Abstract
We have constructed a relaxed mutant of El Tor biotype Vibrio cholerae strain C7258 by disruption of the RelA catalytic domain. The ability of the V. cholerae relaxed mutant to biosynthesize guanosine tetraphosphate and pentaphosphate was severely affected; the mutant showed a reduced growth rate in minimal medium that could be reversed by the addition of Casamino Acids, and it was thermosensitive. Contrary to published findings, the new relA mutant still produced significant cholera toxin and toxin-coregulated pilus. The V. cholerae relA mutant was motile, produced normal biofilms, and colonized the suckling mouse intestine. Our data suggest that levels of basal guanosine nucleotides pppGpp and ppGpp, rather than the availability of a stringent response, could influence expression of virulence factors, depending on strain and culture conditions. Production of hemagglutinin (HA)/protease, which requires HapR, RpoS, and the cyclic AMP receptor protein, was not strongly affected. Nevertheless, overexpression of RelA protein from an isopropyl-β-d-thiogalactopyranoside-inducible promoter posttranscriptionally diminished production of HA/protease.
Vibrio cholerae, the causative agent of Asiatic cholera, colonizes the human small bowel and causes a potentially life-threatening watery diarrhea. Cholera toxin (CT) and the toxin-coregulated pilus (TCP) have been recognized as primary virulence factors. Expression of CT and TCP in V. cholerae is controlled by a complex network of regulatory proteins. The genes required for production of CT and TCP are regulated by two pairs of transmembrane proteins, ToxR/S and TcpP/H, that cooperate to activate the soluble cytoplasmic regulator ToxT (5, 6, 11, 17). ToxT, in turn, activates expression of CT and TCP (5, 6). It has been shown that quorum-sensing regulators LuxO and HapR also act to control CT and TCP expression and biofilm formation in V. cholerae (22, 25, 30). At low cell density, the active form of LuxO (phospho-LuxO) represses hapR (22, 25, 30). In the absence of HapR, regulator AphA, in conjunction with AphB, activates tcpPH to increase production of CT and TCP (14, 15, 22). At high cell density, LuxO is inactive and HapR is expressed. Expression of HapR leads to repression of aphA, diminished expression of TcpP/H (14, 15), and production of hemagglutinin (HA)/protease (13, 20). Mutations in other global regulators, such as the cyclic AMP receptor protein (CRP) (21) and RpoS (16), as well as inactivation of motility and chemotaxis determinants (2) also affect the capacity of V. cholerae to colonize the infant mouse intestine and cause disease.
The relA gene encoding guanosine pentaphosphate synthase I is the genetic determinant of the stringent response. The RelA-catalyzed increase in cellular guanosine nucleotides pppGpp and ppGpp, hereafter referred to as (p)ppGpp, leads to rapid inhibition of stable RNA biosynthesis, ribosome and protein synthesis and, ultimately, growth arrest (4). The stringent response is elicited when the rate of tRNA aminoacylation does not meet the demands of protein synthesis (i.e., during amino acid starvation) (4). Under this condition, the ribosome-bound RelA protein is activated to convert GTP to pppGpp, which is subsequently converted to ppGpp by pppGpp 5′-phosphohydrolase (4). Consequently, during this process, the intracellular level of (p)ppGpp rises to a concentration 10- to 20-fold higher than the low steady-state basal level typical of cells in rapid exponential growth (4). The spoT gene encodes two activities, a 3′-pyrophosphohydrolase that degrades (p)ppGpp to GDP and a weak (p)ppGpp synthase activity. In Escherichia coli, only relAspoT double mutants entirely lack (p)ppGpp (27).
Recently, a V. cholerae relA mutant was shown to produce significantly reduced levels of CT and TCP, altered levels of OmpU and OmpT porins, reduced motility, and a 1,000-fold reduction in infant mouse colonization (10). The reduced expression of CT and TCP was explained by the diminished transcription of toxR in the relA mutant (10). In the course of studies aimed to elucidate the genetic determinants of HA/protease expression, we constructed another relA mutant from the El Tor biotype strain C7258 (20). The (p)ppGpp biosynthesis in the new relA mutant was severely inhibited, and the mutant exhibited numerous relA phenotypes. However, contrary to a previous report (10), relA mutants from C7258 produced CT, TCP, and HA/protease, formed wild-type biofilms, were motile, and colonized the suckling mouse intestine.
Disruption of V. cholerae relA.
The RelA protein from E. coli and V. cholerae contains two functionally independent domains, an N-terminal activity domain (Fig. 1, shadowed box) and a C-terminal regulatory domain (9) (Fig. 1, open box). Residues G251 and H354 in the highly conserved N-terminal domain are essential for enzyme activity (9). To inactivate the V. cholerae RelA activity domain, an internal 1,077-bp relA fragment was amplified from strain C6709-1 (wild type, El Tor, Peru isolate 1991) using primers 5′-GTTCTAGAGGTGATTAAGCTTGCCG and 5′-GTTGCATGCCCCGCTTCGAGATTT and the Advantage 2 PCR kit (BD Biosciences). The amplified fragment was cloned in pUC19 for sequence confirmation and subsequently transferred to suicide vector pCVD442 (7) as a SphI-XbaI fragment to generate plasmid pCVDΔ relA. A PstI fragment containing the Kmr gene from plasmid pUC4K (Amersham Biosciences) was subsequently inserted in a unique NsiI site located within the highly conserved enzyme activity domain (between essential residues G251 and H354) to create plasmid pCVDΔrelAK. Suicide plasmids were constructed in E. coli SY327λpir (18), transferred to E. coli SM10λpir (18), and mobilized by conjugation to V. cholerae C7258 (El Tor, Ogawa, Peru isolate 1991). Exconjugants were selected in LB medium containing ampicillin (100 μg/ml), kanamycin (50 μg/ml), and polymyxin B (100 units/ml). When pCVDΔ relAK was transferred to V. cholerae C7258, two classes of exconjugants containing duplications of the relA locus were detected. Class I exconjugants contained the sequence 3′-truncated relA-vector DNA-5′-truncated relA with a Kmr insertion (data not shown). Since the 3′-truncated allele is known to encode a protein that retains enzyme activity in E. coli (9), these exconjugants were not considered for further studies. Class II exconjugants contained the sequence 3′-truncated relA with a Kmr insertion in the activity domain-vector DNA-5′-truncated relA (Fig. 1, bottom panel). The organization of relA loci was investigated by Southern hybridization using the digoxigenin (DIG)-labeled 1,077-bp relA fragment and a PstI Kmr probe. Figure 1 shows the result for class II exconjugant AJB19 (top panel) and the deduced organization of its relA loci (bottom panel). Since the two relA copies in strain AJB19 contain either an insertion or a deletion in the enzyme activity domain (Fig. 1, bottom panel), we expected it to be defective in (p)ppGpp biosynthesis. Additionally, we plated AJB19 in LB containing 5% sucrose to select for vector elimination. The resulting strain AJB42 contains a Kmr insertion in the relA activity domain (Fig. 1, lane 6 and bottom panel).
FIG. 1.
Southern hybridization analysis of relA mutants. Top panel, Southern hybridization of NsiI (N) digests of genomic DNA. Lanes 1 and 3, C7258; lanes 2 and 4, AJB19; lane 5, AJB41; lane 6, AJB42. Lanes 1, 2, 5, and 6 were hybridized with a DIG-labeled 1,077-bp relA fragment (probe). Lanes 3 and 4 were hybridized with a DIG-labeled Kmr PstI fragment (filled box). The molecular weight marker lane (MW) was DIG-labeled λ DNA digested with EcoRI plus HindIII (Roche Biochemicals). Bottom panel, organization of the relA locus in C7258 and relA mutants. Shaded box, DNA encoding N-terminal activity domain; open box, DNA encoding C-terminal regulatory domain; filled box, Kmr insertion; thick line, flanking chromosomal DNA; thin line, vector DNA.
V. cholerae relA mutants are defective in (p)ppGpp accumulation.
Accumulation of (p)ppGpp was determined with 96-well flat-bottom microtiter plates as described in reference 3. Isolated colonies were picked from LB agar plates and resuspended in 0.1 ml of phosphate-free MOPS (morpholinepropanesulfonic acid) minimal medium plus 0.2% glucose to an optical density at 600 nm (OD600) of 0.4. To induce the stringent response, the suspension was diluted 1:25 in MOPS minimal medium plus 0.2% glucose containing dl-serine hydroxamate (1 mg/ml), l-valine (0.5 mg/ml), and 100 μCi/ml of carrier-free [32P]H3PO4 (Amersham Biosciences). The plates were incubated for 30 min at 37°C. Labeled nucleotides were extracted by the addition of 0.1 ml of 13 M formic acid and two cycles of freezing and thawing. Finally, 5 μl of each sample was applied to polyethyleneimine-cellulose F thin-layer chromatography (TLC) plates (EMD Chemicals, Inc.) and developed with 1.5 M KH2PO4 (pH 3.4). The chromatogram was air dried and exposed to X-ray film (Kodak BioMax Light) for 16 h at −70°C. The (p)ppGpp spots were identified by comparisons to E. coli wild-type strain CF1648 (MG1655) and its isogenic relA mutant CF1652 (MG1655 relA251 Kmr) (3). A ppGpp spot of intensity similar to GTP is considered a positive stringent response in this assay. Figure 2 (left panel) shows that strain AJB19 is defective in boosting (p)ppGpp levels in response to amino acid starvation. Identical results were obtained with strain AJB42 (see Fig. 5, bottom panel). This phenotype could be complemented by the E. coli wild-type relA introduced on plasmid pALS10 (23) (data not shown).
FIG. 2.
Biosynthesis of (p)ppGpp in V. cholerae relaxed mutants. Left panel, the stringent response was induced as described in the text; radiolabeled guanine nucleotides were analyzed by TLC and detected by autoradiography. Lane 1, C7258; lane 2, AJB19; lane 3, CF1648; lane 4, CF1652 (relA251). Right panel, 10 μl of three independent single-colony cultures of C7258 and AJB19 grown in LB medium were spotted in M9 minimal medium, M9 medium supplemented with AT, and M9 medium supplemented with SMGL and incubated 24 h at 37°C.
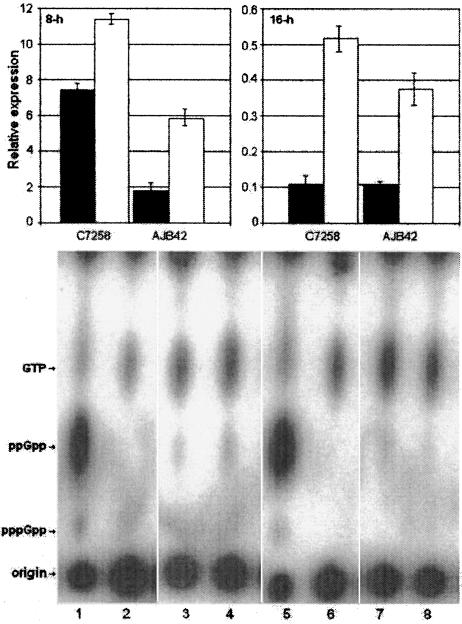
FIG. 5.
Transcription of ctxA and tcpA and (p)ppGpp synthetic capacity of wild-type V. cholerae and relA isogenic mutants in AKI culture. Top panel, relative abundance of ctxA (filled bar) and tcpA mRNA (open bar) (relative to recA mRNA) in C7258 and AJB42 grown 8 and 16 h in AKI cultures. Bottom panel, TLC detection of RelA activity in C7258 and AJB42 grown 8 h (lanes 1 to 4) and 16 h (lanes 5 to 8) in AKI cultures. Lanes 1 and 5, C7258 plus dl-serine hydroxamate and l-valine; lanes 2 and 6, C7258; lanes 3 and 7, AJB42 plus dl-serine hydroxamate and l-valine; lanes 4 and 8, AJB42.
Relaxed mutants are more sensitive to amino acid analogues or amino acid imbalances due to their incapacity to completely derepress amino acid synthesis operons that require (p)ppGpp (19). The sensitivity of strain AJB19 to 3-amino-1,2,4-triazole (AT), a histidine biosynthesis inhibitor, was determined by using M9 minimal medium supplemented with glucose (0.2%), all amino acids except histidine (4 μg/ml), adenine (1 mM), thiamine (1 mM), and AT (15 mM). In addition, the sensitivity to inhibition by serine, methionine, glycine, and leucine (SMGL) was tested with M9 glucose medium containing SMGL (100 μg/ml each), adenine (50 μg/ml), thymine (50 μg/ml), and calcium pantothenate (1 μg/ml). Figure 2 (right panel) shows that AJB19 is sensitive to AT and inhibition by amino acids SMGL. In addition, strain AJB19 showed a reduced growth rate in glucose minimal medium but was indistinguishable from the wild type in the same medium supplemented with vitamin-free Casamino Acids at 37°C (Difco) (Fig. 3). When strains were incubated in MOPS-glucose-Casamino Acids at 45°C, strain AJB19 displayed a thermosensitive phenotype (Fig. 3). Similar results were obtained using strain AJB42. Taken together, these results show that V. cholerae relA mutants were pleiotropic and exhibited phenotypes typical of E. coli and Salmonella enterica serovar Typhimurium relaxed mutants.
FIG. 3.
V. cholerae relaxed mutants show reduced growth rates in minimal medium and are thermosensitive. Overnight cultures of strains C7258 and AJB19 in LB broth were washed twice in MOPS-glucose minimal medium (MOPS-G) and diluted 1:100 in MOPS-glucose medium or MOPS-glucose supplemented with 1% vitamin-free Casamino Acids (MOPS-G-CAS). Cultures were incubated at 37°C or 45°C with shaking (250 rpm). □, C7258 in MOPS-G; ▪, AJB19 in MOPS-G; ▵, C7258 in MOPS-G-CAS; ▴, AJB19 in MOPS-G-CAS; ⋄, C7258 in MOPS-G-CAS at 45°C; ⧫, AJB19 in MOPS-G-CAS at 45°C.
Relaxed mutants derived from C7258 expressed major virulence factors, formed normal biofilm, are motile, and colonized the suckling mouse intestine.
In view of a previous publication showing that a relA mutant of El Tor biotype strain C6709-1 is defective in the expression of virulence factors (10), we investigated the production of key virulence factors in strain AJB19 and AJB42. Production of CT is an indicator of the expression of positive regulators TcpP/H, ToxR/S, and ToxT (5, 6, 11, 17), while production of HA/protease is an indicator of the expression of regulators HapR, RpoS, and CRP (20). For production of CT, V. cholerae strains were grown in 10-ml AKI cultures (12). The amount of CT in culture supernatants was determined by GM1-enzyme-linked immunosorbent assay using rabbit anti-CT (Sigma Chemical Co.) and peroxidase-conjugated anti-rabbit immunoglobulin G (Sigma Chemical Co.) as primary and secondary antibodies, respectively (8). CT concentrations were calculated using a standard curve constructed with pure CT (Sigma Chemical Co.). As shown in Fig. 4A, the inactivation of relA in AJB19 had a small effect on production of CT that did not reach statistical significance (t test, P > 0.05, n = 3). Strain AJB42 produced low levels of CT compared to C7258; these results were statistically significant (t test, P < 0.05, n = 3) (Fig. 4A). Under identical conditions, negative control AJB216, an aphA mutant derived from strain N16961 (20), produced very low levels of CT (<0.01 μg/ml/OD600). Since expression of the TCP pilus is known to be coregulated with CT, we performed a Western blot analysis to determine the production of the TCP major subunit TcpA in the wild type and the relA mutant. Strains C7258, AJB19, AJB42, and KHT52 (ΔtcpA10) were grown in AKI cultures, and the cell pellet of 0.5 OD600 units was boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer. Samples were resolved in a 12% sodium dodecyl sulfate-acrylamide gel, transferred to an Immobilon-P polyvinylidene fluoride membrane (Millipore), and TcpA detected using rabbit anti-TcpA serum and peroxidase-conjugated anti-rabbit immunoglobulin G. Figure 4B shows that C7258, AJB19, and AJB42 produced a 21-kDa antigen that reacted with anti-TcpA serum and was absent in the tcpA deletion mutant KHT52 (24).
FIG. 4.
Expression of virulence factors and biofilm formation in relA mutants derived from strain C7258. (A) Production of cholera toxin in supernatants of V. cholerae strains grown in AKI cultures. (B) Detection of TcpA in the wild type and in relA mutants grown in AKI cultures. (C) Motility of the wild type and of relA mutants. Strain AJB32 (C6709-1, motY::Km) was used as the negative control. (D) Production of HA/protease in supernatants of V. cholerae strains grown in tryptic soy broth. The HapR-defective strain N16961 was used as the negative control. (E) Biofilm formation by V. cholerae strains expressed as OD570/OD600. The error bars indicate the standard deviations of the means for three independent cultures.
Motility was measured in a swarm assay by stabbing LB plates containing 0.3% agar with overnight cultures of V. cholerae strains grown in LB medium and incubation for 12 h at 37°C. We did not detect any differences in the abilities of C7258, AJB19, and AJB42 to swarm away from the inoculation site (Fig. 4C). The above strains were highly motile compared to strain AJB32, which contains a Kmr insertion in motY (Fig. 4C). V. cholerae motility mutants are defective in biofilm formation (26). Therefore, the motile phenotypes of strains AJB19 and AJB42 are in agreement with their capacities to form wild-type biofilms (Fig. 4E).
For HA/protease production, strains were grown to saturation in Bacto tryptic soy broth (Becton Dickinson and Co.) at 37°C and extracellular protease activity was measured using an azocasein assay (1). In this assay, one azocasein unit is defined as the amount of enzyme producing an increase of 0.01 OD442 units per h. Figure 4D shows that strains AJB19 and AJB42 made slightly elevated amounts of HA/protease compared to C7258, suggesting that (p)ppGpp could have a negative effect. To further investigate this effect, we used the relA isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible plasmid pALS10 (Ampr, lacIq) (23) to overexpress RelA in C7258. Plasmid pALS14 is a control plasmid encoding an IPTG-inducible inactive RelA protein (23). Induction of RelA in pALS10 increases (p)ppGpp levels by augmenting the fraction of ribosomes bound to RelA that can be transiently activated to make (p)ppGpp when a hungry codon is encountered (23). Contrary to results seen with E. coli, no growth inhibition was observed in C7258 overexpressing RelA (data not shown). Figure 4D shows that the induction of full-length RelA in pALS10, but not the inactive RelA encoded by pALS14, severely diminished production of HA/protease. Total RNA was isolated using an RNeasy kit (QIAGEN, Inc.), and RNA samples were analyzed by real-time reverse transcription (RT)-PCR using an iScript one-step RT-PCR kit with SYBR Green (Bio-Rad Laboratories) as previously described (20). The primer pairs 5′-GCACGGCGTTCAGTTATGCTTGTA and 5′-AGGTAAAACGCGCGGTTAAACACG and 5′-GTGCTGTGGATGTCATCGTTGTTG and 5′-CCACCACTTCTTCGCCTTCTTTGA were used to assess hapA mRNA encoding HA/protease and recA mRNA levels, respectively. Relative expression values (R) were calculated using the equation R = 2−(ΔCT target−ΔCT reference), where CT is the fractional threshold cycle. The relative abundance of hapA mRNA in IPTG-induced and noninduced pALS10-containing C7258 were 2.7 ± 0.7 and 1.6 ± 0.1 (n = 3), respectively, indicating that overexpression of RelA diminishes HA/protease production by a posttranscriptional mechanism.
For analysis of biofilm formation, strains were grown for 24 h at 30°C in 100 μl of LB medium in 96-well flat-bottom polystyrene microtiter plates. Growth was measured by reading the OD600, the cultures were discarded, and each well was washed three times with 200 μl of phosphate-buffered saline. Biofilm formation was detected as described in reference 29 by staining with 0.1% crystal violet. Plates were incubated 30 min at 25°C and washed thoroughly. The biofilm was dissolved with dimethyl sulfoxide, and the optical density was read at 570 nm (OD570). Biofilm formation was expressed as the OD570/OD600 ratio (29). As shown in Fig. 4E, strains C7258, AJB19, and AJB42 did not differ significantly in their capacity to form biofilms. Under identical conditions, strain N16961 (HapR defective) made thicker biofilms (OD570/OD600 = 4.0) than strain C7258.
Finally, two competitive colonization experiments were performed by administering mixtures of AJB19 plus C7258 and AJB42 plus C7258 to groups of five 4-day-old CD-1 suckling mice. After an 18-h incubation period, mice were sacrificed by cervical dislocation; the small intestine was withdrawn, homogenized in 5 ml of phosphate-buffered saline (pH 7.4), and plated on LB and LBK agar. The competitive index was calculated as the output ratio of mutant to wild-type divided by the input ratio. The competitive indices for strain AJB19 and AJB42 were 4.1 ± 2.3 and 2.5 ± 1.4, respectively. This result shows that strains AJB19 and AJB42 colonized the mouse intestine as well as or better than C7258. This result is in agreement with AJB19 and AJB42 being motile (Fig. 4C) and expressing TcpA (Fig. 4B).
To investigate the differences between our relA mutant and the previously constructed SHK17 (10), we transferred plasmid pCVDΔrelAK from E. coli to C6709-1, the precursor of SHK17, to generate AJB41 (Fig. 1, lane 5). Results of a guanine nucleotide TLC analysis of AJB41 were indistinguishable from results obtained with AJB19 (Fig. 2, lane 2) and AJB42 (Fig. 5, lanes 3 and 7). C6709-1 expressed slightly less CT than C7258, and its mutant AJB41 showed the lowest levels of CT production (Fig. 4A). However, strain AJB41 still produced significantly more CT than that reported for the relA mutant SHK17 (10).
We investigated if transcription of ctxA and tcpA in AKI medium is paralleled by unusually high RelA activity. Strains C7258 and AJB42 were grown in AKI cultures for 8 h and 16 h. At these time points, cells were collected for preparation of total RNA and for determination of their RelA activity. Real-time RT-PCR was conducted as described above, and the relative abundance of ctxA and tcpA mRNA determined using primers pairs 5′-TATGCCAAGAAGACAGAGTGAGTAC-5′-ACCTGCCAATCCATAACCATCTGC and 5′-CACGATAAGAAAACCGGTCAAGAGG-5′-AGCGACAGCAGCGAAAGCACCTT, respectively. In parallel, 1 OD600 unit of cells was centrifuged, washed three times with MOPS-glucose, and resuspended in MOPS-glucose to an OD600 of 0.4, and aliquots (0.1 ml) were transferred to 96-well microtiter plates. These cells were diluted 1:25 in MOPS-glucose containing 100 μCi/ml of carrier-free [32P]H3PO4 with and without dl-serine hydroxamate and l-valine. No incorporation of 32P to (p)ppGpp could be detected without induction of a stringent response by the addition of dl-serine hydroxamate and l-valine (Fig. 5). This experiment demonstrates that at the time V. cholerae cells are expressing ctxA and tcpA, the (p)ppGpp biosynthetic capacity of the cell is at a basal state. Relative expression of ctxA and tcpA was lower for AJB42 than for C7258 at 8 h but diminished to values similar to that for C7258 at 16 h (Fig. 5).
Discussion. V. cholerae relaxed mutants were thermosensitive and showed reduced growth rate in minimal medium that could be alleviated by the addition of Casamino Acids. This observation suggests that relA mutants display reduced amino acid biosynthetic capacity. The temperature sensitivities of some E. coli relA mutants have been associated with decreased thermotolerance (28). These properties of V. cholerae relA mutants could potentially hinder their ability to colonize specific ecological niches in which a required amino acid is in short supply and/or to survive environmental temperature shifts.
Production of CT and TCP in vivo by El Tor biotype V. cholerae has been shown to occur at low cell density, at which expression of HapR is repressed and AphA activates the expression of tcpPH (22, 25, 30). A V. cholerae relA mutant was recently found to be defective for the expression of CT, TCP, motility, and intestinal colonization (10). V. cholerae strains of El Tor biotype produce CT and TCP when cultured in rich peptone-containing medium or in the protein (mucin)-rich environment of the small intestine. These nutrient-rich environments are unlikely to induce a stringent response in a low-cell-density bacterial population. Thus, it is reasonable to assume that CT, pilus, and flagellum are expressed at a growth phase in which intracellular levels of (p)ppGpp are basal. Figure 5 confirmed that transcription of ctxA and tcpA are not associated with an elevated RelA activity. Our results clearly show that while strain AJB19 and AJB42 are defective in stringent response and display numerous relaxed phenotypes (Fig. 2 and 3), they still produce CT, are motile, and make enough TCP to effectively colonize the suckling mouse intestine (Fig. 4A, D, and E). These results demonstrate that availability of a stringent response is not required for expression of major virulence factors and intestinal colonization. We hypothesize that basal levels of (p)ppGpp, rather than availability of a stringent response, can influence virulence gene expression and perhaps other cell functions (e.g., motility). The conflicting phenotypes displayed by relA mutants AJB19, AJB42, and SHK17 (10) could be due to the presence of an unselected mutation affecting the expression of virulence in SHK17. These differences could also arise from the use of different strains differing in residual SpoT (p)ppGpp synthase activities. It is conceivable that in a relA-positive background, the RelA protein could be transiently activated to adjust (p)ppGpp levels without inducing a full-blown stringent response. Consequently, relA mutants might lack the capacity to readjust (p)ppGpp levels in exponentially growing cells facing occasional environmental changes, such as the shift from static to shaken conditions in AKI cultures, or during intraintestinal growth. RelA could be more or less dispensable for exponentially growing cells, depending on culture conditions and strain-to-strain variation in residual SpoT (p)ppGpp synthase activity. We have cloned DNA sequences flanking the V. cholerae spoT gene (VC2710) in suicide vector pCVD442 (7) but have failed in several attempts to construct a (p)ppGpp-null relAspoT double mutant. This result suggests that a V. cholerae (p)ppGpp-null mutant could be lethal or have unusual nutritional demands.
The finding that AJB19 and AJB42 also produced HA/protease and normal biofilms indicates that, under our culture conditions, RelA is not required for the expression of quorum sensing and stationary phase regulators HapR and RpoS, respectively. In fact, no differences in the relative abundance of rpoS and hapR mRNA between cultures of the wild type and relA mutants grown to stationary phase were observed (data not shown). This result does not rule out the possibility that under conditions of stringent response, the accumulation of (p)ppGpp to high levels could modulate the expression of the above or other regulators. Although overexpression of RelA in V. cholerae severely diminished the production of HA/protease, this condition did not affect transcription of hapA, rpoS, and hapR. Further studies are required to understand the mechanism by which overexpression of RelA affects production of HA/protease.
Acknowledgments
This work was supported by research grants RO1AI63187 to J.A.B. and S06GM008248 to A.J.S. from the National Institute of Allergy and Infectious Diseases and the National Institute of General Medical Sciences, respectively.
We thank Michael Cashel (National Institute of Child Health and Human Development, Bethesda, Md.) for providing plasmids and reference E. coli strains with wild-type and mutant relA alleles and Ronald K. Taylor (Dartmouth Medical School, Hanover, N.H.) for TCP antiserum.
REFERENCES
- 1.Benitez, J. A., A. J. Silva, and R. A. Finkelstein. 2001. Environmental signals controlling production of hemagglutinin/protease in Vibrio cholerae. Infect. Immun. 69:6549-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler, S. M., and A. Camilli. 2004. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 101:5018-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cashel, M. 1994. Detection of (p)ppGpp accumulation patterns in Escherichia coli mutants, p. 341-356. In K. W. Adolph (ed.), Methods in molecular genetics, vol. 3, part A. Academic Press, New York, N.Y. [Google Scholar]
- 4.Cashel, M., D. R. Gentry, V. J. Hernandes, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Humbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1, ASM Press, Washington, D.C. [Google Scholar]
- 5.DiRita, V. J., and J. J. Mekalanos. 1991. Periplasmic interaction between two membrane regulatory proteins ToxR and ToxS results in signal transduction and transcriptional activation. Cell 64:29-37. [DOI] [PubMed] [Google Scholar]
- 6.DiRita, V. J., C, Parsot, G. Jander, and J. J. Mekalanos. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:5403-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fando, R., B. L. Rodriguez, J. Campos, A. Robert, J. L. Pérez, L. García, A. Silva, and J. A. Benítez. 1997. Promoter activities in the Vibrio cholerae CTXΦ prophage. Infect. Immun. 65:1561-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gropp, M., Y. Strausz, M. Gross, and G. Glaser. 2001. Regulation or Escherichia coli relA requires oligomerization of the C-terminal domain. J. Bacteriol. 183:570-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haralalka, S., S. Nandi, and R. K. Bhadra. 2003. Mutation in the relA gene of Vibrio cholerae affects in vitro and in vivo expression of virulence factors. J. Bacteriol. 185:4672-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hase, C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 95:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwanaga, M., K. Yamamoto, Y. Ichinose, N. Nakasone, and M. Tanabe. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 30:1075. [DOI] [PubMed] [Google Scholar]
- 13.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 14.Kovacikova, G., and K. Skorupski. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 46:1135-1147. [DOI] [PubMed] [Google Scholar]
- 15.Kovacikova, G., E. Lin, and K. Skorupski. 2003. The virulence factor activator AphA links quorum sensing to pathogenesis and physiology in Vibrio cholerae by repressing the expression of a penicillin amidase gene on the small chromosome. J. Bacteriol. 185:4825-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merrell, D. S., A. D. Tischler, S. H. Lee, and A. Camilli. 2000. Vibrio cholerae requires rpoS for efficient intestinal colonization. Infect. Immun. 68:6691-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, V. L., and J. J. Mekalanos. 1985. Genetic analysis of the cholera toxin-positive regulator gene toxR. J. Bacteriol. 163:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane protein and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudd, K. E., B. R. Bochner, M. Cashel, and J. R. Roth. 1985. Mutations in the spoT gene of Salmonella typhimurium: effects on his operon expression. J. Bacteriol. 163:534-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva, A. J., and J. A. Benitez. 2004. Transcriptional activation of Vibrio cholerae hemagglutinin/protease expression by the cAMP receptor protein and RpoS. J. Bacteriol. 189:6374-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skorupski, K., and R. K. Taylor. 1997. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin co-regulated pilus in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 94:265-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skorupski, K., and R. K. Taylor. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 59:114-118. [DOI] [PubMed] [Google Scholar]
- 23.Svitil, A. L., M. Cashel, and J. W. Zyskind. 1993. Guanosine tetraphosphate inhibits protein synthesis in vivo. J. Biol. Chem. 268:2307-2311. [PubMed] [Google Scholar]
- 24.Thelin, K. H., and R. K. Taylor. 1996. Toxin co-regulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64:2853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vance, R. E., J. Zhu, and J. J. Mekalanos. 2003. A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun. 71:2571-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 28.Yang, X., and E. R. Ishiguro. 2003. Temperature-sensitive growth and decreased thermotolerance associated with relA mutations in Escherichia coli. J. Bacteriol. 185:5765-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, D., C. Rajanna, W. Sun, and D. K. Karaolis. 2003. Analysis of the Vibrio pathogenicity island-encoded Mop protein suggests a pleiotropic role in the virulence of epidemic Vibrio cholerae. FEMS Microbiol. Lett. 225:311-318. [DOI] [PubMed] [Google Scholar]
- 30.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]