Abstract
Mesophilic Aeromonas spp. constitutively express a single polar flagellum that helps the bacteria move to more favorable environments and is an important virulence and colonization factor. Certain strains can also produce multiple lateral flagella in semisolid media or over surfaces. We have previously reported 16 genes (flgN to flgL) that constitute region 1 of the Aeromonas hydrophila AH-3 polar flagellum biogenesis gene clusters. We identified 39 new polar flagellum genes distributed in four noncontiguous chromosome regions (regions 2 to 5). Region 2 contained six genes (flaA to maf-1), including a modification accessory factor gene (maf-1) that has not been previously reported and is thought to be involved in glycosylation of polar flagellum filament. Region 3 contained 29 genes (fliE to orf29), most of which are involved in flagellum basal body formation and chemotaxis. Region 4 contained a single gene involved in the motor stator formation (motX), and region 5 contained the three master regulatory genes for the A. hydrophila polar flagella (flrA to flrC). Mutations in the flaH, maf-1, fliM, flhA, fliA, and flrC genes, as well as the double mutant flaA flaB, all caused loss of polar flagella and reduction in adherence and biofilm formation. A defined mutation in the pomB stator gene did not affect polar flagellum motility, in contrast to the motX mutant, which was unable to swim even though it expressed a polar flagellum. Mutations in all of these genes did not affect lateral flagellum synthesis or swarming motility, showing that both A. hydrophila flagellum systems are entirely distinct.
Flagellar motility represents an important advantage for bacteria in moving toward favorable conditions or in avoiding detrimental environments, and it allows flagellated bacteria to successfully compete with other microorganisms (21). In addition, motility and flagella play a crucial role in adhesion, biofilm formation, and colonization of several pathogenic bacteria, such as Pseudomonas aeruginosa (59), Salmonella enterica (13), Escherichia coli (51), Helicobacter pylori (19), Vibrio cholerae (22), and Aeromonas hydrophila (43, 53).
Mesophilic Aeromonas spp. are ubiquitous waterborne bacteria and pathogens of reptiles, amphibians, and fish (5). They can be isolated as a part of the fecal flora of a wide variety of other animals, including some used for human consumption, such as pigs, cows, sheep, and poultry. In humans, Aeromonas hydrophila belonging to hybridization group 1 (HG1) and HG3, A. veronii biovar sobria (HG8/HG10), and A. caviae (HG4) have been associated with gastrointestinal and extraintestinal diseases, such as wound infections of healthy humans, and less commonly with septicemia of immunocompromised patients (30). The swimming motility of all mesophilic aeromonads has been linked to a single polar unsheathed flagellum, expressed constitutively, which is required for adherence to and invasion of human and fish cell lines (25, 43, 53, 63). Moreover, 50% to 60% of mesophilic aeromonads are able to produce many unsheathed peritrichous lateral flagella when grown in viscous environments or over surfaces (58), which increase bacterial adherence and are required for swarming motility and biofilm formation (23). The expression of two distinct flagellar systems is relatively uncommon, although it has been observed with Vibrio parahaemolyticus (39), Azospirillum brasilense (45), Rhodospirillum centenum (32), Helicobacter mustelae (49), and Plesiomonas shigelloides (29).
Previous reports described two noncontiguous polar flagellum regions for Aeromonas: (i) a polar flagellum region of A. caviae, containing five genes that encode two tandem flagellins (FlaA and FlaB), a protein involved in flagellum filament length control (FlaG), a HAP-2 distal capping protein (FlaH), and a putative flagellin chaperone (FlaJ) (53); and (ii) a polar flagellum region of A. hydrophila, containing 16 genes which encode chemotaxis (CheV and CheR), hook (FlgE, FlgK, and FlgL), rod (FlgB, FlgC, FlgD, FlgF, FlgG, and FlgJ), L ring (FlgH), and P ring (FlgI) proteins, as well as two specific chaperones (FlgA and FlgN) and the anti-σ28 factor (FlgM) (2).
Although some genes have been described, many others are required for the expression and regulation of Aeromonas polar flagella. This work employed transposon mutagenesis and mutant complementation to isolate the A. hydrophila AH-3 chromosomal regions required for polar flagellum expression. Furthermore, we investigated the distribution of these genes among the mesophilic Aeromonas species, characterized several Aeromonas strains with defined mutations in different polar flagellar genes, and studied their motility, presence or absence of both types of flagella, adherence to HEp-2 cells, and ability to form biofilms.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown on Luria-Bertani (LB) Miller broth and LB Miller agar at 37°C, Aeromonas strains were grown in either tryptic soy broth (TSB) or tryptic soy agar (TSA) at 30°C, and Vibrio cholerae strains were grown in either LB or LB agar with 2 mM glutamine at 37°C. When required, ampicillin (50 μg/ml), kanamycin (50 μg/ml), chloramphenicol (25 μg/ml), rifampin (100 μg/ml), spectinomycin (50 μg/ml), and tetracycline (20 μg/ml) were added to the different media.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| A. hydrophila | ||
| AH-3 | A. hydrophila wild type, serogroup O:34 | 42 |
| AH-405 | AH-3; spontaneous Rifr | 2 |
| AH-4422 | AH-405; maf-1::miniTn5Km-1 | This work |
| AH-4423 | AH-405; flaH::Kmr | This work |
| AH-4424 | AH-405; maf-1::Kmr | This work |
| AH-4425 | In-frame AH-405 ΔflaB mutant | This work |
| AH-4426 | AH-405; flaA::Kmr | This work |
| AH-4427 | AH-4425; flaA::Kmr | This work |
| AH-4440 | AH-405; flhA::miniTn5Km-1 | This work |
| AH-4441 | AH-405; fliM::Kmr | This work |
| AH-4442 | AH-405; flhA::Kmr | This work |
| AH-4443 | AH-405; fliA::Kmr | This work |
| AH-4444 | AH-405; pomB::Kmr | This work |
| AH-4460 | AH-405; motX::miniTn5Km-1 | This work |
| AH-4461 | AH-405; motX::Kmr | This work |
| AH-4462 | AH-405; flrC::Kmr | This work |
| E. coli | ||
| DH5α | F−endA hdsR17(rK− mK+) supE44 thi-1 recA1 gyrA96 φ80lacZ | 27 |
| XL1-Blue | endA1 recA1 hsdR17 supE44 thi-1 gyrA96 relA1 lac (F′ proAB lacIZΔM15 Tn10) | Stratagene |
| S17-1λpir | thi thr-1 leu tonA lacy supE recA::RP4-2 (Tc::Mu) Kmr λpir with miniTn5Kml | 16 |
| MC1061λpir | thi thr-1 leu-6 proA2 his-4 argE2 lacY1 galK2 ara-14 xyl-5 supE44 λpir | 55 |
| Vibrio cholerae | ||
| KKV98 | ΔflrC1 ΔlacZ | 35 |
| KKV59 | ΔflrA1 ΔlacZ | 35 |
| Plasmids | ||
| pGEMT | Cloning vector; Apr | Promega |
| pBCSK | Cloning vector with lacZ gene; Cmr | Stratagene |
| pLA2917 | Cosmid vector; Tcr Kmr | 1 |
| pACYC184 | Plasmid vector; Cmr Tcr | 56 |
| pRK2073 | Helper plasmid; Spr | 55 |
| pFS100 | pGP704 suicide plasmid, pir dependent; Kmr | 55 |
| pDM4 | Suicide plasmid, pir dependent with sacAB genes, oriR6K; Cmr | 44 |
| COS-FLG | pLA2917 with AH-3 polar flagellum region 1 (flgA to flgL); Tcr | 2 |
| pLA-FLA | pLA2917 with AH-3 flaA to maf-1; Tcr | This work |
| pLA-FLIP1 | pLA2917 with AH-3 fliE to cheY; Tcr | This work |
| pLA-FLIP2 | pLA2917 with AH-3 fliQ to orf29; Tcr | This work |
| pLA-MOTX | pLA2917 with AH-3 motX; Tcr | This work |
| pLA-FLR | pLA2917 with AH-3 flrABC; Tcr | This work |
| pACYC-MAF | pACYC184 with AH-3 maf-1; Tcr | This work |
| pACYC-FLR1 | pACYC184 with AH-3 flrBC; Tcr | This work |
| pFS-FLAA | pFS100 with internal fragment of AH-3 flaA gene; Kmr | This work |
| pFS-MAFP | pFS100 with internal fragment of AH-3 maf-1 gene; Kmr | This work |
| pFS-POMB | pFS100 with internal fragment of AH-3 pomB gene; Kmr | This work |
| pFS-MOTX | pFS100 with internal fragment of AH-3 motX gene; Kmr | This work |
| pFS-FLRC | pFS100 with internal fragment of AH-3 flrC gene; Kmr | This work |
| pDM-FLAH | pDM4 with AH-3 flaH::Km; Cmr Kmr | This work |
| pDM-FLIM | pDM4 with AH-3 fliM::Km; Cmr Kmr | This work |
| pDM-FLHA | pDM4 with AH-3 flhA::Km; Cmr Kmr | This work |
| pDM-FLIA | pDM4 with AH-3 fliA::Km; Cmr Kmr | This work |
| pDM-FLAB | pDM4 ΔflaB of AH-3; Cmr | This work |
Tcr, tetracycline resistant; Kmr, kanamycin resistant; Apr, ampicillin resistant; Rifr, rifampin resistant; Cmr, chloramphenicol resistant; Spr, spectinomycin resistant.
Motility assays (swarming and swimming).
Freshly grown bacterial colonies were transferred with a sterile toothpick into the center of swarm agar (1% tryptone, 0.5% NaCl, 0.6% agar) or swim agar (1% tryptone, 0.5% NaCl, 0.25% agar). Agar plates consisting of LB with 0.3% agar and 2 mM glutamine were used to measure Vibrio cholerae motility. The plates were incubated face up for 16 to 24 h at 30°C, and motility was assessed by examining the migration of bacteria through the agar from the center towards the periphery of the plate. Moreover, swimming motility was assessed by light microscopy observations with liquid media.
Transmission electron microscopy (TEM).
Bacterial suspensions were placed on Formvar-coated grids and negative stained with a 2% solution of uranyl acetate, pH 4.1. Preparations were observed with a Hitachi 600 transmission electron microscope.
MiniTn5Km-1 mutagenesis.
Conjugal transfer of transposition element miniTn5Km-1 from E. coli S17-1λpirKm-1 (16) to A. hydrophila AH-405 (AH-3, rifampin resistant) was carried out in a conjugal drop incubated for 6 h at 30°C with the ratio 1:5:1, corresponding to S17-1λpirKm-1, AH-405, and HB101 pRK2073 (helper plasmid), respectively. Serial dilutions of the mating mix were plated on TSA supplemented with rifampin and kanamycin in order to select mutants.
DNA techniques.
DNA manipulations were carried out essentially as previously described (56). DNA restriction endonucleases, T4 DNA ligase, E. coli DNA polymerase Klenow fragment, and alkaline phosphatase were used as recommended by the suppliers. PCR was performed using Taq DNA polymerase (Invitrogen) in a gene amplifier PCR system 2400 thermal cycler (Perkin-Elmer). Colony hybridizations were carried out by colony transfer onto positive nylon membranes (Roche) and then lysed according to the manufacturer's instructions. Probe labeling with digoxigenin, hybridization, and detection (Amersham) were carried out as recommended by the suppliers.
Cloning of DNA flanking miniTn5Km-1 insertions.
Chromosomal DNA of miniTn5Km-1 mutants was digested with EcoRI, PstI, and EcoRV, purified, ligated into the vector pBCSK (Stratagene), and introduced into E. coli XL1-Blue. Recombinant plasmids containing the transposon with flanking insertions were selected in LB plates supplemented with kanamycin and chloramphenicol. The miniTn5Km-1 flanking sequences were obtained by using primers specific to the I and O ends of miniTn5Km-1 (5′-AGATCTGATCAAGAGACAG-3′ and 5′-ACTTGTGTATAAGAGTCAG-3′, respectively), as well as M13for and T3 primers from the plasmid vector used.
Nucleotide sequencing and computer sequence analysis.
Plasmid DNA for sequencing was isolated with a QIAGEN plasmid purification kit (QIAGEN, Inc., Ltd.) as recommended by the suppliers. Double-stranded DNA sequencing was performed by using the Sanger dideoxy-chain termination method (57) with an ABI Prism dye terminator cycle sequencing kit (Perkin-Elmer). Custom-designed primers used for DNA sequencing were purchased from Amersham Biosciences.
The DNA sequence was translated in all six frames, and all open reading frames (ORFs) greater than 100 bp were inspected. Deduced amino acid sequences were inspected in the GenBank, EMBL, and SwissProt databases by using the BLASTX, BLASTP, or PSI-BLAST network service at the National Center for Biotechnology Information (NCBI) (3). A protein family profile was performed using the Protein Family Database Pfam at the Sanger Center (6). Determination of possible terminator sequences was done by using the Terminator program from the Genetics Computer Group package (Madison, Wisconsin). Other online sequence analysis services were also used.
RT-PCR.
Total RNA was isolated from A. hydrophila AH-3 grown in liquid medium (TSB) and plates by Trizol reagent (Invitrogen). To ensure that RNA was devoid of contaminating DNA, the preparation was treated with RNase-free DNase I, amplification grade (Invitrogen). The isolated RNA was used as a template in reverse transcriptase (RT) PCR, utilizing a Thermoscript RT-PCR system (Invitrogen) according to the manufacturer's instructions. PCR without reverse transcriptase was also performed to confirm the absence of contaminating DNA in the RNA samples. RT-PCR amplifications were performed at least twice with total RNA preparations obtained from minimum of two independent extractions. The RT-PCR and PCR products were analyzed by gel electrophoresis.
Mutant construction.
To obtain single defined insertion mutants with mutations in genes flaA, maf-1, pomB, motX, and flrC, we used a method based on suicide plasmid pFS100 (55). Briefly, an internal fragment of the selected gene was amplified by PCR, ligated into pGEM-Teasy (Promega), and transformed into E. coli XL1-Blue. The DNA insert was recovered by EcoRI restriction digestion and was ligated into EcoRI-digested and phosphatase-treated pFS100. The ligation product was transformed into E. coli MC1061 (λpir) and selected for kanamycin resistance (Kmr). Triparental mating with the mobilizing strain HB101/pRK2073 was used to transfer the recombinant plasmid into A. hydrophila AH-405 rifampin-resistant (Rifr) strains to obtain defined insertion mutants, selecting for Rifr and Kmr.
To obtain strains with mutations in flaH, fliM, flhA, and fliA, the genes were amplified by PCR, ligated into vector pGEMTeasy (Promega), and transformed into E. coli XL1-Blue. The Tn5-derived kanamycin resistance cartridge (nptll) from pUC4-KIXX was inserted into each of these genes. The cartridge contains an outward-reading promoter that ensures the expression of downstream genes when inserted in the correct orientation (9); however, such an insertion will alter the regulation of such genes. The SmaI-digested cassette was inserted into a restriction site internal to each gene, and the presence of a single HindIII site in the SmaI-digested cassette allowed its orientation to be determined. Constructs containing the mutated genes were ligated into suicide vector pDM4 (44), electroporated into E. coli MC1061 (λpir), and plated on chloramphenicol plates at 30°C. Plasmids with mutated genes were transferred into A. hydrophila AH-405 rifampin-resistant (Rifr) strains by triparental mating using E. coli MC1061 (λpir) containing the insertion constructs and the mobilizing strain HB101/pRK2073. Transconjugants were selected on plates containing chloramphenicol, kanamycin, and rifampin. PCR analysis confirmed that the vector had integrated correctly into the chromosomal DNA. To complete the allelic exchange, the integrated suicide plasmid was forced to recombine out of the chromosome by addition of 5% sucrose to the agar plates. The pDM4 vector contains sacB, which produces an enzyme that converts sucrose into a product that is toxic to gram-negative bacteria. Transconjugants surviving on plates with 5% sucrose (Rifr, Kmr, and chloramphenicol sensitive [Cms]) were chosen and confirmed by PCR.
The chromosomal in-frame flaB deletion mutant A. hydrophila AH-4425 was constructed by allelic exchange as described by Milton et al. (44). Briefly, DNA regions flanking the flaB gene were amplified using the primer pairs A (5′-CGCGGATCCCCTGGTCAAGAGATGGAA-3′) and B (5′-CCCATCCACTAAACT TAAACAGGCGTTCAGTGATGAAGT-3′) and C (5′-TGTTTAAGTTTAGTGGATGGGTCCCTGCTGGGTTAACAG-3′) and D (CGCGGATCCCTGGGCATTGGTCTT TTT-3′) in two sets of asymmetric PCRs to amplify DNA fragments of 592 (AB) and 646 (CD) bp, respectively. DNA fragment AB contains nucleotide 1194, outside flaB, to nucleotide 1786, corresponding to the 14th codon of flaB. DNA fragment CD contains nucleotide 2636, corresponding to the first base in the 328 codon of flaB, to nucleotide 3282, inside the flaH gene. DNA fragments AB and CD were annealed at their overlapping regions (underlined letters in primers B and C) and amplified as a single fragment by using primers A and D. The fusion product was purified, BamHI digested (the BamHI site is double underlined in primers A and D), ligated into BglII-digested and phosphatase-treated pDM4 vector (44), electroporated into E. coli MC1061 (λpir), and plated on chloramphenicol plates at 30°C to obtain plasmid pDM-FLAB. Introduction of plasmid pDM-FLAB into the A. hydrophila AH-405 Rifr strain was performed as previously described. Transconjugants were selected on plates containing chloramphenicol and rifampin. PCR analysis confirmed that the vector had integrated correctly into the chromosomal DNA. After sucrose treatment, transformants (Rifr and Cms) were chosen and confirmed by PCR.
Plasmid construction.
Plasmid pACYC-MAF, containing the complete maf-1 gene, and plasmid pACYC-FLR1, containing flrB and flrC genes from A. hydrophila AH-3, were obtained by PCR amplification of genomic DNA using oligonucleotides 5′-GGGGTGGGATTAGGATACC-3′ and 5′-GGGATACATGAGCCTACGG-3′ to generate a band of 1,829 bp and 5′-TGGAGCACAGTGCCAGTTA-3′ and 5′-TCCCCCTGCTCTACCAATA-3′ to generate a band of 2,857 bp, respectively. The amplified bands were ligated into pGEM-Teasy (Promega) and transformed into E. coli XL1-Blue. The DNA insert was recovered by EcoRI restriction digestion and ligated into EcoRI phosphatase-treated pACYC184 vector (56), introduced into E. coli DH5α to generate plasmid pACYC-MAF.
Whole-cell protein preparation and immunoblotting.
Whole-cell proteins were obtained from Aeromonas strains grown at 30°C. Equivalent numbers of cells were harvested by centrifugation, and the cell pellet was suspended in 50 to 200 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer and boiled for 5 min. Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transfer to nitrocellulose membranes, the membranes were blocked with bovine serum albumin (3 mg/ml) and probed with either polyclonal rabbit anti-polar or anti-lateral flagellin antibody (1:1,000), previously obtained (23). The unbound antibody was removed by three washes in phosphate-buffered saline (PBS), and a goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:1,000) was added. The unbound secondary antibody was removed by three washes in PBS. The bound conjugate was then detected by the addition of 2 ml 0.5% 4-chloro-1-naphthol (Sigma) prepared in methanol and diluted in 8 ml PBS containing 50 μl of 30% H2O2.
Adherence assay to HEp-2 cells.
Tissue culture was maintained as described by Thornley et al. (63). The adherence assay was conducted as a slight modification of that described by Carrello et al. (12). Bacteria were grown statically in brain heart infusion broth at 37°C, harvested by gentle centrifugation (1,600 × g for 5 min), and resuspended in PBS (pH 7.2) at approximately 107 CFU/ml (A600 of ∼0.07). The monolayer was infected with 1 ml of the bacterial suspension for 90 min at 37°C in 5% CO2. Following infection, the nonadherent bacteria were removed from the monolayer by three washes with PBS. The remaining adherent bacteria and the monolayers were then fixed in 100% methanol for 5 min. Methanol was removed by washing monolayers and bacteria with PBS, and the HEp-2 cells with the adherent bacteria were stained for 45 min in 10% (vol/vol) Giemsa stain (BDH) prepared in Giemsa buffer. The coverslips were air dried, mounted, and viewed by oil immersion under a light microscope. Twenty HEp-2 cells/coverslips were randomly chosen, and the number of bacteria adhering to HEp-2 cells was recorded. Assays were carried out in duplicate or triplicate.
Biofilm formation.
Quantitative biofilm formation was performed in a microtiter plate as described previously (51), with minor modifications. Briefly, bacteria were grown on TSA and several colonies were gently resuspended in TSB (with or without the appropriate antibiotic); 100-μl aliquots were place in a microtiter plate (polystyrene) and incubated for 48 h at 30°C without shaking. After the bacterial cultures were poured out, the plate was washed extensively with water, fixed with 2.5% glutaraldehyde, washed once with water, and stained with 0.4% crystal violet solution. After solubilization of the crystal violet with ethanol-acetone (80/20, vol/vol), the absorbance was determined at 570 nm.
Statistical analysis.
The differences in adherence to HEp-2 cells or biofilm formation in vitro between the wild-type and mutant strains were analyzed by the t test using Microsoft Excel software.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to the GenBank/EMBL database under accession numbers DQ119104, DQ124696, DQ124697, and DQ124698.
RESULTS
Isolation and characterization of A. hydrophila AH-3 mutants with reduced swimming phenotype in plates.
In order to find the chromosomal regions of A. hydrophila AH-3 involved in polar flagellum biogenesis, we performed miniTn5Km-1 mutagenesis using A. hydrophila AH-405 (AH-3, rifampin resistant) as the recipient strain, and transconjugants were screened for reduced swimming motility in swim agar. From 7,000 transconjugants analyzed by light microscopy, 23 transposon insertion mutants exhibited a reproducible reduction in swim agar and inability to move in liquid media. Transposon insertion mutants unable to swim were analyzed by transmission electron microscopy after growth in liquid media and on plates; these were subsequently divided in two groups on the basis of their ability to produce polar flagella. The first group was unable to produce polar flagella but was able to produce lateral flagella, and the second group was unable to swim but showed both flagellum types.
As no EcoRV restriction sites were present in the transposon, all altered-motility mutants were analyzed for the presence of the transposon by Southern hybridization of EcoRV chromosomal DNA digestions. A single band was detected in every mutant, indicating that each mutant had a single copy of the mini-transposon in its genome.
Sequence analysis of miniTn5Km-1 interrupted genes.
The DNA flanking the transposon was isolated and cloned into pBCSK (see Materials and Methods). From 22 mutants of the first transposon insertion mutant group, 18 ORFs that shared homology with different A. hydrophila AH-3 polar flagellum structural genes in the flg locus (2) were revealed: two mutants showed the transposon inserted in flgN, two in flgM, four in flgB, three in flgE, three in flgK, and four in flgL (2). However, the encoded product of sequences flanking the insertion of four more mutations had not previously been described for Aeromonas. The predicted amino acid sequences from mutant AH4422 encoded a product similar to an uncharacterized protein of Shewanella oneidensis (GenBank no. SO3259), Clostridium acetobutylicum (CAC2196), and Campylobacter jejuni (Cj1337), a group of proteins thought to be involved in phase-variable flagellum-mediated motility and flagellin modification (33). Of the other ORF products, AH-4440 shared homology with FlhA of Shewanella oneidensis and V. parahaemolyticus, a component of the polar flagellar export machinery (60). The only representative mutant of the second mutant group, AH-4460, revealed an ORF whose predicted amino acid sequence shared homology with the sodium-type flagellar motor component MotX of Vibrio alginolyticus.
Organization of A. hydrophila AH-3 polar flagellum region 2.
A cosmid genomic library of A. hydrophila AH-3 (41) was screened by colony blotting using a DNA probe to the maf-like transposon flanking sequences from mutant AH-4422. Several positive recombinant clones were identified, of which clone pLA-FLA was chosen because it was able to complement mutant AH-4422.
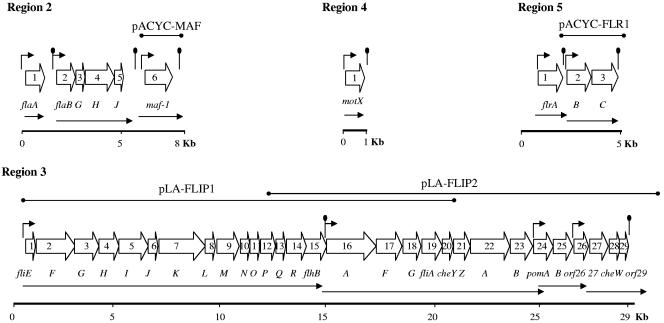
Nucleotide sequencing of pLA-FLA revealed six complete ORFs (Fig. 1) transcribed in the same direction, five of which are related to Aeromonas salmonicida and A. caviae polar flagella (53, 65). Flanking the 3′ end of fla polar flagella loci, 70 bp downstream of ORF6, and transcribed in the opposite direction was the stop codon of a incomplete ORF whose amino acid sequence exhibited homology to the OrfB transposase of Pseudomonas putida, as well as transposases of other bacteria. ORF1 (flaA) was separated from ORF2 (flaB) by 585 bp, and ORF5 (flaJ) was separated from ORF6 (maf-1) by 1,060 bp. The other ORFs were located one behind the other, with intergenic regions less than 80 bp. Sequences defining putative ribosome binding sites were found upstream of each of the ORF's start codons. Data summarizing the locations of the six complete ORFs are shown in Table 2. Sequence analysis in silico showed three possible transcriptional terminator rho-independent sequences downstream of ORF1 (flaA), ORF5 (flaJ), and ORF6 (maf-1) and putative σ28 promoter sequences upstream of ORF1 (flaA), ORF2 (flaB), and ORF6 (maf-1), as well as putative σ54 promoter sequences upstream of ORF2 (flaB) (Fig. 1). RT-PCR using specific primers and total RNA from A. hydrophila AH-3 grown in liquid (TSB) and solid (TSA) media demonstrated amplifications between ORFs 2 and 5 (flaB to flaJ) under both conditions. However, no amplifications were obtained with oligonucleotide pairs from ORFs 1 (flaA) to 2 (flaB) and ORFs 5 (flaJ) to 6 (maf-1), confirming that flaA and maf-1 are transcribed independently (Fig. 1).
FIG. 1.
Genetic organization of A. hydrophila AH-3 polar flagellum regions 2, 3, 4, and 5. ORFs and their transcriptional directions are indicated by large open arrows and named after their homologues in other bacterial species. Black arrows above the ORFs indicate the locations of putative promoter sequences. Lollipop structures depict the approximate positions of the putative transcriptional rho-independent terminators. We also show the clusters determined by RT-PCR in the sequenced regions (black arrows below the ORFs) and genes contained in different plasmids.
TABLE 2.
Characteristics of A. hydrophila AH-3 polar flagellum gene regions 2, 4, and 5
| ORF and region | Nucleotide positions | Protein size (aa)a | Molecular mass (kDa) | pI | Predicted function | Homologous gene | Identity/ similarity (%) |
|---|---|---|---|---|---|---|---|
| Region 2 | |||||||
| 1 | 248-1160 | 304 | 32.4 | 8.99 | Flagellin | flaA of Aeromonas salmonicida | 81/84 |
| 2 | 1745-2648 | 301 | 31.8 | 8.86 | Flagellin | flaB of Aeromonas salmonicida | 83/85 |
| 3 | 2689-3127 | 146 | 16 | 4.85 | Filament length control | flaG of Aeromonas salmonicida | 69/71 |
| 4 | 3157-4564 | 469 | 49.9 | 8.89 | HAP-2 | flaH of Aeromonas caviae | 60/76 |
| 5 | 4592-5012 | 140 | 15.9 | 5.29 | Chaperone | flaJ of Aeromonas caviae | 58/79 |
| 6 | 6072-7404 | 444 | 50.9 | 5.61 | Motility accessory factor | maf-1 of Campylobacter jejuni | 26/41 |
| Region 4 | |||||||
| 1 | 171-1104 | 311 | 32.8 | 5.31 | Stator | motX of Vibrio alginolyticus | 54/73 |
| Region 5 | |||||||
| 1 | 811-2235 | 475 | 53.2 | 5.24 | Flagellum regulator | flrA of Vibrio cholerae | 53/65 |
| 2 | 2345-3379 | 344 | 37.2 | 6.18 | Two-component sensor kinase | flrB of Vibrio cholerae | 40/53 |
| 3 | 3376-4716 | 447 | 49.4 | 5.16 | Two-component response kinase | flaM of Vibrio parahaemolyticus | 60/73 |
aa, amino acids.
Table 2 summarizes the characteristics of the individual proteins and predicted functions analyzed using the BLASTP program (3) of the NCBI database. ORFs 1 to 5 (flaA to flaJ) had the same organization and were similar to those previously reported for the polar flagellins of A. salmonicida and A. caviae (53, 65). However, downstream of ORF5 (flaJ) was ORF6, whose deduced amino acid sequence contained the DUF115 protein family domain and exhibited homology to the motility accessory factor Maf-1 of Campylobacter jejuni (26% identity), thought to be involved in a slipped-strand mispairing mechanism of phase variation (33).
Organization of A. hydrophila AH-3 polar flagellum region 3.
The cosmid library of A. hydrophila AH-3 (48) was screened by colony blotting using DNA probes to the A. hydrophila AH-3 flhA-like gene. Several positive recombinant clones were identified, of which pLA-FLIP1 and pLA-FLIP2 were selected, as they were able to complement the AH-4440 mutant.
Nucleotide sequencing of the overlapping cosmids pLA-FLIP1 (ORFs 1 to 20) and pLA-FLIP2 (ORFs 13 to 29) revealed 29,799 bp which contained 29 ORFs transcribed in the same direction (Fig. 1), most of which encoded homologues ofproteins involved polar flagellum biosynthesis and chemotaxis in different bacteria, such as Photobacterium profundum, Shewanella oneidensis, and V. parahaemolyticus (41, 60). ORFs 1 to 23 (fliE to cheB) and ORFs 26 to 29 had the same organization as region 2 of the V. parahaemolyticus polar flagellum system (Fig. 2). However, unlike V. parahaemolyticus, A. hydrophila lacked the flaF to flaM genes in this chromosomal region but contained the pomAB genes (ORFs 24 to 25) which are located in region 3 of the V. parahaemolyticus polar flagellum system (Fig. 2). Upstream of A. hydrophila ORF1 (fliE) was a truncated ORF transcribed in the opposite direction, with homology to different bacterial regulatory proteins related to the AraC family, and downstream of ORF29 was another truncated ORF transcribed in the same direction, which shared homology with the CcmA ATP-binding subunit of a putative ABC-type heme exporter. Sequences defining putative ribosome binding sites were found upstream of each ORF's start codons. Most ORFs were overlapping or with intergenic regions less than 80 bp. Data summarizing the locations of the 29 complete ORFs are shown in Table 3. Sequence analysis in silico showed possible transcriptional terminator rho-independent sequences downstream of ORF15 (flhB) and ORF29. Putative σ54 promoter sequences were found upstream of ORF1 (fliE), ORF16 (flhA), and ORF24 (pomA), and a putative promoter sequence was found upstream of ORF26 (Fig. 1). RT-PCRs using specific primers showed amplifications between ORFs 1 and 15 (fliE to flhB), ORFs 16 and 27 (flhA to cheB), ORFs 24 and 25 (pomAB), and ORFs 26 to 29. However, no amplifications were obtained with oligonucleotide pairs from ORFs 15 (flhB) to 16 (flhA), ORFs 23 (cheB) to 24 (pomA), and ORFs 25 (pomB) to 26. The RT-PCR results showed that fli and flh A. hydrophila polar genes are distributed in four clusters, although no rho-independent terminator sequences were found downstream of ORF23 (cheB) and ORF25 (pomB).
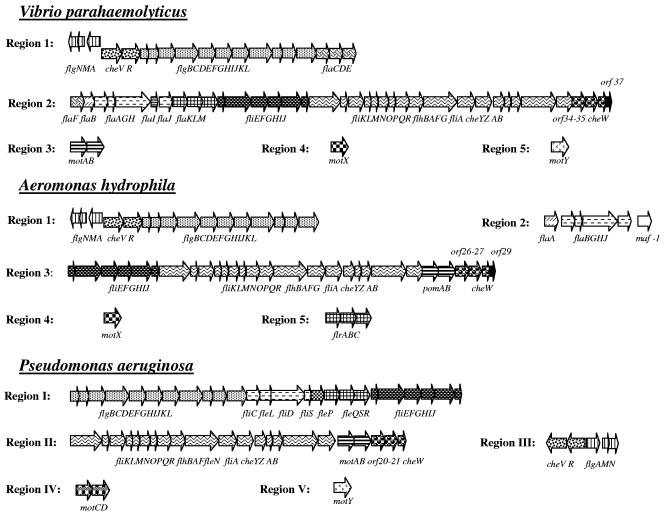
FIG. 2.
A. hydrophila AH-3 polar flagellum genetic regions compared to polar flagellum genetic regions of V. parahaemolyticus and P. aeruginosa. Arrows with the same shading correspond to homologous genes among these bacteria.
TABLE 3.
Characteristics of A. hydrophila AH-3 polar flagellum gene region 3
| ORF | Nucleotide positions | Protein size (aa)a | Molecular mass (kDa) | pI | Predicted function | Homologous protein | Identity/ similarity (%) |
|---|---|---|---|---|---|---|---|
| 1 | 1607-1922 | 105 | 11.4 | 4.76 | MS ring/rod adapter | FliE of Vibrio vulnificus | 57/76 |
| 2 | 1937-3647 | 570 | 62.1 | 4.88 | M ring | FliF of Shewanella oneidensis | 50/67 |
| 3 | 3642-4767 | 375 | 41.7 | 4.78 | Switch | FliG of Shewanella oneidensis | 71/85 |
| 4 | 4773-5584 | 271 | 29.6 | 4.66 | Export/assembly | FliH of Photobacterium profundum | 38/56 |
| 5 | 5620-6949 | 443 | 47.7 | 6.04 | Export ATP synthase | FliI of Vibrio parahaemolyticus | 68/82 |
| 6 | 6978-7416 | 146 | 17.0 | 9.47 | Export/assembly | FliJ of Pseudomonas syringae | 39/62 |
| 7 | 7514-9452 | 646 | 66.5 | 4.97 | Hook length | FliK of Vibrio parahaemolyticus | 43/69 |
| 8 | 9512-10025 | 171 | 18.4 | 6.37 | Flagellar protein | FliL of Photobacterium profundum | 55/73 |
| 9 | 10037-11105 | 356 | 40.4 | 4.81 | Switch | FliM of Photobacterium profundum | 80/91 |
| 10 | 11159-11549 | 130 | 14.1 | 4.39 | Switch | FliN of Photobacterium profundum | 80/90 |
| 11 | 11511-11994 | 161 | 13.7 | 6.69 | Export/assembly | FliO of Vibrio parahaemolyticus | 43/70 |
| 12 | 11983-12760 | 259 | 28.3 | 5.83 | Export/assembly | FliP of Vibrio parahaemolyticus | 63/78 |
| 13 | 12790-13057 | 89 | 9.7 | 4.35 | Export/assembly | FliQ of Vibrio vulnificus | 67/82 |
| 14 | 13112-13934 | 274 | 29.5 | 5.49 | Export/assembly | FliR of Shewanella oneidensis | 51/73 |
| 15 | 14023-15151 | 376 | 41.9 | 9.42 | Export/assembly | FlhB of Shewanella oneidensis | 46/63 |
| 16 | 15261-17361 | 700 | 75.4 | 5.87 | Export/assembly | FlhA of Shewanella oneidensis | 60/73 |
| 17 | 17380-18796 | 472 | 51.6 | 6.71 | Polar flagellar site determinant | FlhF of Vibrio vulnificus | 50/65 |
| 18 | 18791-19673 | 294 | 32 | 8.73 | Flagellar number regulator | FlhG of Shewanella oneidensis | 73/87 |
| 19 | 19668-20385 | 239 | 26.6 | 5.56 | σ28 | FliA of Shewanella oneidensis | 66/80 |
| 20 | 20511-20880 | 123 | 13.8 | 6.73 | Chemotaxis | CheY of Shewanella oneidensis | 91/95 |
| 21 | 20895-21444 | 183 | 21 | 6.42 | Chemotaxis | CheZ of Shewanella oneidensis | 58/77 |
| 22 | 21601-23743 | 714 | 76.1 | 4.70 | Chemotaxis | CheA of Photobacterium profundum | 61/69 |
| 23 | 23778-24891 | 371 | 39.8 | 8.94 | Chemotaxis | CheB of Vibrio vulnificus | 67/78 |
| 24 | 24896-25631 | 245 | 26.9 | 5.03 | Motor | MotA of Pseudomonas aeruginosa | 50/69 |
| 25 | 25636-26545 | 303 | 33 | 6.77 | Motor | MotB of Pseudomonas putida | 30/52 |
| 26 | 26552-27344 | 264 | 29.6 | 9.17 | Unknown | Soj-like protein of Photobacterium profundum | 63/83 |
| 27 | 27343-28213 | 290 | 32.7 | 4.53 | Unknown | VP2226 of Vibrio parahaemolyticus | 50/67 |
| 28 | 28285-28771 | 162 | 17.9 | 4.23 | Chemotaxis | CheW of Vibrio parahaemolyticus | 78/93 |
| 29 | 28786-29172 | 130 | 14.4 | 5.28 | Unknown | VC2058 of Photobacterium profundum | 50/73 |
aa, amino acids.
Table 3 shows the characteristics of the individual proteins and their protein homologues analyzed using the BLASTP program (3) of the NCBI database. ORF1, ORF2, ORF7, and ORF8 encoded proteins that exhibited more than 40% identity to different polar flagellum basal body and hook proteins FliE (57% identity) of Vibrio vulnificus, FliF (50% identity) of S. oneidensis, FliK (43% identity) of V. parahaemolyticus, and FliL (55% identity) of P. profundum, respectively. ORF3, ORF9, and ORF10 predicted amino acid sequences exhibited high homology (>70% identity) to the polar flagellum motor switch proteins FliG, FliM, and FliN of different bacteria, respectively. ORFs 4 to 6 and ORFs 11 to 16 exhibited homologies to different export and assembly polar flagellum proteins, with ORF4 and ORF6 having 38% identity to the ATP synthase-negative regulator FliH of Photobacterium profundum and 39% identity to a potential nonspecific chaperone FliJ of Pseudomonas syringae, respectively. ORF5, which is homologous to FliI, a polar flagellum ATP synthase, and ORFs 11 to 16 (FliOPQR and FlhBA homologues) displayed homologies greater than 43% identity to Vibrio spp., S. oneidensis, and P. profundum export and assembly proteins. ORFs 17 and 18 matched with a pair of proteins that could be involved in the control of site selection of flagellum insertion as well as flagellum number. ORF17 (FlhF homologue) showed homology to FlhF of V. vulnificus and V. parahaemolyticus (50% identity), a GTP-binding signal recognition particle pathway protein required for flagellar synthesis in Bacillus subtilis (38), Campylobacter jejuni (28), and Helicobacter pylori (47) and required for polar flagellar placement in P. putida (50). Furthermore, it has recently been demonstrated with V. cholerae that FlhF can regulate class III flagellar transcription (46). ORF18 (FlhG homologue) encoded a protein with a CbiA domain and is a MinD-related protein involved in septum site determination in Bacillus subtilis (11, 38) as well as in regulation of flagellum number in P. aeruginosa (14). The predicted amino acid sequence of ORF19 (FliA homologue) exhibited 66% identity to the RNA polymerase σ27 factor of S. oneidensis and 62% identity to the RNA polymerase σ28 factor of V. vulnificus. ORFs 20 to 23 and ORF28 shared 58 to 91% identity to different chemotaxis proteins (7, 10, 37). ORFs 20 to 23 and ORF28 matched with CheY, CheZ, CheA, CheB, and CheW, respectively (Table 3). Proteins encoded by ORF24 and ORF25 showed homology (50% and 30% identity, respectively) to the cytoplasmic membrane proteins MotA and MotB (of Pseudomonas spp.), respectively, which constitute the stator of the flagellum motor. Together, these proteins are involved in the formation of a proton or sodium-conducting channel to generate rotational motion in the proton-type or sodium-type flagellum motor (31, 61). ORF25 (MotB) contained an OmpA domain at the C-terminal sequence, which is probably involved in peptidoglycan interaction (17). Although ORF26, ORF27, and ORF29 appear not to be involved in polar flagellum formation, their locations, downstream of chemotaxis genes, are conserved in different bacteria that possess polar flagella. ORF26 encodes a putative protein with a CbiA domain that exhibits high homology to Soj-like proteins as well as other ATPase proteins involved in chromosome partitioning (36). The predicted protein for ORF27 had a CheW-like domain which is found in proteins involved in the two-component signaling systems regulating bacterial chemotaxis (7) and matched to VP2226 of V. parahaemolyticus (Table 3). ORF29 showed sequence homology (41 to 50% identity) (Table 3) to hypothetical proteins located at the end of polar flagellum regions of different bacteria (41).
Sequence analysis of A. hydrophila AH-3 polar flagellum region 4.
The A. hydrophila AH-3 genomic library (48) was screened by colony blotting using a DNA probe to the motX-like gene of A. hydrophila AH-3, leading to the identification of clone pLA-MOTX, which was able to complement the AH-4460 mutant. By sequence analysis of pLA-MOTX, we found a complete ORF of 933 bp, encoding a protein of 311 amino acids and a predicted molecular weight of 32.8 kDa, that was homologous to MotX of V. alginolyticus (54% identity), involved in the sodium-type motor formation in V. parahaemolyticus (35) (Table 2). A putative Shine-Dalgarno sequence is positioned 7 bp upstream of the ATG start codon, and there isalso a putative σ28 promoter sequence (CTAAG-N15-GCCGATAA) 26 bp upstream of the start codon. The nucleotide sequence GACCCACAGATCTGCATCTGTGGGTC downstream of the motX-like TGA stop codon could provide a termination stem-loop structure (Fig. 1). Moreover, 86 nucleotides upstream of the motX-like start codon is found a putative terminator sequence, AAGGGGAGCTTCGGCTCCCCTT, and the stop codon (125 bp upstream) of an incomplete ORF whose deduced amino acid sequence is similar to that of the malate dehydrogenase of V. cholerae. Downstream of motX, transcribed in the opposite direction, is an incomplete ORF which showed 55% identity to a Yersinia pestis flavohemoprotein which plays a central role in the inducible response to nitrosative stress.
Cloning and sequence analysis of A. hydrophila AH-3 polar flagellum regulatory region 5.
The A. hydrophila AH-3 genomic library (48) was transferred by mating into the rifampin-resistant Vibrio cholerae flrC in-frame deletion mutant strain KKV98 (35). Transconjugants were selected for rifampin and tetracycline resistance and inoculated onto 0.3% agar LB plates supplemented with 2 mM glutamine and tetracycline. The complemented colonies which spread on the plates were isolated, and the plasmid pLA-FLR was recovered. Sequence analysis of pLA-FLR revealed three complete ORFs transcribed in the same direction, which encoded proteins related to the polar flagellum master regulatory proteins of different Vibrio species (35, 41, 52, 60). ORF2 (flrB) began 106 bp downstream of the ORF1 (flrA) stop codon, and ORF2 and ORF3 (flrC) are overlapping sequences. Moreover, 220 nucleotides upstream of the ORF1 (flrA) start codon is found a putative rho-independent terminator sequence and the stop codon (260 bp upstream) of a incomplete ORF whose encoded protein has a topoisomerase DNA binding C4 zinc finger domain.
Nucleotide analysis in silico shows putative Shine-Dalgarno sequences upstream of each ORF, two putative σ54 promoter sequences upstream of ORF1 and ORF2 (70 and 35 bp, respectively), and two rho-independent terminator sequences downstream of ORF1 and ORF3 (3 and 31 bp, respectively). RT-PCR using specific primers showed amplifications between ORFs 2 and 3 (flrB to flrC), and no amplifications were obtained with oligonucleotide pairs from ORFs 1 (flrA) and 2 (flrB) (Fig. 1). A search of the protein databases revealed that ORF1 (flrA) encoded a cytoplasmic protein of 475 amino acids which displayed more than 50% identity to the polar flagellum proteins FlrA of V. cholerae and FlaK of V. parahaemolyticus. ORF2 (flrB) encoded a transmembrane protein of 344 amino acids and ORF3 (flrC) a cytoplasmic protein of 447 amino acids, which are homologous to two-component flagellar regulatory proteins. ORF2 (flrB) encoded a protein which exhibited 38 to 40% identity to different two-component sensor kinases, such as FlrB of V. cholerae and FlaK of V. parahaemolyticus, and ORF3 (flrC) encoded a protein which shared high homology with the two-component response regulator proteins FlaM of V. parahaemolyticus (60% identity) and FlrC of V. fischeri (58% identity) (Table 2).
Construction of polar flagellum mutants and complementation studies.
We constructed several mutants of A. hydrophila AH-3 to study the Aeromonas polar flagellum regions and to determine if they had any role in lateral flagellum formation. We obtained an in-frame flaB deletion mutant (AH-4425) and the following defined insertion mutants: flaA, flaH, and maf-1, with mutations in polar flagellum region 2; fliM, flhA, fliA, and pomB, with mutations in polar flagellum region 3; motX, with a mutation in polar flagellum region 4; and flrC, with a mutation in polar flagellum region 5. Amplified internal fragments of flaA, maf-1, pomB, motX, and flrC were ligated into pFS100 to construct plasmids pFS-FLAA (flaA), pFS-MAFP (maf-1), pFS-POMB (pomB), pFS-MOTX (motX), and pFS-FLRC (flrC), respectively. The Km cassette was inserted into flaH, fliM, flhA, and fliA and constructs ligated into pDM4, resulting in pDM-FLAH (flaH), pDM-FLIM (fliM), pDM-FLHA (flhA), and pDM-FLIA (fliA), respectively. Plasmids were independently introduced into A. hydrophila AH-405 by mating. Using this method, we obtained the following A. hydrophila mutant strains, with mutations in the genes in parentheses: AH-4426 (flaA), AH-4423 (flaH), AH-4424 (maf-1), AH-4441 (fliM), AH-4442 (flhA), AH-4443 (fliA), AH-4444 (pomB), AH-4461 (motX), AH-4462 (flrC), and AH-4427 (flaA flaB double mutant). The correct construction of all mutants was verified by Southern blot hybridization (data not shown). Swarming motility of A. hydrophila mutant strains was then assessed on swarm agar, swimming motility was assessed by light microscopy after growth in liquid media, and the presence/absence of both flagellum types (lateral and polar) was analyzed by TEM after growth in solid and liquid media. The motility phenotypes of the mutant strains are shown in Table 4. Mutations of either flaA (AH-4426) or flaB (AH-4425), located in A. hydrophila polar flagellum region 2, did not abolish motility and produced only a minor reduction in the swarm size on semisolid plates compared to the wild type, such as with flaA or flaB A. caviae mutant strains (53). Because single insertions in flaA or flaB donot abolish motility, we introduced the suicide plasmid pFS-FLAA into the A. hydrophila mutant AH-4425 in order to construct a double mutant strain (AH-4427), in which both genes were mutated in tandem. Motility assays showed that the double mutation abolished swimming motility and also caused a large decrease in swarming motility. TEM analysis and specific immunoblotting of the single mutant and double mutant whole cells, after growth in solid or liquid media, using lateral flagellin- or polar flagellin-specific antibodies, showed that flaA and flaB single mutant strains possess both flagellum types; however, the double mutant strain is unable to form polar flagella but is able to form lateral flagella (Fig. 3). This suggests that either of the flagellin proteins is able to substitute for the other to a certain extent, causing only a slight loss of motility, and that these flagellins are not involved in lateral flagellum formation. The flaH (AH-4423), maf-1 (AH-4424), fliM (AH-4441), flhA (AH-4442), fliA (AH-4443), and flrC (AH-4462) defined insertion mutants were unable to swim (examined by microscopy) and exhibited a 60% decreased swarming motility, and under TEM, all of these mutant strains showed lateral flagella but no polar flagella, as seen previously for the miniTn5 mutants AH-4422 and AH-4440 (Fig. 4). In contrast, the pomB (AH-4444) defined insertion mutant did not show any differences in swimming or swarming motility from the wild-type strain. Finally, the motX defined insertion mutant (AH-4461) was unable to swim and had a 60% decrease in swarming motility, and TEM assays showed both flagellum types.
TABLE 4.
Phenotypes of in-frame and defined insertion mutants
| Strain | Result for:
|
Gene defect | |||
|---|---|---|---|---|---|
| Motility
|
Flagellation typec
|
||||
| Swarming (cm)a | Swimmingb | Lateral | Polar | ||
| AH-3 | 3.5 | + | + | + | |
| AH-4423 | 0.8 | − | + | − | flaH |
| AH-4424 | 0.7 | − | + | − | maf-1 |
| AH-4425 | 2.1 | + | + | + | flaB |
| AH-4426 | 2.3 | + | + | + | flaA |
| AH-4427 | 0.7 | − | + | − | flaA flaB |
| AH-4441 | 0.9 | − | + | − | fliM |
| AH-4442 | 0.8 | − | + | − | flhA |
| AH-4443 | 0.7 | − | + | − | fliA |
| AH-4444 | 3.3 | + | + | + | pomB |
| AH-4461 | 0.9 | − | + | + | motX |
| AH-4462 | 0.6 | − | + | − | flrC |
Migration in centimeters of bacteria through the swarm agar plates from the center towards the periphery of the plate.
−, no motility in swim plates and liquid media; +, motility equivalent to that of the wild-type strain in swim plates or liquid media.
−, absence of flagella; +, flagella equivalent to amount in the wild type.
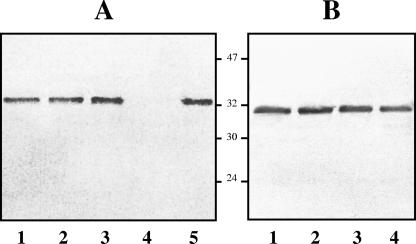
FIG. 3.
(A) Western blot analysis with anti-polar flagellin (1:1,000) polyclonal antibodies of whole-cell preparations of A. hydrophila AH-3 (wild type) and mutants AH-4425 (flaB), AH-4426 (flaA), AH-4427 (flaA flaB), and AH-4427; bacteria were complemented with plasmid pLA-FLA (lanes 1, 2, 3, 4, and 5, respectively), grown at 30°C in TSB. (B) Western blot analysis with anti-lateral flagellin (1:1,000) polyclonal antibodies of whole-cell preparations of A. hydrophila AH-3 (wild type) and mutants AH-4425 (flaB), AH-4426 (flaA), and AH-4427 (flaA flaB) (lanes 1, 2, 3, and 4, respectively), grown at 30°C in TSA. Molecular mass markers (in kilodaltons) are noted between the blots.
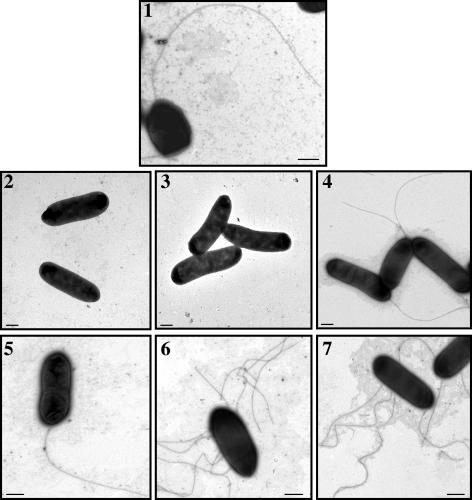
FIG. 4.
TEM of A. hydrophila AH-3 (wild type) and mutants AH-4424 (maf-1), AH-4462 (flrC), and AH-4424 complemented with plasmid pACYC-MAF and AH-4462 complemented with plasmid pACYC-FLR1 (panels 1, 2, 3, 4, and 5, respectively), grown at 30°C in TSB. Lanes 6 and 7 show AH-4424 and AH-4462 mutants, respectively, grown at 30°C on TSA. Bacteria were gently placed onto Formvar-coated copper grids and negatively stained using 2% uranyl acetate. Bar = 0.5 μm.
Complementation studies were undertaken to determine if wild-type motility could be restored to the mutants by providing polar flagellum genes in trans. The plasmid pLA-FLA, containing A. hydrophila polar flagellum region 2, was introduced into the mutant strains AH-4423, AH-4424, AH-4425, AH-4426, and AH-4427. Swimming motility and polar flagellum production were restored in AH-4423, AH-4424, and AH-4427 (Fig. 3), whereas in AH-4425 and AH-4426, full swarming motility was recovered to the level of the wild type (data not shown). Furthermore, plasmid pACYC-MAF (see Materials and Methods) was able to fully complement mutant AH-4424 (Fig. 4) and the mini-Tn5 mutant AH-4422. Plasmids pLA-FLIP1 and pLA-FLIP2, containing the A. hydrophila fliE to cheY and fliQ to orf29 genes, respectively, of polar flagellum region 3, were introduced into AH-4441, AH-4442, and AH-4443 mutant strains. The flhA and fliA mutants (AH-4442 and AH-4443, respectively) were able to swim and to produce polar flagella when pLA-FLIP1 or pLA-FLIP2 was introduced independently. In contrast, AH-4441 (fliM) recovered the wild-type swimming phenotype and the ability to produce polar flagella only when plasmid pLA-FLIP1 was introduced (data not shown). When plasmid pLA-MOTX, containing the A. hydrophila motX gene of polar flagellum region 4, was introduced into AH-4461, the mutant strain was fully able to swim (data not shown). Introduction of plasmid pLA-FLR (containing A. hydrophila polar flagellum region 5) and plasmid pACYC-FLR1 (containing the flrBC genes [see Materials and Methods]) into the AH-4462 mutant strain rescued the swimming phenotype and the ability to synthesize polar flagella, as determined by TEM (Fig. 4). Moreover, V. cholerae KKV59 (ΔflrA) and KKV98 (ΔflrC) (35) mutant strains complemented with plasmid pLA-FLR spread on Vibrio motility plates as fast as the wild type, and the formation of polar flagella was confirmed by TEM (data not shown). No complementation was observed when only the vector pLA2917 or pACYC184 was introduced into the mutants.
Distribution of polar flagellum genes in mesophilic Aeromonas strains.
The distribution of polar flagellum genes in mesophilic Aeromonas strains (n = 50) was analyzed by dot blot hybridization experiments against total genomic DNA, using independent PCR probes. The distribution of polar flagellum region 2 was analyzed using three PCR probes (flaB to flaH, flaA, and maf-1); region 3 was analyzed using three PCR probes (fliK to fliM, flhA to fliA, and pomAB); region 4 was determined by an intragenic motX probe; and master regulatory genes on region 5 were analyzed using two PCR probes (flrA and flrBC).
Polar flagellum probes hybridized to the chromosomal DNA of all mesophilic Aeromonas strains tested, whether or not the strains were able to produce lateral flagella.
Adhesion to HEp-2 cells and biofilm formation.
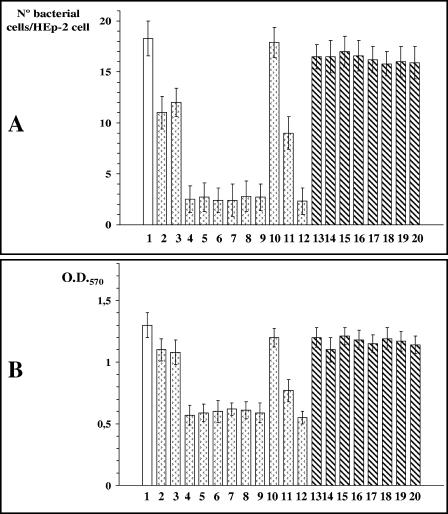
In order to correlate polar flagellum presence and motility with adherence to mammalian cells, we examined the interaction of polar flagellum mutants with cultured monolayers of HEp-2 cells. Differences in adherence were calculated by determining the average numbers of bacteria adhering to HEp-2 cells (Fig. 5A). Also, we compared the abilities of the wild-type and polar mutant strains to form biofilms in microtiter plates (Fig. 5B). The A. hydrophila wild-type strain, AH-3, exhibited an adhesion value of 18.3 (±1.7) bacteria adhered per HEp-2 cell and a biofilm formation ability with an optical density at 570 nm of 1.3 (±0.1). Polar flagellum mutant strains AH-4423 (flaH), AH-4424 (maf-1), AH-4427 (flaA flaB), AH-4441 (fliM), AH-4442 (flhA), AH-4443 (fliA), and AH-4462 (flrC), which produce lateral flagella but are unable to produce polar flagella, showed approximately an 86% reduction in adhesion to HEp-2 cells and a 55% reduction in biofilm formation. These results were similar to those for A. hydrophila flg polar flagellum mutants described previously (2). Mutant strains AH-4425 (flaB) and AH-4426 (flaA), which possess both flagellum types but show a small reduction in motility phenotype, exhibit a slight reduction in adhesion to HEp-2 cells, as well as in ability to form biofilms (approximately 35% and 10%, respectively). Finally, AH-4461 (motX), which has both flagellum types but is unable to swim, exhibited approximately half of the adhesion capacity to HEp-2 cells compared to the wild-type strain and had a 37% reduction in its ability to form biofilms. No changes in adhesion or biofilm formation were observed for the AH-4444 (pomB) mutant in comparison to the wild-type strain. Complementation studies using polar flagellum mutants AH-4423, AH-4425, AH-4426, and AH-4427 with plasmid pLA-FLA; AH-4424 with plasmid pLA-FLA or pACYC-MAF; AH-4441 with plasmid pLA-FLIP1; AH-4442 and AH-4443 with plasmid pLA-FLIP1 or pLA-FLIP2; AH-4461 with plasmid pLA-MOTX; and AH-4462 with plasmid pLA-FLR or pACYC-FLR1 showed values similar to those for the wild-type strain regarding adhesion to HEp-2 cells and biofilm formation. These adhesion and biofilm formation assays showed that polar flagella have an important role in cellular adhesion and biofilm formation, as was previously reported (2).
FIG. 5.
(A) Adhesion of A. hydrophila AH-3 and its lateral flagellum mutants to HEp-2 cells. Mean numbers of adherent bacteria per HEp-2 cell are shown. (B) Biofilm formation ability of A. hydrophila AH-3 and its lateral flagellum mutants. The optical density at 570 nm (O.D.570) quantifies the amount of crystal violet retained by the biofilm on the microtiter plates after staining. Columns: 1, AH-3 (wild type); 2, AH-4425 (flaB); 3, AH-4426 (flaA); 4, AH-4427 (flaA flaB); 5, AH-4423 (flaH); 6, AH-4424 (maf-1); 7, AH-4441 (fliM); 8, AH-4442 (flhA); 9, AH-4443 (fliA); 10, AH-4444 (pomB); 11, AH-4461 (motX); 12, AH-4462 (flrC); 13, AH-4427 (flaA flaB) with plasmid pLA-FLA; 14, AH-4423 (flaH) with plasmid pLA-FLA; 15, AH-4424 (maf-1) with plasmid pACYC-MAF; 16, AH-4441 (fliM) with plasmid pLA-FLIP1; 17, AH-4442 (flhA) with plasmid pLA-FLIP2; 18, AH-4443 (fliA) with plasmid pLA-FLIP2; 19, AH-4461 (motX) with plasmid pLA-MOTX; and 20, AH-4462 (flrC) with plasmid pACYC-FLR1. The averages of three independent experiments (each experiment performed in duplicate) are shown.
DISCUSSION
Mesophilic Aeromonas spp. produce a single polar flagellum that is expressed in both liquid and solid media; in addition, 50 to 60% of strains also have an inducible lateral flagellum system that is expressed in high viscosity. Previously, only a few genes required for polar flagellum formation have been published (2, 53). The isolation of A. hydrophila AH-3 transposon mutants unable to swim allowed us to genetically characterize three of five A. hydrophila AH-3 chromosomal regions (regions 2, 3, and 4) containing polar flagellum genes (Fig. 1). Region 2 is composed of fla and maf genes; fli, flh, chemotaxis, and motor genes are in region 3; and the motor gene motX is alone in region 4. Polar flagellum loci that have been sequenced in other bacteria, such as Vibrio or Pseudomonas, indicated that the gene organization seems highly conserved, although their distributions in chromosomal regions are different between organisms (Fig. 2). Genes coding for flagellins and capping protein (fla), as well as genes coding for the basal body, hook length regulator, switch, export apparatus, flagellum placement determinant, flagellum number regulator, σ28 factor, chemotaxis, and motor proteins (fli and flh) of A. hydrophila AH-3, are distributed in two different chromosomal regions (2 and 3). In contrast, V. parahaemolyticus (Fig. 2) showed all of these genes grouped in a single chromosomal region (34, 41).
Organization of the A. hydrophila fla genes in region 2 is identical to that of A. caviae (53). A similar gene organization is also observed with V. parahaemolyticus polar flagellum region 2 (Fig. 2); however, this organism possesses one more flagellin gene (flaF) and a gene encoding a putative chaperone (flaI) between flaH and flaJ (30, 36). The A. hydrophila insertional flaH mutant and flaA flaB double mutant showed lateral flagella, absence of polar flagella, and a dramatic reduction in adhesion to HEp-2 cells and ability to form biofilms. All of these effects were rescued by the introduction of pLA-FLA in the mutant strains, suggesting that the fla genes distributed in this chromosomal region are involved only in polar flagellum biosynthesis or assembly, and both flagellin genes (flaA and flaB) are required for optimal polar flagellum function. Recent unpublished work performed with A. caviae has shown that nonpolar mutants (flaA flaB and flaH) do in fact express lateral flagella, in contrast to the previously reported results (53). The A. hydrophila AH-3 strains with mutations in flaH, flaA, flaB, and flaA flaB demonstrated the same phenotypes as their A. caviae counterparts (53). In addition we found, downstream of the A. hydrophila polar flagellum fla loci and preceded by a σ28 promoter sequence, an independently transcribed gene not described before for Aeromonas spp. or Vibrio spp. This gene encoded a homologue of the Maf proteins reported for Helicobacter pylori, Clostridium acetobutylicum, and Campylobacter jejuni. In all of these bacteria, the genes encoding Maf proteins are linked to either flagellum biosynthesis genes and/or genes involved in sugar biosynthesis and transport (26, 33). The inactivation of the A. hydrophila maf-1 gene abolished only polar flagellum formation and did not affect lateral flagella; the wild-type phenotype was restored by introduction of pACYC-MAF (Fig. 4). This fact together with the knowledge that A. hydrophila AH-3 polar flagellins are glycosylated (unpublished observation), similarly to the A. caviae polar flagellins (25, 53), may suggest that the encoded protein (Maf-1) is involved in posttranslational polar flagellum glycosylation, but their exact role in flagellar biosynthesis remains unknown.
The A. hydrophila polar flagellum region 3 (Table 3) showed an organization similar to that of the genes downstream of flaM in V. parahaemolyticus polar flagellum region 2, with the absence of the motor genes (34, 41) (Fig. 2). No master regulatory genes encoding homologues of V. parahaemolyticus FlaK, FlaL, and FlaM, V. cholerae FlrA, FlrB, and FlrC (52), or P. aeruginosa FleQ, FleS, and FleR (4, 54) were found upstream of A. hydrophila fliE. In contrast to the Vibrio polar flagellum systems, we found two genes (pomA and pomB) which encode orthologues of the MotA and MotB motor proteins of Pseudomonas (15). The A. hydrophila pomA and pomB genes are transcribed independently, according to the RT-PCR results, while in P. aeruginosa, these genes are cotranscribed with some chemotaxis genes (15).
Strains with mutations in the fliM, flhA, and fliA genes were able to produce lateral flagella but were unable to produce polar flagella, and they also showed a large reduction in adhesion to HEp-2 cells and ability to form biofilms. The data obtained from these mutants suggest that they are required only for the production of polar flagella but not for production of lateral flagella. The MotA-MotB complex constitutes the stator of the flagellum motor and is involved in the formation of a proton or sodium-conducting channel to generate the force necessary to drive the flagella (31, 61). The pomB insertion mutant had both flagellum types and was fully able to swim, adhere, and form biofilms as well as the wild-type strain. This situation was similar for P. aeruginosa motB mutants (18, 64) but not for V. cholerae or V. parahaemolyticus motB mutants, which were able to produce polar flagella but were nonmotile (8, 24). In Pseudomonas, there are two sets of motAB-like genes, motAB and motCD, distributed in different chromosomal regions, as well as another gene, motY, which contributes to proton-driven flagellar motility (Fig. 2). Loss of either motAB-like gene still resulted in motile bacteria in aqueous environments, and only mutations of both sets of genes encoding the MotA or MotB homologue were sufficient to abolish motility (18, 64). The data obtained from the pomB mutant suggest that PomB is not essential for swimming motility, leading to two different possible explanations: the stator of lateral flagella can supply its function, or another pomAB-like locus is present in A. hydrophila. Further studies are required in order to completely understand the motility process in A. hydrophila AH-3.
Region 4 of A. hydrophila AH-3 polar flagella includes a gene that encodes a homologue of the sodium-driven motor MotX of V. alginolyticus and V. parahaemolyticus, which is involved with MotA, MotB, and MotY in torque generation of polar flagella (8, 40), although its exact function is unknown. This Aeromonas gene has both a putative σ28 promoter sequence and a putative terminator sequence, suggesting that it is transcribed independently, as in Vibrio spp. Inactivation of the A. hydrophila motX gene did not affect either lateral or polar flagellum formation, but mutants were unable to swim in liquid media, as described for V. parahaemolyticus, and showed a reduction (50%) in adhesion to HEp-2 cells and ability to form biofilms (37% reduction) in comparison with the wild-type strain.
In contrast to master regulatory genes for other polar flagellated bacteria, such as Vibrio and Pseudomonas (Fig. 2), the Aeromonas flrA to flrC genes are located in an independent chromosomal region (region 5). The central domain of Aeromonas FlrB and FlrC homologues contains a σ54 interaction domain, which is present in σ54 activators, and carboxyl- and amino-terminal domains of FlrB and FlrC homologues, respectively, which define these two proteins as members of the two-component family of bacterial signal transducers. A characteristic histidine kinase domain was found in the FlrB homologue protein, and their N termini contain a PAS domain, usually associated with proteins that play a role in detection and adaptation to environmental change (62). A PAS domain was also found in V. cholerae FlrB and P. aeruginosa FleS N termini, in contrast to FlaL of V. parahaemolyticus. The Aeromonas flrC mutant was able to produce lateral flagella but unable to produce polar flagella and had a large reduction in adhesion to HEp-2 cells and ability to form biofilms. The data obtained from this mutant suggest that it is required only for the production of polar flagella but not for production of lateral flagella. Plasmids pLA-FLR and pACYC-FLR1 were able to fully complement the flrC defects when introduced independently into this mutant strain. With the possible exception of the motor genes, which require further investigation, the rest of the structural genes for polar and lateral flagellum formation in A. hydrophila AH-3 are clearly independent, and further studies are required in order to completely understand the motility process in A. hydrophila AH-3.
By comparing previous results with results described in this work for mutants unable to produce polar and lateral flagella (25), mutants able to produce polar but not lateral flagella (23), and mutants able to produce lateral but not polar flagella, we can conclude that both types of flagella contribute to HEp-2 cell adhesion and biofilm formation in A.hydrophila AH-3. In V. parahaemolyticus, only the polar flagella seem to be involved in these pathogenic features (20).
Acknowledgments
This work was supported by grants from Plan Nacional de I+D (Ministerio de Ciencia y Tecnología, Spain), Generalitat de Catalunya, and the Wellcome Trust. R.C. and S.R. were supported by fellowships from Universidad de Barcelona and the Ministerio de Educación, Ciencia y Deporte (Spain), respectively.
We thank Maite Polo for her technical assistance and K. E. Klose for providing strains KKV59 and KKV98.
REFERENCES
- 1.Allen, L. N., and R. S. Hanson. 1985. Construction of broad-host-range cosmid cloning vectors: identification of genes necessary for growth of Methylobacterium organophilum on methanol. J. Bacteriol. 161:955-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altarriba, A., S. Merino, R. Gavín, R. Canals, A. Rabaan, J. G. Shaw, and J. M. Tomás. 2003. A polar flagella operon (flg) of Aeromonas hydrophila contains genes required for lateral flagella expression. Microb. Pathog. 34:249-259. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, F. S., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zang, W. Miller, and J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austin, B., and C. Adams. 1996. Fish pathogens, p. 197-243. In B. Austin, M. Altwegg, P. J. Gosling, and S. W. Joseph (ed.), The genus Aeromonas. John Wiley and Sons, New York, N.Y.
- 6.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boin, M. A., M. J. Austin, and C. C. Hase. 2004. Chemotaxis in Vibrio cholerae. FEMS Microbiol. Lett. 239:1-8. [DOI] [PubMed] [Google Scholar]
- 8.Boles, B. R., and L. L. McCarter. 2000. Insertional inactivation of genes encoding components of the sodium-type flagellar motor and switch of Vibrio parahaemolyticus. J. Bacteriol. 182:1035-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bott, M., M. Meyer, and P. Dimroth. 1995. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol. Microbiol. 18:533-546. [DOI] [PubMed] [Google Scholar]
- 10.Bren, A., and M. Eisenbach. 2000. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J. Bacteriol. 182:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpenter, P. B., D. W. Hanlon, and G. W. Ordal. 1992. flhF, a Bacillus subtilis flagellar gene that encodes a putative GTP-binding protein. Mol. Microbiol. 6:2705-2713. [DOI] [PubMed] [Google Scholar]
- 12.Carrello, A., K. A. Silburn, J. R. Budden, and B. J. Chang. 1988. Adhesion of clinical and environmental Aeromonas isolates to HEp-2 cells. J. Med. Microbiol. 26:19-27. [DOI] [PubMed] [Google Scholar]
- 13.Ciacci-Woolwine, F., I. C. Blomfield, S. H. Richardson, and S. B. Mizel. 1998. Salmonella flagellin induces tumor necrosis factor alpha in a human promonocytic cell line. Infect. Immun. 66:1127-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dasgupta, N., S. K. Arora, and R. Ramphal. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 182:357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasgupta, N., M. C. Wolfgang, A. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809-824. [DOI] [PubMed] [Google Scholar]
- 16.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeMot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 18.Doyle, T. B., A. C. Hawkins, and L. L. McCarter. 2004. The complex flagellar torque generator of Pseudomonas aeruginosa. J. Bacteriol. 186:6341-6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enos-Berlage, J. L., Z. T. Guvener, C. E. Keena, and L. L. McCarter. 2005. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol. Microbiol. 55:1160-1182. [DOI] [PubMed] [Google Scholar]
- 21.Fenchel, T. 2002. Microbial behavior in a heterogeneous world. Science 296:1068-1071. [DOI] [PubMed] [Google Scholar]
- 22.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gavín, R., A. A. Rabaan, S. Merino, J. M. Tomás, I. Gryllos, and J. G. Shaw. 2002. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 43:383-397. [DOI] [PubMed] [Google Scholar]
- 24.Gosink, H. K., and C. C. Häse. 2000. Requirements for conversion of the Na+-drive flagellar motor of Vibrio cholerae to the H+-drive motor of Escherichia coli. J. Bacteriol. 182:4234-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gryllos, I., J. G. Shaw, R. Gavín, S. Merino, and J. M. Tomas. 2001. Role of flm operon in mesophilic Aeromonas species adherence. Infect. Immun. 69:65-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerry, P., P. Doig, R. A. Alm, D. H. Burr, N. Kinsella, and T. J. Trust. 1996. Identification and characterization of genes required for post-translational modification of Campylobacter coli VC167 flagellin. Mol. Microbiol. 19:369-378. [DOI] [PubMed] [Google Scholar]
- 27.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 28.Hendrixson, D. R., and V. J. DiRita. 2003. Transcription of sigma54-dependent but not sigma28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50:687-702. [DOI] [PubMed] [Google Scholar]
- 29.Inoue, K., Y. Kosako, K. Suzuki, and T. Shimada. 1991. Peritrichous flagellation in Plesiomonas shigelloides strains. Jpn. J. Med. Sci. Biol. 44:141-146. [DOI] [PubMed] [Google Scholar]
- 30.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentation and unanswered questions. Clin. Infect. Dis. 27:332-344. [DOI] [PubMed] [Google Scholar]
- 31.Jaques, S., Y. K. Kim, and L. L. McCarter. 1999. Mutations conferring resistance to phenamil and amiloride, inhibitors of sodium-driven motility of Vibrio parahaemolyticus. Proc. Natl. Acad. Sci. USA 96:5740-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang, Z.-Y., and C. E. Bauer. 1997. Analysis of a chemotaxis operon from Rhodospirillum centenum. J. Bacteriol. 179:5712-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlinshev, A. V., D. Linton, N. A. Gregson, and B. W. Wren. 2002. A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology 148:473-480. [DOI] [PubMed] [Google Scholar]
- 34.Kim, Y.-K., and L. L. McCarter. 2000. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 182:3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28:501-520. [DOI] [PubMed] [Google Scholar]
- 36.Leonard, T. A., P. J. Butler, and J. Löwe. 2005. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer—a conserved biological switch. EMBO J. 24:270-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 38.Marston, A. L., H. B. Thomaides, D. H. Edwards, M. E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12:3419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarter, L. L., and M. Silverman. 1990. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol. Microbiol. 4:1057-1062. [DOI] [PubMed] [Google Scholar]
- 40.McCarter, L. L. 1994. MotX, a channel component of the sodium-type flagellar motor. J. Bacteriol. 176:5988-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merino, S., S. Camprubi, and J. M. Tomas. 1991. The role of lipopolysaccharide in complement-killing of Aeromonas hydrophila strains of serotype O:34. J. Gen. Microbiol. 137:1583-1590. [DOI] [PubMed] [Google Scholar]
- 43.Merino, S., X. Rubires, A. Aguilar, and J. M. Tomás. 1997. The role of flagella and motility in the adherence and invasion to fish cell lines by Aeromonas hydrophila serogroup O:34 strains. FEMS Microbiol. Lett. 151:213-217. [DOI] [PubMed] [Google Scholar]
- 44.Milton, D. L., R. O'Toole, P. Hörstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moens, S., K. Michiels, V. Keijer, F. Van Leuven, and J. Vanderleyden. 1995. Cloning, sequencing, and phenotypic analysis of laf1, encoding the flagellin of the lateral flagella of Azospirillum brasilense Sp7. J. Bacteriol. 177:5419-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nidia, E. C., F. Peng, and K. E. Klose. 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J. Bacteriol. 187:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niehus, E., H. Gressmann, F. Ye, R. Schlapbach, M. Dehio, C. Dehio, A. Stack, T. F. Meyer, S. Suerbaum, and C. Josenhans. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52:947-961. [DOI] [PubMed] [Google Scholar]
- 48.Nogueras, M. M., S. Merino, A. Aguilar, V. J. Benedí, and J. M. Tomas. 2000. Cloning, sequencing and role in serum susceptibility of porin II from mesophilic Aeromonas hydrophila. Infect. Immun. 68:1849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Rouke, J., A. Lee, and J. G. Fox. 1992. An ultrastructural study of Helicobacter mustelae and evidence of a specific association with gastric mucosa. J. Med. Microbiol. 36:420-427. [DOI] [PubMed] [Google Scholar]
- 50.Pandza, S., M. Baetens, C. H. Park, T. Au, M. Keyhan, and A. Matin. 2000. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol. Microbiol. 36:414-426. [DOI] [PubMed] [Google Scholar]
- 51.Pratt, L. A., and R. Kotler. 1998. Genetic analysis of Escherichia coli biofilm formation: role of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 52.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 53.Rabaan, A. A., I. Gryllos, J. M. Tomás, and J. G. Shaw. 2001. Motility and the polar flagellum are required for Aeromonas caviae adherence to HEp-2 cells. Infect. Immun. 69:4257-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchings, B. W., E. C. Almira, S. Lory, and R. Ramphal. 1995. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 63:4868-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubirés, X., F. Saigi, N. Piqué, N. Climent, S. Merino, S. Albertí, J. M. Tomás, and M. Regué. 1997. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 179:7581-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 57.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimada, T., R. Sakazaki, and K. Suzuki. 1985. Peritrichous flagella in mesophilic strains of Aeromonas. Jpn. J. Med. Sci. Biol. 38:141-145. [DOI] [PubMed] [Google Scholar]
- 59.Stanley, P. M. 1983. Factors affecting the irreversible attachment of Pseudomonas aeruginosa to stainless steel. Can. J. Microbiol. 29:1493-1499. [DOI] [PubMed] [Google Scholar]
- 60.Stewart, B. J., and L. L. McCarter. 2003. Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 185:4508-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stolz, B., and H. C. Berg. 1991. Evidence for interaction between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol. 173:7033-7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thornley, J. P., J. G. Shaw, I. Gryllos, and A. Eley. 1996. Adherence of Aeromonas caviae to human cell lines Hep-2 and Caco-2. J. Med. Microbiol. 45:445-451. [DOI] [PubMed] [Google Scholar]
- 64.Toutain, C. M., M. E. Zegans, and G. A. O'Toole. 2005. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol. 187:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Umelo, E., and T. J. Trust. 1997. Identification and molecular characterization of two tandemly located flagellin genes from Aeromonas salmonicida A449. J. Bacteriol. 179:5292-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]