEF protein standards as described by the supplier.
RESULTS
Identification and sequence analysis of the sulfite oxidase gene from D. radiodurans.
Sequence analysis of the D. radiodurans genome (38) using the A. thaliana SO coding sequence as the query allowed identification of a 1,068-bp open reading frame (annotated as DRA0225) which was designated draSO and potentially codes for a putative sulfite oxidase. The derived amino acid sequence predicted a polypeptide consisting of 356 residues with a calculated molecular mass of 38,937 Da and a pI of 6.09. All nonredundant databases were screened for entries showing similarity to DraSO with the BLASTP program (1), and partial high levels of similarity were found with several bacterial and eukaryotic sulfite oxidase/dehydrogenases. In this search only small proteins (about 200 kDa) were found in archaeal genomes, and they appeared to consist of only a MoCo domain and hence to belong to a different, uncharacterized family, as pointed out previously by Kletzin et al. (24). Therefore, the new protein sequence was compared to orthologs that have been characterized biochemically (Fig. 1) by multiple-sequence alignment with hierarchical clustering, available at the website http://prodes.toulouse.inra.fr/multalin/multalin.html, and significant similarity was found with eukaryotic and bacterial representatives. In particular, alignment with the SOs from chicken liver and from the plant A. thaliana produced the highest identity scores (37% and 32%, respectively), which surprisingly were higher than the score obtained with the bacterial sequence from T. novellus.
FIG. 1.
Multiple alignment of the D. radiodurans R1 sulfite oxidase sequence with the sequences of SOs from different sources. The protein sequence translated from the DRA0225 open reading frame was compared to both eukaryotic and bacterial counterparts that have been biochemically characterized, and significant similarity was found with the sequences indicated. D. ra, D. radiodurans SO; G. ga, chicken liver SO; A. th, A. thaliana SO; T. no, T. novellus SO. The search for structural motifs involved in known biochemical functions revealed the presence of a conserved central region, namely, an oxidoreductase molybdopterin binding domain (open bar) and a C-terminal MoCo oxidoreductase dimerization domain (dark gray bar) and the absence of the N-terminal heme binding domain (light gray bar) and signal peptide for secretion.
Moreover, the search for structural motifs involved in known biochemical functions, performed with the PFAM (Protein Families database of Alignments and HMMs) program available at the website www.sanger.ac.uk, revealed a general catalytic oxidoreductase molybdopterin binding domain (residues 1 to 209) and a MoCo oxidoreductase dimerization domain (residues 226 to 349) that are present in all of the sulfite oxidases compared. Interestingly, as in the plant enzyme, no heme binding domain common to animal and T. novellus SOs was found in the N-terminal region of the protein.
In particular, the residues of the catalytic site (with Cys76 corresponding to Cys98 of the plant enzyme, which was identified as crucial for catalysis) and most of the hydrogen bond-forming amino acids necessary for pterin binding (His32, Asp130, Lys192, Tyr28, Arg30, His174, and Arg179, corresponding to A. thaliana His53, Asp161, Lys220, Tyr49, Arg51, His202, and Arg207) and for interaction with Mo oxo groups (Arg30, Ala77, and Tyr213, corresponding to residues Arg51, Ala99, and Tyr216 in the plant sequence) are conserved.
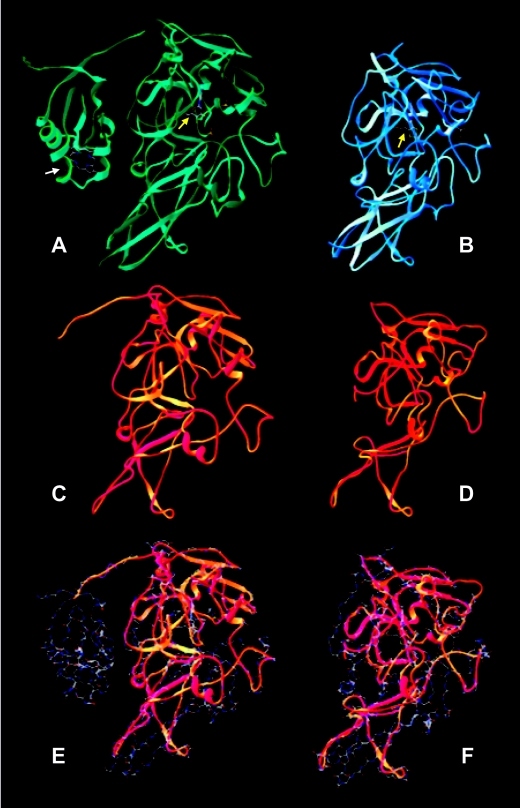
The homology in the primary sequences is reflected by high levels of structural similarity with both animal and plant proteins and by the overall three-dimensional architecture conserved for the secondary structure and general folding (Fig. 2), as clearly indicated by the superimposition models with both the available chicken SO (23) and plant SO (34) structures obtained with the Deep View Swiss-PdbViewer, version 3.7. In fact, significant high-confidence E values of 0.000323 and 0.00522 for the comparisons with the chicken and plant SO sequences, respectively, were calculated using the specific program available on the 3D-PSSM fold recognition server of the Structural Bioinformatics Group at the Imperial College of Science, Technology and Medicine (http://www.sbg.bio.ic.ac.uk).
FIG. 2.
Structural prediction for DraSO based on A. thaliana and chicken liver SO models. Superimposition models with both the available chicken SO and plant SO (At-SO) structures were obtained with the Deep View Swiss-PdbViewer, version 3.7. (A and B) Three-dimensional structures of bird and plant SOs. The view shows the heme binding domain in chicken SO (indicated by a white arrow) that is absent in At-SO and the binding to molybdopterin coenzyme (indicated by yellow arrows) in both structures. (C and D) Predicted structure of DraSO using chicken SO and At-SO as superimposition models, respectively. (D and F) Superposition of DraSO (ribbons) on chicken SO and At-SO (wire frame).
Heterologous gene expression and purification of recombinant His6-DraSO.
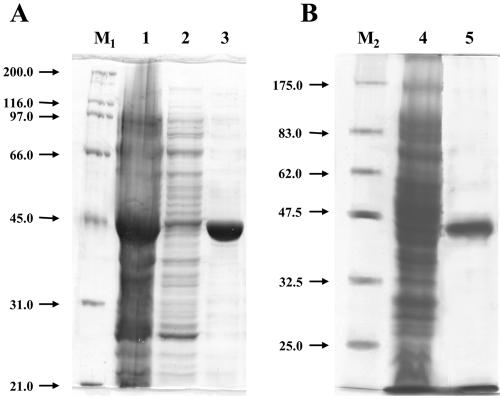
Two different plasmids able to drive gene expression with a six-His-tagged fusion protein in E. coli were constructed to verify that the gene product of the draSO sequence catalyzes the oxidation of sulfite to sulfate. The PCR-amplified coding sequence of the draSO gene was cloned into pET-28(c)+ and overexpressed in E. coli BL21-CodonPlus(DE3)-RIL. Transformed cells were grown in the presence of Mo ions and exposed to IPTG for induction of gene expression at 22°C in order to reduce accumulation of the protein in the inclusion bodies, which was observed when induction was performed at 37°C. The tagged protein was detectable after nickel affinity chromatography of the cleared protein extracts. The purified recombinant enzyme had an apparent molecular mass of 43 kDa as determined by SDS-PAGE, which is in good agreement with the value calculated for the tagged sequence (Fig. 3A). The protein was expressed at a high level, but it exhibited very low specific activity compared to other characterized SOs. In order to increase the catalytic efficiency of the enzyme, the His-tagged draSO gene was cloned into the pTrc99A vector, which allowed T7 polymerase-independent expression, and transferred into E. coli strain TP1000 (ΔmobAB), which contained higher levels of molybdopterin. Figure 3B shows the SO purification results for pTrcHis-draSO-transformed TP1000 cells; as hypothesized, a 10-fold increase in the specific activity was obtained, although the yield was lower, compared to the pET28/BL21 system. The enzyme could be purified by nickel affinity gel chromatography with a level of purity of 80% as judged by SDS-PAGE analysis and by gel isoelectric focusing; the pI determined (pI 6.3) was in perfect agreement with the value deduced from the amino acid sequence.
FIG. 3.
SDS-PAGE analysis of His6-tagged DraSO expression and purification by affinity chromatography. E. coli BL21-CodonPlus(DE3)-RIL transformed with plasmid pET-draSO (A) and TP1000 (ΔmobAB) transformed with plasmid pTrcHis-draSO (B) were grown and subjected to induction of gene expression at 22°C. The His6-tagged protein was detectable after nickel affinity chromatography of the cleared protein extracts (lanes 1 and 4) and was recovered in 0.25 mM imidazole fractions (lanes 3 and 5) after washing in 10 mM imidazole (lane 2). The molecular mass, about 43.0 kDa, was determined with reference to molecular weight standards (lanes M1 and M2).
Similar to A. thaliana SO, size exclusion chromatography of DraSO resulted in two distinct peaks that accounted for 65 and 35% of the protein, as estimated by the relative areas. The major and minor peaks corresponded to molecular masses of 80.5 and 44.30 kDa, respectively. Since the molecular mass of His-tagged DraSO calculated from the amino acid sequence is 41.1 kDa, the protein is mainly dimeric, although there is a considerable monomeric fraction. Both dimeric protein and monomeric protein displayed SO activity, and the dimer exhibited threefold-higher catalytic efficiency.
Wild-type draSO gene expression.
An enzyme staining analysis performed with the crude extract of D. radiodurans R1 after separation on native gels revealed a unique band that exhibited mobility indistinguishable from that of the recombinant DraSO (Fig. 4).
FIG. 4.
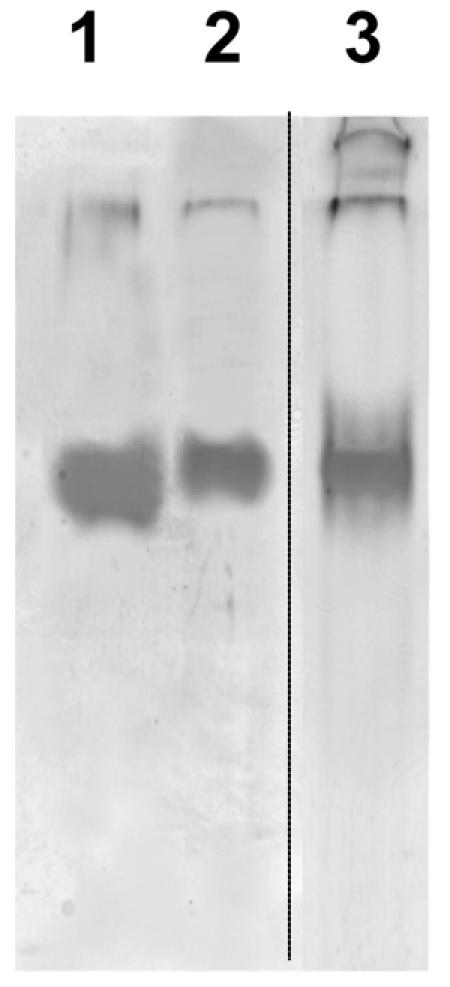
Detection of native and recombinant DraSO activities on a 7.5% nondenaturing gel. Samples were stained for sulfite oxidase activity after electrophoresis under alkaline conditions and equilibration of the gel in the assay buffer. Lane 1, recombinant His6-tagged DraSO after purification on nickel affinity gel; lane 2, crude extract of D. radiodurans; lane 3, Coomassie brilliant blue staining of purified recombinant DraSO.
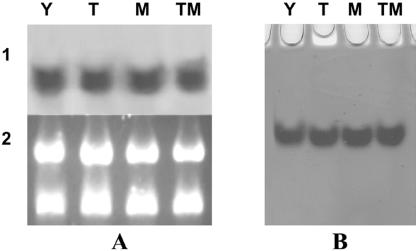
Regulation of the draSO gene expression was analyzed in D. radiodurans cells grown in yeast extract-tryptone medium supplemented with sodium molybdate or thiosulfate or both, and RNA synthesis levels were evaluated in exponentially grown cultures. Northern blot analysis using the draSO coding sequence as the probe revealed a single hybridization band that exhibited intensity that was independent of the growth conditions (Fig. 5A) (namely, the intensity was independent of the individual or combined salts in the single cultures). The molecular size was compared to the sizes of RNA molecular weight standards and was calculated to be about 1,100 nucleotides, which is in agreement with the size expected for a monocistronic mRNA from the gene coding sequence (1,068 nucleotides). Protein expression in the same cultures exhibited an identical trend, as demonstrated by specific sulfite oxidase activity detected on nondenaturing gels by enzyme staining (Fig. 5B).
FIG. 5.
Northern analysis of draSO gene expression and enzyme activity on gels. Cells of D. radiodurans were cultured in yeast-glucose minimal medium (lane Y) supplemented with sodium thiosulfate (lane T) or sodium molybdate (lane M) or both (lane TM) and harvested in the mid-exponential phase of growth. Total RNA (A) was extracted and analyzed by electrophoresis on a denaturing agarose gel stained with ethidium bromide (panel 1) and by Northern blot using the draSO coding sequence as the probe (panel 2). Aliquots of the cells from the same cultures were disrupted by ultrasonic treatment, and the protein extract was analyzed on a nondenaturing gel with enzyme staining (B).
Spectral and kinetic determinations.
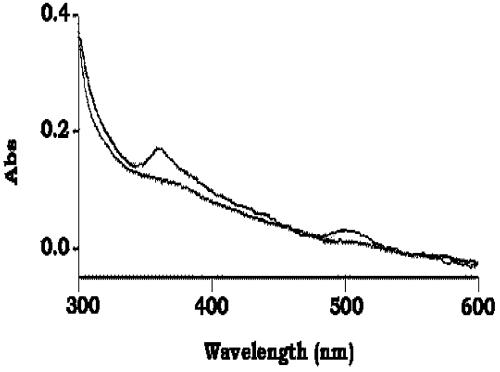
The presence of molybdopterin and the absence of the prosthetic heme group in the enzyme structure were ascertained by obtaining UV-visible absorption spectra for purified DraSO samples (Fig. 6). The analysis also revealed an absorption maximum at about 350 nm with a broad shoulder between 400 and 500 nm (which can be assigned to the electronic transition molybdenum(VI)-to-cysteine and enedithiolate-to-molybdenum charge transfer bands, respectively), which was greatly decreased in the partially reduced enzyme, as in the case of A. thaliana SO (10).
FIG. 6.
UV-visible spectra of oxidized and partially reduced DraSO. Absorption spectra of oxidized (lower line) and sulfite-reduced (upper line) enzymes (1.0 mg/ml) were recorded in 10 mM Tris-HCl (pH 8.0). The 360- and 480-nm absorption bands of the oxidized enzyme are due to cysteine-to-molybdenum and enedithiolate-to-molybdenum charge transfer bands, respectively.
In the temperature range from 30 to 60°C the affinity-purified DraSO protein had sulfite-oxidizing activity when ferricyanide was used as an electron acceptor. As expected, no activity was found with cytochrome c as the electron acceptor, since the heme domain, which is known to mediate electron transfer between the MoCo domain and cytochrome c in animal (13, 23) and T. novellus (19) SOs, is missing in the protein. The Km for sulfite was determined to be 94.5 μM, which is the same order of magnitude as the Km values for plant A. thaliana and Nicotiana tabacum SOs (10) and among the lowest Km values determined for bacterial SOs. The enzyme also exhibited moderate thermophilicity, with the highest catalytic efficiency at 55°C, and thermal deactivation at temperatures far above the optimum growth temperature of D. radiodurans (30°C), with half-lives of 10 min at 60°C, 30 min at 55°C, and 3 h at 45°C. These features were also confirmed for the wild-type enzyme in crude extracts of D. radiodurans.
Like plant SO (10) but with different trends, DraSO is sensitive to increased ionic strength. The effects of various concentrations of salts and the substrate analogue nitrate in 50 mM Tris-HCl (pH 8.0) and buffers were tested; the concentrations that produced 50% inhibition compared with the activity calculated in the standard assay were 100 mM Tris-HCl (pH 8.0), 80 mM Tris-acetate (pH 8.0), 20 mM potassium phosphate (pH 8.0), 150 mM NaCl, 30 mM KCl, 3 mM potassium nitrate, 20 mM EDTA, 7 mM sodium arsenate, and 20 mM sodium sulfate. Nevertheless, some salts and buffers tested, such as NaCl up to a concentration of 100 mM, sodium arsenate up to a concentration of 5 mM, Tris-HCl up to a concentration of 70 mM, and Tris-acetate up to a concentration of 70 mM, resulted in up to twofold activation before inhibition occurred at higher concentrations. Complete inhibition was obtained with 0.1 mM NiCl2 and 0.1 mM imidazole.
DISCUSSION
By using bioinformatic screening of the D. radiodurans genome sequence (38), the draSO gene was identified as the gene that encodes a sulfite oxidase belonging to the molybdopterin-containing oxidase family, which includes representatives characterized from the Eukarya and Bacteria (7, 10, 19, 22). Interestingly, the N-terminal heme binding domain was missing in the D. radiodurans SO, as it is in the At-SO enzyme from A. thaliana (10). For this reason, At-SO was used in this work as a structural and biochemical reference for comparison and as a probe for more general screening of different prokaryotic genome sequences, including that of D. radiodurans; no other bacterial SO that lacks the heme binding domain and has been biochemically characterized previously was found. The significant sequence similarity with the plant enzyme also allowed prediction of a conserved general architecture in the three-dimensional structure of DraSO. Moreover, the strategy adopted for heterologous gene expression in E. coli, similar to that described for At-SO (10), confirmed that higher-level expression of active SO could be obtained using the TP1000 strain that is deficient in the genes controlling the biosynthetic pathway of the guanylyl version of molybdopterin and hence is able to accumulate a higher level of available unmodified cofactor (29).
Therefore, the biochemical function of the enzyme could be confirmed by specific assays performed at a higher temperature, with the activity showing higher thermophilicity, as expected, compared to the mesophilic plant enzyme but having a similar pH optimum, similar catalytic efficiency, and similar inhibitors (10).
DraSO was the major sulfite-oxidizing enzyme in D. radiodurans soluble extracts obtained from cells grown under different medium composition conditions when activity detection was performed with ferricyanide as the electron acceptor on zymograms; gene expression appeared to be unaffected by the common SO inducers, such as thiosulfate and the molybdenum ion, at the level of relative mRNA abundance.
Similar to At-SO, the lack of a heme binding domain resulted in the inability of the bacterial enzyme to mediate electron transfer from sulfite to cytochrome, indicating that the electron acceptor in vivo could be oxygen. The genome location of the draSO open reading frame, which is not surrounded by a cytochrome-encoding gene and/or genes involved in thiosulfate oxidation or linked to respiratory chain oxidation, suggests a different role for this enzyme. The absence of a direct association with these metabolic pathways was also clearly demonstrated by the ineffective action of thiosulfate alone or in combination with the molybdenum ion, at both the mRNA and enzyme activity levels. Furthermore, an in-depth inspection of the genome indicated the presence of only one more putative SO-like gene, annotated as DR2536 and a homolog of the E. coli YedY gene, whose product has nevertheless been shown to possess sulfoxide reductase activity and no SO activity (26). In addition, no homolog of periplasmic and cell-bound SOs (SoxC) that cooperate with other membrane cytochromes and flavoproteins (gene products of the soxABCDEF operon) to utilize sulfite from the environment or, more likely, to participate in the reaction sequence for thiosulfate metabolism in gram-negative bacteria (24, 39) was identified in the gram-positive organism D. radiodurans (28). Since no sulfite oxidase activity was found to be membrane associated in vegetative or thiosulfate-induced cultures and since wild-type and draSO-transformed E. coli TP100 exhibited identical sensitivities to sulfite (data not shown), we concluded that DraSO is the only SO enzyme in D. radiodurans and that it is involved in the intracellular dissimilatory metabolism of sulfite oxidation.
Moreover, the catalytic properties and structural features investigated, which so far are similar only to the catalytic properties and structural features described for the plant counterpart, make this enzyme an interesting model for evolutionary studies of sulfur metabolism.
Acknowledgments
This work was supported by grants from MIUR (Ministero dell'Istruzione dell'Università e della Ricerca) Decreto Direttoriale prot. N. 1105/2002 and by Centro Regionale di Competenza BioTekNet. The project was also partially funded by Eurochem S.p.A.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, A., H. Nordan, R. Cain, G. Parrish, and D. Duggan. 1956. Studies on a radoresistant micrococcus. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol. 10:575-578. [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brody, M. S., and R. Hille. 1999. The kinetic behavior of chicken liver sulfite oxidase. Biochemistry 38:6668-6677. [DOI] [PubMed] [Google Scholar]
- 5.Brune, D. C. 1995. Sulfur compounds as photosynthetic electron donors, p. 847-870. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 6.Brüser, T., P. Lens, and H. G. Trüper. 2000. The biological sulfur cycle, p. 47-86. In Environmental technologies to treat sulfur pollution. IWA Publishing, London, United Kingdom.
- 7.Cohen, H. J., and I. Fridovich. 1971. Hepatic sulfite oxidase. Purification and properties. J. Biol. Chem. 246:359-366. [PubMed] [Google Scholar]
- 8.Cohen. H. J., and I. Fridovich. 1971. Hepatic sulfite oxidase. The nature and function of the heme prosthetic groups. J. Biol. Chem. 246:367-373. [PubMed] [Google Scholar]
- 9.Coury, L. A., J. Yang, and L. Murray. 1993. Electrochemical study of the rate of activation of the molybdoheme protein sulfite oxidase by organic electron acceptors. Anal. Chem. 65:242-246. [Google Scholar]
- 10.Eilers, T., G. Schwarz, H. Brinkmann, C. Witt, T. Richter, J. Nieder, B. Koch, R. Hille, R. Hänsch, and R. R. Mendel. 2001. Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase. A new player in plant sulfur metabolism. J. Biol. Chem. 276:46989-46994. [DOI] [PubMed] [Google Scholar]
- 11.Fischer, U. 1988. Sulfur in biotechnology, p. 463-496. In H. J. Rehm and G. Reed (ed.), Biotechnology. VCH Verlagsgesellschaft, Weinheim, Germany.
- 12.Friedrich, C. G. 1998. Physiology and genetics of sulfur-oxidizing bacteria. Adv. Microb. Physiol. 39:235-289. [DOI] [PubMed] [Google Scholar]
- 13.Garrett, R. M., and K. V. Rajagopalan. 1996. Site-directed mutagenesis of recombinant sulfite oxidase. J. Biol. Chem. 271:7387-7391. [PubMed] [Google Scholar]
- 14.Garrett, R. M., J. L. Johnson, T. N. Graf, A. Feigenbaum, and K. V. Rajagopalan. 1998. Human sulfite oxidase R160Q: identification of the mutation in a sulfite oxidase-deficient patient and expression and characterization of the mutant enzyme. Proc. Natl. Acad. Sci. USA 95:6394-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 16.Hille, R. 1996. The mononuclear molybdenum enzymes. Chem. Rev. 96:2757-2816. [DOI] [PubMed] [Google Scholar]
- 17.Hille, R. 2002. Molybdenum and tungsten in biology. Trends Biochem. Sci. 27:360-367. [DOI] [PubMed] [Google Scholar]
- 18.Hille, R. 2003. Plants have SOX: the structure of sulfite oxidase from Arabidopsis thaliana. Structure 11:1189-1190. [DOI] [PubMed] [Google Scholar]
- 19.Kappler, U., B. Bennett, J. Rethmeier, G. Schwarz, R. Deutzmann, A. G. McEwan, and C. Dahl. 2000. Sulfite:cytochrome c oxidoreductase from Thiobacillus novellus. Purification, characterization, and molecular biology of a heterodimeric member of the sulfite oxidase family. J. Biol. Chem. 275:13202-13212. [DOI] [PubMed] [Google Scholar]
- 20.Kappler, U., and C. Dahl. 2001. Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS Microbiol. Lett. 203:1-9. [DOI] [PubMed] [Google Scholar]
- 21.Kelly, D. P., J. K. Shergill, W. P. Lu, and A. P. Wood. 1997. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Leeuwenhoek 71:95-107. [DOI] [PubMed] [Google Scholar]
- 22.Kessler, D. L., and K. V. Rajagopalan. 1972. Purification and properties of sulfite oxidase from chicken liver. Presence of molybdenum in sulfite oxidase from diverse sources. J. Biol. Chem. 247:6566-6573. [PubMed] [Google Scholar]
- 23.Kisker, C. 2001. Sulphite oxidase, p. 1121-1135. In A. Messerschmidt, R. Huber, K. Wieghardt, and T. Poulos (ed.), Handbook of metalloproteins, vol. 1. Wiley, New York, N.Y. [Google Scholar]
- 24.Kletzin, A., T. Urich, F. Müller, T. M. Bandeiras, and C. M. Gomes. 2004. Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J. Bioenerg. Biomembr. 36:77-91. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Loschi, L., S. J. Brokx, T. L. Hills, G. Zhang, M. G. Bertero, A. L. Lovering, J. H. Weiner, and N. C. Strynadka. 2004. Structural and biochemical identification of a novel bacterial oxidoreductase. J. Biol. Chem. 279:50391-50400. [DOI] [PubMed] [Google Scholar]
- 27.Lyric, R. M., and I. Suzuki. 1970. Enzymes involved in the metabolism of thiosulfate by Thiobacillus thioparus. II. Properties of adenosine-5′-phosphosulfate reductase. Can. J. Biochem. 48:344-354. [DOI] [PubMed] [Google Scholar]
- 28.Makarova, K. S., L. Aravind, Y. I. Wolf, R. L. Tatusov, K. W. Minton, E. V. Koonin, and M. J. Daly. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65:44-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer, T., C. L. Santini, C. Iobbi-Nivol, D. J. Eaves, D. H. Boxer, and G. Giordano. 1996. Involvement of the narJ and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol. Microbiol. 20:875-884. [DOI] [PubMed] [Google Scholar]
- 30.Pedone, E., R. Cannio, M. Saviano, M. Rossi, and S. Bartolucci. 1999. Prediction and experimental testing of Bacillus acidocaldarius thioredoxin stability. Biochem. J. 339:309-317. [PMC free article] [PubMed] [Google Scholar]
- 31.Rajagopalan, K. V. 1980. Sulfite oxidase, p. 241-271. In M. P. Coughlan (ed.),Molybdenum and molybdenum-containing enzymes. Pergamon, Oxford, United Kingdom.
- 32.Rajagopalan, K. V., and J. L. Johnson. 1992. The pterin molybdenum cofactors. J. Biol. Chem. 267:10199-10202. [PubMed] [Google Scholar]
- 33.Saiki, R. K. 1990. Amplification of genomic DNA, p. 13-20. In M. A. Innis, D. A. Gelfand, J. J. Sninski, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, Calif.
- 34.Schrader, N., K. Fischer, K. Theis, R. R. Mendel, G. Schwarz, and C. Kisker. 2003. The crystal structure of plant sulfite oxidase provides insights into sulfite oxidation in plants and animals. Structure 11:1251-1263. [DOI] [PubMed] [Google Scholar]
- 35.Sorokin, D. Y. 1995. Sulfitobacter pontiacus gen. nov., sp. nov., a new heterotrophic bacterium from the Black Sea specialized on sulfite oxidation. Microbiology 64:295-305. [Google Scholar]
- 36.Southerland, W. M., and F. Toghrol. 1983. Sulfite oxidase activity in Thiobacillus novellus. J. Bacteriol. 156:941-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toghrol, F., and W. M. Southerland. 1983. Purification of Thiobacillus novellus sulfite oxidase. Evidence for the presence of heme and molybdenum. J. Biol. Chem. 258:6762-6766. [PubMed] [Google Scholar]
- 38.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, K. W. Minton, R. D. Fleischmann, K. A. Ketchum, K. E. Nelson, S. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wodara, C., F. Bardischewsky, and C. G. Friedrich. 1997. Cloning and characterization of sulfite dehydrogenase, two c-type cytochromes, and a flavoprotein of Paracoccus denitrificans GB17: essential role of sulfite dehydrogenase in lithotrophic sulfur oxidation. J. Bacteriol. 179:5014-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida, K., K. Kobayashi, Y. Miwa, C. M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]