Abstract
In the present work, the metabolic consequences of the deletion of the methionine and cysteine biosynthesis repressor protein (McbR) in Corynebacterium glutamicum, which releases almost all enzymes of methionine biosynthesis and sulfate assimilation from transcriptional regulation (D. A. Rey, A. Pühler, and J. Kalinowski, J. Biotechnol. 103:51-65, 2003), were studied. C. glutamicum ATCC 13032 ΔmcbR showed no overproduction of methionine. Metabolome analysis revealed drastic accumulation of a single metabolite, which was not present in the wild type. It was identified by isotopic labeling studies and gas chromatography/mass spectrometry as l-homolanthionine {S-[(3S)-3-amino-3-carboxypropyl]-l-homocysteine}. The accumulation of homolanthionine to an intracellular concentration of 130 mM in the ΔmcbR strain was accompanied by an elevated intracellular homocysteine level. It was shown that cystathionine-γ-synthase (MetB) produced homolanthionine as a side reaction. MetB showed higher substrate affinity for cysteine (Km = 260 μM) than for homocysteine (Km = 540 μM). The cell is able to cleave homolanthionine at low rates via cystathionine-β-lyase (MetC). This cleavage opens a novel threonine-independent pathway for isoleucine biosynthesis via 2-oxobutanoate formed by MetC. In fact, the deletion mutant exhibited an increased intracellular isoleucine level. Metabolic flux analysis of C. glutamicum ΔmcbR revealed that only 24% of the O-acetylhomoserine at the entry of the methionine pathway is utilized for methionine biosynthesis; the dominating fraction is either stored as homolanthionine or redirected towards the formation of isoleucine. Deletion of metB completely prevents homolanthionine accumulation, which is regarded as an important step in the development of C. glutamicum strains for biotechnological methionine production.
The gram-positive bacterium Corynebacterium glutamicum is an important organism for the industrial production of fine chemicals, such as the amino acids lysine and glutamate (3). Methionine, applied in large amounts for animal nutrition, is, however, produced by a chemical process yielding the racemic dl mixture and employing rather hazardous chemicals (19). In this regard, C. glutamicum appears as a promising candidate for future biotechnological production of l-methionine. Accordingly, the biosynthetic pathway for this amino acid has been the focus of research in recent years (9, 10, 26-28). In C. glutamicum, methionine biosynthesis is carried out by parallel pathways of transsulfuration and direct sulfhydrylation (10, 18). The corresponding genes were recently identified by targeted gene deletion and homologous complementation (28). Moreover, the regulation of the methionine pathway was studied by proteome (27) and transcriptome (26) analyses. Hereby, a transcriptional repressor (McbR) that controls 86 genes was identified. Among these genes are several of methionine and cysteine metabolism, including hom (homoserine dehydrogenase), metX (homoserine O-acetyltransferase), metB (cystathionine-γ-synthase), metY (O-acetylhomoserine sulfhydrylase), metE and metH (cysteine synthases I and II, respectively), metK (S-adenosylmethionine synthetase), cysJ and cysI (putative sulfite reductases), and cysK (O-acetylserine sulfhydrylase). Additionally, McbR regulates its own expression and that of at least two other putative transcriptional regulators, indicating a complex regulatory network with McbR as the master regulator (26). Different enzymes of the methionine pathway are further regulated by feedback inhibition (17). This involves homoserine dehydrogenase (controlled by threonine), homoserine O-acetyltransferase (methionine, S-adenosylmethionine, and O-acetylhomoserine), cystathionine-γ-synthase (S-adenosylmethionine), cystathionine-β-lyase (methionine, cys-teine, and glycine), and O-acetylhomoserine sulfhydrylase (methionine and O-acetylserine). It has been suggested that the deletion of McbR might be an important step in constructing a methionine-overproducing organism. In the present work, the effect of deleting this repressor in C. glutamicum was investigated from a metabolic perspective.
MATERIALS AND METHODS
Bacterial strains.
The wild-type Corynebacterium glutamicum ATCC 13032 was obtained from the American Type Culture Collection (Manassas, VA). The C. glutamicum ΔmcbR strain was derived from the wild type. C. glutamicum ΔmcbR ΔmetB was an McbR knockout strain in which cystathionine γ-synthase was also deleted. C. glutamicum ΔmcbR Δhom Δhsk was a threonine and homoserine auxotrophic strain also derived from the wild type. The knockout mutants were constructed as follows. The deletion of mcbR in C. glutamicum ATCC 13032 was performed by using the primer pairs BK1987/BK1968 and BK1967/BK1988 for crossover PCR (Table 1). This resulted in a DNA construct containing the upstream and downstream sequences of the mcbR gene but not its coding sequence. The obtained PCR construct containing the deleted mcbR allele was then integrated into C. glutamicum ATCC 13032 for first and second recombinants. As vector, the plasmid pClik, which carried kanamycin resistance and the sacB gene as selection markers, was used. Since pClik cannot replicate in C. glutamicum, transformation of the organism with the plasmid and subsequent selection for the kanamycin resistance plasmid marker yielded transformants which had integrated the plasmid DNA into the genome via a single-crossover homologous-recombination event. Subsequently, each kanamycin-resistant integrant was grown for 1 day without kanamycin to allow a second recombination event to take place. The sacB-positive selection system (11) was used to select for the second recombination event. Since the expression of integrated plasmid-borne sacB in the presence of sucrose is lethal to C. glutamicum, cells can grow only on the selective plate if sacB has been deleted as a consequence of the second homologous recombination. By this recombination, either the wild-type mcbR gene or the shortened DNA fragment with the deleted coding region of mcbR remains in the genome. Clones positive for the deletion of mcbR were identified using primers BK1987 and BK1988. These two primers flank the 5′ and the 3′ regions of the insert. In the case of the mcbR knockout mutant, PCR amplification leads to a shortened PCR fragment length compared to that obtained for the wild type. The subsequent knockout of metB in C. glutamicum ΔmcbR, leading to C. glutamicum ΔmcbR ΔmetB, was performed with plasmid pSL315 as described by Hwang et al. (10). For the construction of C. glutamicum ΔmcbR Δhom Δhsk, the primer pairs HS605/HS606 and HS607/HS608 were used for crossover PCR on chromosomal DNA of the C. glutamicum wild type to delete approximately 1,780 internal nucleotides of the hom and hsk loci. The yielded PCR fragment contained a small 5′ fragment of hom and a small 3′ fragment of hsk together with the hsk downstream region. It was cloned into the pCLIK5A sacB vector using the BamHI sites. Subsequently, it was introduced into C. glutamicum ΔmcbR for first and second recombinants. Clones positive for the deletion of hom and hsk were identified, as described above, using primers HS605 and HS608, which yielded a shortened PCR product compared to that for the wild type.
TABLE 1.
Primer sequences used for the construction of C. glutamicum ΔmcbR, C. glutamicum ΔmcbR ΔmetB, and C. glutamicum ΔmcbR Δhom Δhsk
| Primer | Sequence |
|---|---|
| BK1987 | GAGAGAGACTCGAGCTCTCCAATCTCCACTGAGG |
| BK1967 | ACTCTTGCCTGAAGCGCTAGCAGCCACGTT |
| BK1968 | GAGAGAGGCTAGCTAATCCTTGATGGTGGA |
| BK1988 | CTCTCTCTACGCGTCAGCAACAACCTGTGGACGC |
| HS 605 | BamHI GCGGGATCCATGACCTCAGCATCT |
| HS 606 | CCCATCCACTAAACTTAAACACCGCTGCATCAGCAA |
| HS 607 | TGTTTAAGTTTAGTGGATGGGGATGCTCGTGAGTCT |
| HS 608 | BamHI GCGGGATCCATCTTCCAAACACGC |
Chemicals.
l-Homocysteine was prepared from l-homocysteine thiolactone (1). Casamino Acids, beef extract, polypeptone, and yeast extract were supplied by Difco (Detroit, MI). The tracer substrates, 99% [13C6]glucose and 98% [13C4]threonine, were supplied by Cambridge Isotopes, Inc. (Andova, MA). Ammonium sulfate labeled with 15N (99%) was purchased from Campro Scientific (Veenendaal, The Netherlands), and [34S]sulfate was kindly provided by BASF AG (Ludwigshafen, Germany). All other chemicals were of analytical grade and purchased from Grüssing (Filsum, Germany), Acros Organics (Geel, Belgium), Merck (Darmstadt, Germany), Aldrich (Steinheim, Germany), or Fluka (Buchs, Switzerland).
Media and growth conditions.
Cells for inoculation were grown at 30°C on agar plates with rich medium containing 10.0 g/liter glucose, 2.5 g/liter NaCl, 2.0 g/liter urea, 5.0 g/liter yeast extract, 5.0 g/liter beef extract, 5.0 g/liter polypeptone, 20.0 g/liter Casamino Acids, and 20.0 g/liter agar. Single colonies served as inoculum for the precultures, which were grown overnight in 250-ml baffled shake flasks with 25 ml rich medium as described above without agar. Cells were harvested by centrifugation (2 min, 10,000 × g, 4°C), washed twice with 0.9% NaCl, and used for inoculation of the second preculture on minimal medium. The second preculture growth was harvested as described above and used as inoculum for the main cultivations, which were carried out in 500-ml baffled shake flasks with 50 ml minimal medium. The minimal medium contained the following per liter: 20 g glucose, 16 g K2HPO4, 4 g KH2PO4, 20 g (NH4)2SO4, 300 mg 3,4-dihydroxybenzoic acid, 10 mg CaCl2, 250 mg MgSO4 · 7H2O, 10 mg FeSO4 · 7H2O, 10 mg MnSO4 · H2O, 2 mg ZnSO4 · 7H2O, 200 μg CuSO4 · 5H2O, 20 μg NiCl2 · 6H2O, 20 μg Na2MoO4 · 2H2O, 100 μg cyanocobalamin, 300 μg thiamine, 4 μg pyridoxal phosphate, and 100 μg biotin. Tracer experiments were performed with 5 ml minimal medium in 50-ml baffled shake flasks. Hereby, selected medium constituents were replaced by the corresponding compounds labeled with stable isotopes. All cultivations were carried out at 30°C on a rotary shaker (250 rpm; shaking diameter, 5 cm) (Multitron; Infors AG, Bottmingen, Switzerland).
Quantification of intermediates of the methionine pathway.
Intracellular metabolites were extracted by quick filtration and subsequent boiling in water (15 min) as described previously (16). The obtained cell extract was used to quantify metabolites related to methionine biosynthesis in C. glutamicum (cysteine, homocysteine, homoserine, O-acetylhomoserine, cystathionine, and homolanthionine). For this purpose, high-performance liquid chromatography (HPLC) with precolumn derivatization by o-phthaldialdehyde was utilized (15). Quantification of homolanthionine, for which no external standard was commercially available, was based on the calibration factor for cystathionine, which had a highly similar structure. In this context, serine and homoserine differing by one methylene group, as did cystathionine and homolanthionine, showed only a 5% difference in the calibration factor. Due to this, the calibration via cystathionine should not significantly affect the quantification of homolanthionine.
GC/MS labeling analysis.
Determination of labeling patterns of metabolites in cell extracts or amino acids in the cell protein was carried out by gas chromatography/mass spectrometry (GC/MS). For the analysis of amino acids from the cell protein, biomass (1 mg) was harvested from an exponentially growing culture and washed twice with H2O. The washed cell pellet was hydrolyzed for 48 h (50 μl 6 M HCl, 105°C). The obtained hydrolysate was neutralized (6 M NaOH), clarified by centrifugation (5 min, 16,000 × g) (ultrafree-MC filter units, 0.22-μm Durapore membrane; Millipore, Bedford, MA), and subsequently freeze-dried. GC/MS analysis of the amino acids was performed after derivatization into the t-butyl-dimethylsilyl (TBDMS) derivate (31). For the analysis of intracellular metabolites, 400 μl cell extract was freeze-dried and subsequently derivatized as described above for the hydrolysate.
Overexpression and purification of cystathionine-γ-synthase and cystathionine-β-lyase.
Cystathionine-γ-synthase (MetB) and cystathionine-β-lyase (MetC) of C. glutamicum were separately cloned and overexpressed in Escherichia coli XL1-Blue. For this purpose, the corresponding genes were cloned into vector pQE30 (QIAGEN, Inc., Chatsworth, CA) comprising the addition of a His tag to the N terminus of the expressed protein. The vector carrying either the metB or the metC gene was transformed into Escherichia coli using the CaCl2 method. Transformed E. coli cells were cultivated at 37°C and 250 rpm on Terrific broth (20) with 100 mg/liter ampicillin. At a cell optical density at 600 nm of 1 unit, protein expression was induced by the addition of 1 mM isopropylthiogalactoside (final concentration). After 16 h of induced growth, cells (4 g wet weight) were harvested by centrifugation (4,225 × g, 15 min, 2°C) and resuspended in 16 ml phosphate buffer (100 mM; 100 μM pyridoxal phosphate, 1 g/liter DNase I [pH 7.4]; 4°C), including a washing step in the same buffer. Subsequently, cells were extracted by sonication on ice (five times for 15 s each time; 20 μm). The crude extracts were separated from cell debris by centrifugation (30 min, 2°C, 20,000 × g). The recombinant enzymes were finally purified by affinity chromatography (ÄKTA purifier 900; Amersham Biosciences, Little Chalfont, England) equipped with a chelating nickel-Sepharose column (5 ml) (HiTrap; Amersham Biosciences, Little Chalfont, England). The column was equilibrated with 0.02 M sodium phosphate buffer (pH 7.4) containing 0.5 M NaCl. After the cell extract was applied to the column, the column was washed with 10 volumes of 0.02 M sodium phosphate buffer (pH 7.4). Elution was carried out with a linear gradient of 0.02 M sodium phosphate buffer (pH 7.4; 0.5 M NaCl; 0.5 M imidazole). The fractions containing MetB or MetC were checked for purity with sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then pooled together. Proteins were concentrated and separated from imidazole by three cycles of filtration (molecular weight cutoff, 20,000) (Centrisart; Sartorius, Göttingen, Germany). The protein content (Bio-Rad protein assay; Munich, Germany) finally obtained was 12 mg ml−1 for MetB and 0.3 mg ml−1 for MetC.
In vitro assay of cystathionine-γ-synthase.
The in vitro activity of cystathionine-γ-synthase (MetB) was determined photometrically (Helios α; Thermo Electron, Dreieich, Germany). The assay was based on the quantification of free SH groups using Ellman's reagent with detection at 412 nm (4). The assay mixture (final volume, 1 ml) contained 1.25 mM cysteine (or homocysteine), 3 mM O-acetylhomoserine, and 10 μM pyridoxal-5-phosphate in phosphate buffer (100 mM, pH 7.5). The reaction was started by the addition of 1 μl MetB solution (12 mg ml−1). During the incubation, samples (65 μl) were taken from the assay mixture and immediately injected into 935 μl of a stop solution (100 mM phosphate buffer (pH 7.5) with 38% ethanol and 1 mM dithionitrobenzoic acid. Dithionitrobenzoic acid formed a yellow complex with the remaining homocysteine or cysteine. The extinction of the yellow complex at 412 nm was linearly correlated with free SH groups, e.g., from cysteine or from homocysteine, up to 1.5 mM. The Km values of MetB for cysteine and homocysteine were determined from double-reciprocal Lineweaver-Burk plots.
In vitro assay of cystathionine-β-lyase.
The assay for cystathionine-β-lyase (MetC) was based on the same principle as that for MetB. Due to that fact that the reaction catalyzed by MetC leads to the generation of free SH groups, the activity of the enzyme was monitored photometrically (Helios α; Thermo Electron, Dreieich, Germany) by the increase of extinction at 412 nm using Ellman's reagent (4). The assay mixture (final volume, 1 ml) contained 1.25 mM cystathionine and 10 μM pyridoxal 5-phosphate in phosphate buffer (100 mM, pH 7.5). The reaction was started by the addition of 100 μl MetC solution (0.3 mg ml−1). It should be noted that homolanthionine was not commercially available. Due to this, homolanthionine was added from the assay mixture of the MetB assay after removal of the MetB enzyme by three cycles of filtration (molecular weight cutoff, 20,000) (Centrisart; Sartorius, Göttingen, Germany). This solution contained primarily homolanthionine and to some extent the remaining homocysteine and O-acetylhomoserine. The lack of MetB activity in this solution was checked by the MetB assay. The exact concentration of homolanthionine in the MetC assay was determined by HPLC. During the incubation, samples (65 μl) were taken from the assay mixture and immediately injected into 935 μl of a stop solution as described above. The Km value of MetC for cystathionine was determined from a double-reciprocal Lineweaver-Burk plot.
RESULTS
Metabolomic response to deletion of McbR.
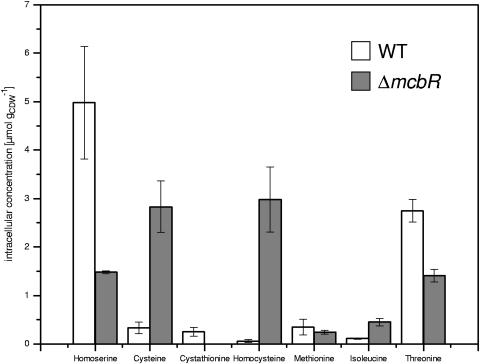
The deletion of the transcriptional repressor McbR in Corynebacterium glutamicum had a significant impact on the growth rate (μ). In contrast to that of the wild type (μ = 0. 40 h−1), the specific growth of the mutant (μ = 0. 18 h−1) was significantly decreased. Additionally, the intracellular level of intermediates of the methionine pathway was severely affected by the McbR deletion (Fig. 1). C. glutamicum ATCC 13032 ΔmcbR exhibited a decreased level of homoserine in comparison to that of the parent strain, C. glutamicum ATCC 13032, which indicated a higher flux into the methionine pathway. The intracellular level of homocysteine, however, was increased by a factor of 50, i.e., from 0.06 to 2.98 μmol g of cells (dry weight)−1 (gCDW−1). The intracellular methionine pool was not affected by the deletion. O-Acetylhomoserine could not be detected in any of the strains. The most pronounced difference between the two strains, however, was the strong accumulation of a single unknown metabolite in the cell extract, which was observed by HPLC as well as by GC/MS analysis.
FIG. 1.
Intracellular pools of methionine intermediates in exponentially growing C. glutamicum ATCC 13032 (wild type [WT]) and C. glutamicum ATCC 13032 ΔmcbR. The concentrations are given in μmoles per gCDW. Mean values for four cell extracts from two different time points with corresponding standard deviations are shown. The level of cystathionine in the ΔmcbR strain was below the detection limit (around 0.1 μmol gCDW−1).
Identification of the novel metabolite as homolanthionine.
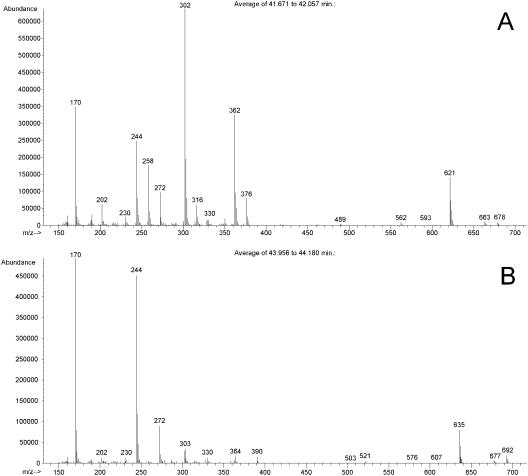
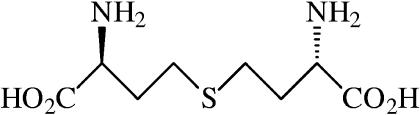
The metabolite significantly accumulating as a consequence of mcbR deletion was identified from cell extracts of C. glutamicum by its GC/MS mass fragment pattern (Fig. 2A and B). After conversion into the TBDMS derivate, the metabolite eluted from the GC column after 44.0 min. The metabolite exhibited a mass spectrum with high similarity to that of cystathionine, which eluted a bit earlier (42.1 min). Characteristic ions typically observed for the performed derivatization, such as the molecular ion [M] at m/z = 692 and fragment ions [M-15] at m/z = 677, [M-57] at m/z = 635, or [M-85] at m/z = 607, all exhibited a mass shift of 14 compared to the corresponding fragments in cystathionine (Fig. 2A and B). The mass shift of 14 indicated the presence of an additional methylene group in the new metabolite compared to cystathionine. Additionally, fragments from the lower mass range, such as m/z=170, m/z = 244, and m/z = 272, were also observed for cystathionine (Fig. 2A), homocysteine, and methionine (data not shown). These findings were the first indication that the new metabolite observed was linked to the methionine metabolism in C. glutamicum and that its accumulation was therefore a direct consequence of the deletion of the methionine repressor McbR. Subsequent tracer cultivations of C. glutamicum ΔmcbR with either [13C6]glucose, [15N2]ammonium sulfate, or [34S]sulfate led to mass shifts in the molecular ion of the metabolite of 8, 2, and 2, respectively. This showed that the carbon, nitrogen, and sulfur contents of the observed metabolite were C8N2S1. Based on these data, the new metabolite was identified as homolanthionine {S-[(3S)-3-amino-3-carboxypropyl]-l-homocysteine} (Fig. 3). Its mass isotopomer distribution determined by GC/MS showed excellent agreement with the theoretical value calculated from the natural abundance of stable isotopes present in the molecule. The homolanthionine structure is highly similar to that of cystathionine and differs by only the content of an additional methylene group. Quantification of homolanthionine in cell extracts of exponentially growing C. glutamicum ΔmcbR revealed a substantial accumulation of 250 μmol gCDW−1. With a cytoplasmic volume of C. glutamicum of 1.9 μl mg−1 (6), this equals an intracellular level of 130 mM. It should be noted that homolanthionine is not commercially available and that therefore its structure could not be confirmed by the analysis of the pure compound. The GC/MS data combined with feeding of labeled precursor substrates, however, provide a very strong indication for the identity of the compound. It is now interesting to see where homolanthionine originates in the metabolism of C. glutamicum and how this is related to the role of mcbR.
FIG. 2.
GC/MS of TBDMS-derivatized l-cystathionine (A) and l-homolanthionine (B). The molecular ions have m/z values of 678 (cystathionine) and 692 (homolanthionine). The mass difference of 14 is due to the additional methylene group present in homolanthionine. Characteristic fragments at [M-15], [M-57], [M-85], and [M-302] typically found for TBDMS-derivatized compounds also exhibited a mass difference of 14. The ions at m/z 170, m/z 244, and m/z 272 are characteristic fragments of the homocysteine residue in both molecules. l-Cystathionine was applied as a pure compound, and l-homolanthionine was from a cell extract of C. glutamicum ATCC 13032 ΔmcbR.
FIG. 3.
Structure of l-homolanthionine {S-[(3S)-3-amino-3-carboxypropyl]-l-homocysteine; CAS 102044-65-5}.
Metabolic origin of homolanthionine.
The first insight into the metabolic origin of homolanthionine was obtained via a triple-deletion mutant, C. glutamicum ΔmcbR Δhom Δhsk. This mutant cannot synthesize homoserine from aspartate semialdehyde, and additionally, it cannot convert homoserine into homoserine phosphate. It was found that this mutant is auxotrophic for threonine and homoserine as expected. Subsequently, cultivation of C. glutamicum ΔmcbR Δhom Δhsk was performed on minimal medium with 10 g liter−1 [13C6]glucose, 10 mM [13C4]threonine, and 10 mM nonlabeled homoserine. An additional cultivation on nonlabeled glucose, threonine, and homoserine was carried out in parallel. Homolanthionine was present in significant amounts in cell extracts of this strain. GC/MS labeling analysis of homolanthionine from both cultivations revealed identical mass spectra, i.e., that of the nonlabeled molecule. This clearly indicated that homoserine was exclusively the precursor of homolanthionine. Taking the number of carbons of homolanthionine (C8) into account, this also indicated that two homoserine C4 units are probably required to yield one molecule of homolanthionine. Thus, homolanthionine originates directly from the methionine pathway of C. glutamicum.
In additional experiments, C. glutamicum ΔmcbR Δhom Δhsk was cultivated under the same conditions, except that methionine, cystathionine, or homocysteine was fed as an additional substrate instead of homoserine. The triple mutant grew on all of these substrates. However, no significant accumulation of homolanthionine was observed. Obviously, the formation of homolanthionine required one of the intermediates at the beginning of the methionine pathway, either homoserine or O-acetylhomoserine.
Combining all the findings, a potential route for the formation of homolanthionine in C. glutamicum could be postulated. It was very likely that an enzyme from the methionine pathway is involved. One possibility could be a side reaction of cystathionine-γ-synthase (MetB), which might accept homocysteine instead of the normal substrate cysteine and thus produces homolanthionine instead of cystathionine, via linkage to O-acetylhomoserine. To further address this question, MetB was overexpressed in E. coli and characterized.
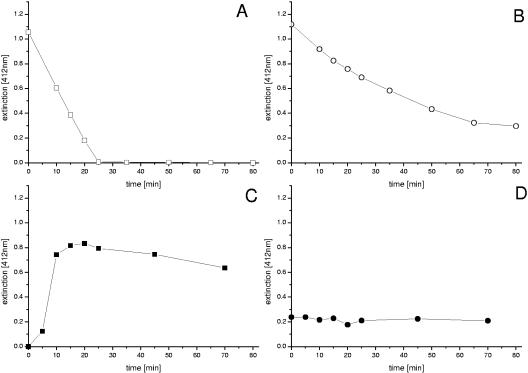
Isolation and characterization of cystathionine-γ-synthase (MetB).
MetB exhibited high activity when incubated with its normal substrates O-acetylhomoserine and cysteine (Fig. 4A). However, MetB could also use homocysteine instead of cysteine (Fig. 4B). Analysis of samples (0 min, 80 min) from the enzyme assays by HPLC and GC/MS showed that the product formed by MetB from O-acetylhomoserine and homocysteine was indeed homolanthionine. In the control incubation, MetB converted cysteine and O-acetylhomoserine into cystathionine. Additional studies on the kinetics of MetB yielded a Km value for cysteine of 260 μM, whereas the Km observed for homocysteine (540 μM) was significantly higher. The MetB activity in crude cell extracts of C. glutamicum ΔmcbR was about threefold higher than that of the wild type, showing that the expression of this enzyme is also probably controlled by McbR.
FIG. 4.
Characterization of cystathionine-γ-synthase (MetB) and cystathionine-β-lyase (MetC) from C. glutamicum after overexpression as His-tagged proteins in E. coli and subsequent purification. The conversion of O-acetylhomoserine and l-cysteine (A) and O-acetylhomoserine and l-homocysteine (B) by MetB and the cleavage of cystathionine (C) and l-homolanthionine (D) by MetC were monitored by photometric measurement of free SH groups via Ellman's reagent at 412 nm. l-Homolanthionine was added from the assay mixture of the MetB assay after removal of the MetB enzyme by three cycles of filtration (molecular weight cutoff, 20,000) (Centrisart; Sartorius, Göttingen, Germany). This solution contained primarily homolanthionine and to some extent remaining homocysteine and O-acetylhomoserine, which explains the initial levels of free SH groups in the assay (D).
Isolation and characterization of cystathionine-β-lyase (MetC).
The incubation of MetC with its natural substrate cystathionine led to the effective release of free SH groups, i.e., via the accumulation of homocysteine (Fig. 4C). MetC revealed slight activity when incubated with homolanthionine, as indicated by the decrease of the homolanthionine level from 200 μM to 170 μM during the incubation and a corresponding increase of the level of homocysteine. This activity, however, was too weak to be observed in the photometric assay (Fig. 4D). Thus, we conclude that MetC is capable of cleaving homolanthionine at low rates. Analogous to the cleavage of cystathionine by MetC, which leads to homocysteine and pyruvate, the cleavage of homolanthionine should result in homocysteine and 2-oxobutanoate. The latter is a precursor of isoleucine. Due to this, further studies were performed to investigate the influence of McbR deletion on the isoleucine biosynthetic pathway in C. glutamicum.
Impact on isoleucine metabolism.
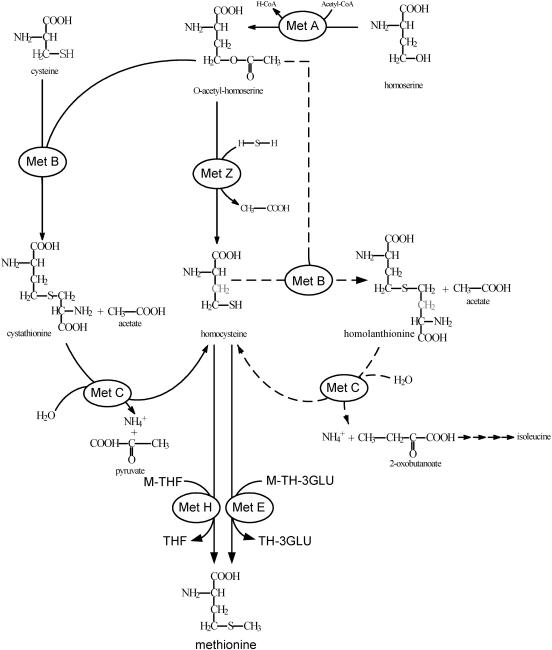
In order to investigate the biosynthesis of isoleucine, C. glutamicum ΔmcbR Δhom Δhsk was cultivated on [13C6]glucose, [13C4]threonine, and nonlabeled homoserine. During the exponential phase, cells were harvested and hydrolyzed for 13C labeling analysis of amino acids from the cell protein, especially isoleucine, threonine, and alanine. For threonine and alanine, the fully labeled mass isotopomer displayed the major fraction. The threonine in the cell protein showed exactly the labeling pattern of the threonine added to the medium. The labeling degree of alanine agreed with that of the glucose added. In the performed tracer study, threonine and pyruvate, the two precursors of the known pathway for isoleucine biosynthesis, were thus completely labeled. The same was expected for isoleucine synthesized via this pathway. However, only 76.3% of the isoleucine in the cell protein was fully labeled. In contrast, the fraction of fully labeled threonine in the protein (88.8%) was significantly higher. This clearly showed that an alternative pathway for isoleucine formation was active in vivo. A closer inspection of the mass isotopomer distribution of isoleucine sheds light on this novel route. Obviously, 13% of the isoleucine formed contained two 13C atoms (Table 2). This situation is possible only if the C4 precursor is unlabeled and pyruvate is labeled, since the latter adds exactly two carbon atoms to the final isoleucine molecule. Due to the deletions present in C. glutamicum ΔmcbR Δhom Δhsk, it is clear that the nonlabeled C4 precursor can come only from homoserine and thus via the methionine pathway. To summarize, C. glutamicum is able to generate isoleucine by a so-far-undescribed pathway which is independent from threonine. The findings are complemented by the fact that the intracellular isoleucine level was indeed significantly increased in C. glutamicum ΔmcbR. Obviously, the activation of the novel pathway leads to enhanced isoleucine supply. Combining previous knowledge of the methionine pathway and the new findings of the present work, we suggest an extended pathway for methionine and isoleucine biosynthesis in C. glutamicum (Fig. 5). In addition to the reactions known before, this extended scheme includes the newly found side reactions of MetB and MetC leading to the formation and cleavage of homolanthionine, respectively. Moreover, the novel biosynthetic pathway for isoleucine that branches off from the methionine pathway is shown.
TABLE 2.
GC/MS labeling analysis of t-butyl-dimethylsilyl-derivatized amino acids from the cell protein of C. glutamicum ΔmcbR Δhom Δhsk cultivated on 99% [13C6]glucose, 98% [13C4]threonine, and nonlabeled homoserine
| Mass isotopomer | Relative mass fraction fora:
|
||
|---|---|---|---|
| Isoleucine | Threonine | Alanine | |
| M | 0.5 | 1.1 | 0.7 |
| M + 1 | 0.5 | 0.4 | 0.6 |
| M + 2 | 13.3 | 1.1 | 5.2 |
| M + 3 | 1.4 | 8.5 | 93.5 |
| M + 4 | 8.0 | 88.8 | |
| M + 5 | 76.3 | ||
The relative mass isotopomer fractions of the ion clusters of m/z 200 (isoleucine, C2 to C6), m/z 404 (threonine, C1 to C4), and m/z 260 (alanine, C1 to C3) are given as percentages. The nonlabeled mass isotopomer fraction is denoted as M and the single-labeled mass isotopomer fraction as M + 1; corresponding terms represent higher labeling.
FIG. 5.
Extended pathway for methionine and isoleucine biosynthesis in C. glutamicum. In addition to the reactions known before, the newly found side reactions of cystathionine-β-synthase (MetB) and cystathionine-β-lyase (MetC) leading to the formation and cleavage of homolanthionine, respectively, are shown. Further enzymes involved are MetA (homoserine transacetylase) and MetZ (O-acetylhomoserine sulfhydrolase).
Effect of MetB deletion on accumulation of homolanthionine.
In contrast to C. glutamicum ΔmcbR, which accumulated high levels of homolanthionine as described above, homolanthionine was not present in cell extracts of C. glutamicumΔmcbR ΔmetB. This gives evidence that MetB is the only source for homolanthionine in C. glutamicum.
In vivo carbon flux through the methionine pathway in C. glutamicum ΔmcbR.
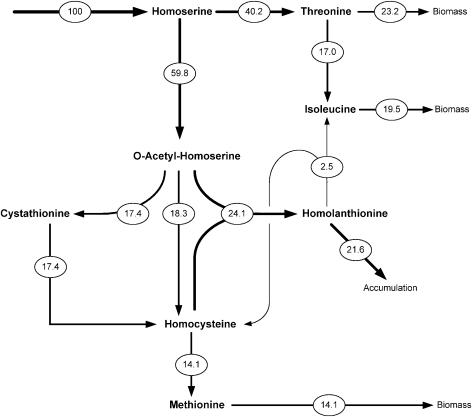
The data from the present work provided information on the relative supply of isoleucine from threonine and from homolanthionine (Table 2). The intracellular homolanthionine pool was so large that the demand for its supply (250 μmol g−1) was considered. In contrast, the demand for the other intermediates could be neglected due to their small intracellular levels (Fig. 1). Together with the anabolic demand for methionine (146 μmol g−1), isoleucine (202 μmol g−1), and threonine (275 μmol g−1) in C. glutamicum (30) and with previous data on the relative flux through the parallel pathways of transsulfuration and direct sulfhydrylation (10), this study provides a detailed insight into the relative carbon flux through the methionine pathway in the McbR deletion mutant (Fig. 6). Hereby, the new findings, the side reaction of MetB leading to homolanthionine, and the novel route towards isoleucine are considered. At the homoserine node, the carbon flux is almost equally directed towards homoserine phosphate into the threonine biosynthesis and towards O-acetylhomoserine into the methionine pathway. It can be further seen that O-acetylhomoserine sulfhydrolase (MetZ) consumes only a small fraction of O-acetylhomoserine. The major flux consuming O-acetylhomoserine is catalyzed by MetB. MetB, however, shows two in vivo activities. A flux of 17.4% is assigned to its natural reaction leading to cystathionine, whereas an even higher flux of 24.1% is found for the formation of homolanthionine. Due to this latter reaction, a significant backflux results from homocysteine towards homolanthionine. In fact, the major fraction of homocysteine is not channeled towards methionine but is utilized for the formation of homolanthionine. It can be further seen that homolanthionine is to some extent cleaved and that its products are recycled back into the methionine and the isoleucine pathways. The major fraction of homolanthionine is accumulating inside the cell. Overall, from the flux of 59.8% entering the methionine pathway at the level of O-acetylhomoserine, only a relatively small flux (14.1%) is converted into methionine. The remaining fraction is either stored as homolanthionine or directed towards the synthesis of isoleucine.
FIG. 6.
Relative flux distribution in the methionine pathway of C. glutamicum ΔmcbR. Fluxes are given as relative fluxes with respect to the homoserine influx. Data on the split ratio between transsulfuration and direct sulfhydrylation are taken from the work of Hwang et al. (10).
DISCUSSION
Recently, the transcriptional repressor McbR, regulating the expression of a number of genes from methionine biosynthesis and sulfur assimilation, was identified in C. glutamicum. It was suggested that the deletion of McbR is an important step towards the development of C. glutamicum strains for the biotechnological production of methionine. In the present work, the effects of deleting McbR in C. glutamicum were investigated from a metabolic perspective. This included the comparison of the wild type, C. glutamicum ATCC 13032, and the corresponding deletion mutant, C. glutamicum ATCC 13032 ΔmcbR. As a response to the McbR deletion, almost all intracellular pools of intermediates from the methionine pathway were changed. This comprised a decreased homoserine level, which indicates an enhanced flux into the methionine pathway compared to that of the wild type. Homocysteine, however, was increased by a factor of 50. The elevated homocysteine level in C. glutamicum ΔmcbR is probably caused by the deregulated expression of homoserine dehydrogenase (Hom), O-acetylhomoserine sulfhydrolase (MetZ), and S-adenosylmethionine synthase (MetK), which are all controlled by McbR (27). Homocysteine is the direct product of the reactions catalyzed by Hom and MetZ, whereas MetK converts methionine into S-adenosylmethionine, which is then recycled via S-adenosylhomocysteine back to homocysteine (26). Interestingly, the intracellular methionine titer remained constant in comparison to that of the C. glutamicum wild type. In combination with the increase of the homocysteine level in C. glutamicum ΔmcbR, this indicates that the preceding step, the reaction catalyzed by the two methionine synthases (MetE and MetH), is limiting. The increased expression of these two enzymes due to the deletion of McbR (26) is obviously not sufficient. In other organisms, homocysteine is directly involved in the regulation of the expression of methionine synthase on the transcriptional level (12, 22). Such a regulation, which additionally could help the cell to avoid undesired accumulation of homocysteine, however, has not been described for C. glutamicum. The limitation on the level of the methionine synthases could have different reasons. Possibly, methionine itself causes a feedback inhibition of both methionine synthases. Another possibility could be a limited supply with 5-methyltetrahydrofolate or 5-methyltetrahydropteroyltri-l-glutamate as the donor of the terminal methyl group in methionine.
The most pronounced change on the metabolic level was the drastic accumulation of a metabolite that could be identified as homolanthionine. Homolanthionine accumulation has been described for other organisms, such as E. coli and Aspergillus nidulans (8, 23). The strains showing this behavior were auxotrophic for methionine due to a knockout of methionine synthase. Methionine synthase, as the final enzyme of the methionine pathway, converts homocysteine into methionine. It seems likely that the lack of a functioning methionine synthase leads to an elevated intracellular homocysteine level, similar to the effects resulting from the deletion of McbR in the present study. The accumulation of homolanthionine observed in these mutants might therefore also be due to an increased pool of homocysteine, activating the side reaction of MetB. Also, cystathionase from human liver (29) and from Streptomyces phaeochromogenes (13) and cystathionine-γ-synthase from Arabidopsis thaliana (25) can catalyze the formation of homolanthionine from homocysteine and O-acetylhomoserine.
It was further shown in the present work that MetB is responsible for the formation of homolanthionine in C. glutamicum. Due to obviously low substrate specificity, MetB uses homocysteine instead of cysteine as a substrate. This reaction yields homolanthionine instead of cystathionine, the natural product of MetB. Although the substrate affinity of MetB for cysteine (Km = 260 μM) is higher than that for homocysteine (Km = 540 μM), the 50-fold increase in the intracellular homocysteine pool from 30 μM to 1,500 μM (calculated from the intracellular pool size and the cytoplasmic volume) (6) can explain the significant formation of homolanthionine in C. glutamicum ΔmcbR.
MetC from C. glutamicum was shown to cleave homolanthionine at a low rate. The natural reaction of MetC, the cleavage of cystathionine, was clearly the preferred route. The Km of MetC for cystathionine was 110 μM. In comparison, this enzyme shows different affinities for cystathionine in other organisms (Table 3). The Km value of the E. coli cystathionase for homolanthionine was previously determined to be 4,540 μM (Table 3). The affinity of the cystathionase from E. coli for homolanthionine is thus about 100-fold lower than that for cystathionine. Concerning the similarity of the Km values of MetC for cystathionine, it appears likely that the affinity of MetC from C. glutamicum for homolanthionine is in the same range as that of MetC E. coli. The intracellular level of homolanthionine in C. glutamicum ΔmcbR was 130 mM. This concentration is still far above the probable Km value, so the cleavage of homolanthionine is probably limited by the specific reaction rate of MetC but not by the availability of the substrate. It remains possible, however, that other enzymes are also involved in the cleavage of homolanthionine.
TABLE 3.
Affinity constants (Km) for cystathionine-γ-synthase (MetB) and cystathonine-β-lyase (MetC) from C. glutamicum (this study) and from data for other bacteria and a plant taken from the literature
| Enzyme | Substrate | Km (μM) (organism or plant) | References |
|---|---|---|---|
| Cystathionine-γ-synthase | l-Cysteine | 260 (C. glutamicum) | This study |
| l-Cysteine | 180 (Spinach) | 24 | |
| l-Cysteine | 50 (E. coli) | 7 | |
| l-Homocysteine | 540 (C. glutamicum) | This study | |
| Cystathionine-β-lyase | l-Cystathionine | 110 (C. glutamicum) | This study |
| l-Cystathionine | 40 (E. coli) | 2 | |
| l-Cystathionine | 220 (Serovar Typhimuriuma) | 2 | |
| l-Cystathionine | 70 (Bordetella avium) | 5 | |
| l-Homolanthionine | 4,540 (E. coli) | 2 |
Salmonella enterica serovar Typhimurium.
In C. glutamicum ΔmcbR, a novel route for the biosynthesis of isoleucine, branching off from the methionine pathway, was identified. This pathway accounted for about 13% of the total isoleucine supply. From the enzymatic studies with MetC, we conclude that the cleavage of homolanthionine by MetC supplies 2-oxobutanoate, a direct isoleucine precursor. Other enzymes, potentially producing 2-oxobutanoate from different intermediates of the methionine pathway, are methionine methanethiol lyase (EC 4.4.1.11), homocysteine hydrogen sulfide lyase (EC 4.4.1.2), and cystathionine cysteine lyase (EC 4.4.1.1). Feeding nonlabeled methionine, homocysteine, or cystathionine, the substrates of these enzymes, together with [13C6]glucose and [13C4]threonine, led to completely labeled isoleucine, so that their contribution to the formation of 2-oxobutanoate could be excluded (data not shown). Further evidence comes from the fact that none of these enzymes has been found in C. glutamicum (KEGG database, http://www.genome.jp/kegg/metabolism.html). On the basis of previous findings and results of the present work, we suggest an extended pathway for methionine and isoleucine biosynthesis in C. glutamicum (Fig. 5). Metabolic flux analysis of C. glutamicum ΔmcbR revealed that only 25% of the O-acetylhomoserine at the entry of the methionine pathway is utilized for methionine biosynthesis and that the dominating fraction of 75% is either stored inside the cell as homolanthionine or redirected towards the formation of isoleucine. It appears very likely that the effects observed here, i.e., the formation of homolanthionine and increased isoleucine synthesis via the methionine pathway, generally occur at increased levels of homocysteine. Therefore, we think that they are not restricted to ΔmcbR mutants. Also, other strains of C. glutamicum exhibiting an engineered methionine metabolism, e.g., targeted overexpression of enzymes of the methionine pathway, might show increased intracellular levels of homolanthionine and isoleucine. We can imagine that, under certain conditions, isoleucine could even be a secreted by-product in methionine-deregulated strains. The increased intracellular isoleucine level could trigger the export of this compound into the medium (14, 21).
The deletion of the transcriptional repressor McbR was previously suggested as the first important step towards the overproduction of methionine by C. glutamicum for a future biotechnological application (27). The deletion strain, however, does not secrete any methionine. The high homocysteine level caused by the deregulated expression of enzymes from the methionine pathway in the ΔmcbR mutant and the resulting enormous accumulation of homolanthionine are, however, highly undesired. C. glutamicum ΔmcbR ΔmetB does not exhibit homolanthionine formation. The deletion of MetB in McbR strains could therefore be a promising strategy for the development and optimization of methionine-overproducing strains. In this regard, the amplification of flux through the direct sulfhydrylation pathway in C. glutamicum ΔmcbR ΔmetB could be an interesting next step.
REFERENCES
- 1.Duerre, J. A., and C. H. Miller. 1966. Preparation of l-homocysteine from l-homocysteine thiolactone. Anal. Biochem. 17:310-315. [DOI] [PubMed] [Google Scholar]
- 2.Dwivedi, C. M., R. C. Ragin, and J. R. Uren. 1982. Cloning, purification, and characterization of beta-cystathionase from Escherichia coli. Biochemistry 21:3064-3069. [DOI] [PubMed] [Google Scholar]
- 3.Eggeling, L., and H. Sahm. 1999. l-Glutamate and l-lysine: traditional products with impetuous developments. Appl. Microbiol. Biotechnol. 52:146-153. [Google Scholar]
- 4.Ellman, G., and H. Lysko. 1979. A precise method for the determination of whole blood and plasma sulfhydryl groups. Anal. Biochem. 93:98-102. [PubMed] [Google Scholar]
- 5.Gentry-Weeks, C. R., J. M. Keith, and J. Thompson. 1993. Toxicity of Bordetella avium beta-cystathionase toward MC3T3-E1 osteogenic cells. J. Biol. Chem. 268:7298-7314. [PubMed] [Google Scholar]
- 6.Gutmann, M., C. Hoischen, and R. Krämer. 1992. Carrier-mediated glutamate secretion by Corynebacterium glutamicum under biotin limitation. Biochim. Biophys. Acta 1112:115-123. [DOI] [PubMed] [Google Scholar]
- 7.Holbrook, E. L., R. C. Greene, and J. H. Krueger. 1990. Purification and properties of cystathionine gamma-synthase from overproducing strains of Escherichia coli. Biochemistry 29:435-442. [DOI] [PubMed] [Google Scholar]
- 8.Huang, H. T. 1963. Accumulation of 1-homolanthionine by an Escherichia coli mutant. Biochemistry 2:296-298. [DOI] [PubMed] [Google Scholar]
- 9.Hwang, B. J., Y. Kim, H. B. Kim, H. J. Hwang, J. H. Kim, and H. S. Lee. 1999. Analysis of Corynebacterium glutamicum methionine biosynthetic pathway: isolation and analysis of metB encoding cystathionine gamma-synthase. Mol. Cells 9:300-308. [PubMed] [Google Scholar]
- 10.Hwang, B.-J., H.-J. Yeom, Y. Kim, and H.-S. Lee. 2002. Corynebacterium glutamicum utilizes both transsulfuration and direct sulfhydrylation pathways for methionine biosynthesis. J. Bacteriol. 184:1277-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jäger, W., A. Schäfer, A. Pühler, G. Labes, and W. Wohlleben. 1992. Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not in Streptomyceslividans. J. Bacteriol. 174:5462-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kacprzak, M. M., I. Lewandowska, R. G. Matthews, and A. Paszewski. 2003. Transcriptional regulation of methionine synthase by homocysteine and choline in Aspergillus nidulans. Biochem. J. 376:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanzaki, H., M. Kobayashi, T. Nagasawa, and H. Yamada. 1986. Synthesis of S-substituted l-homocysteine derivatives by cystathionine γ-lyase of Streptomyces phaeochromogenes. Agric. Biol. Chem. 50:391-397. [Google Scholar]
- 14.Kennerknecht, N., H. Sahm, M.-R. Yen, M. Pátek, M. H. Saier, Jr., and L. Eggeling. 2002. Export of l-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J. Bacteriol. 184:3947-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krömer, J. O., M. Fritz, E. Heinzle, and C. Wittmann. 2005. In vivo quantification of intracellular amino acids and intermediates of the methionine pathway in Corynebacterium glutamicum. Anal. Biochem. 340:171-173. [DOI] [PubMed] [Google Scholar]
- 16.Krömer, J. O., O. Sorgenfrei, K. Klopprogge, E. Heinzle, and C. Wittmann. 2004. In-depth profiling of lysine-producing Corynebacterium glutamicum by combined analysis of the transcriptome, metabolome, and fluxome. J. Bacteriol. 186:1769-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, H. S. 2005. Sulfur metabolism and its regulation, p. 351-376. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, Fla.
- 18.Lee, H. S., and B. J. Hwang. 2003. Methionine biosynthesis and its regulation in Corynebacterium glutamicum: parallel pathways of transsulfuration and direct sulfhydrylation. Appl. Microbiol. Biotechnol. 62:459-467. [DOI] [PubMed] [Google Scholar]
- 19.Leuchtenberger, W. 1996. Amino acids: technical production and use, p. 465-502. In H. J. Rehm, G. Reed, A. Pühler, and P. Stadler (ed.), Biotechnology, vol. 6. VCH, Weinheim, Germany. [Google Scholar]
- 20.Losen, M., B. Frölich, M. Pohl, and J. Büchs. 2004. Effect of oxygen limitation and medium composition on Escherichia coli fermentation in shake-flask cultures. Biotechnol. Prog. 20:1062-1068. [DOI] [PubMed] [Google Scholar]
- 21.Morbach, S., H. Sahm, and L. Eggeling. 1996. l-Isoleucine production with Corynebacterium glutamicum: further flux increase and limitation of export. Appl. Environ Microbiol. 62:4345-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortiou, S., J. M. Alberto, J. L. Gueant, and M. Merten. 2004. Homocysteine increases methionine synthase mRNA level in Caco-2 cells. Cell. Physiol. Biochem. 14:407-414. [DOI] [PubMed] [Google Scholar]
- 23.Paszewski, A., and J. Grabski. 1975. Homolanthionine in fungi: accumulation in the methionine-requiring mutants of Aspergillus nidulans. Acta Biochim. Pol. 22:263-268. [PubMed] [Google Scholar]
- 24.Ravanel, S., M. Droux, and R. Douce. 1995. Methionine biosynthesis in higher plants. I. Purification and characterization of cystathionine gamma-synthase from spinach chloroplasts. Arch. Biochem. Biophys. 316:572-584. [DOI] [PubMed] [Google Scholar]
- 25.Ravanel, S., B. Gakiere, D. Job, and R. Douce. 1998. Cystathionine gamma-synthase from Arabidopsis thaliana: purification and biochemical characterization of the recombinant enzyme over expressed in Escherichia coli. Biochem. J. 331:639-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rey, D. A., S. S. Nentwich, D. J. Koch, C. Rückert, A. Pühler, A. Tauch, and J. Kalinowski. 2005. The McbR repressor modulated by the effector substance s-adenosylhomocysteine controls directly the transcription of a regulon involved in sulphur metabolism of Corynebacterium glutamicum ATCC 13032. Mol. Microbiol. 56:871-887. [DOI] [PubMed] [Google Scholar]
- 27.Rey, D. A., A. Pühler, and J. Kalinowski. 2003. The putative transcriptional repressor McbR, member of the TetR-family, is involved in the regulation of the metabolic network directing the synthesis of sulfur containing amino acids in Corynebacterium glutamicum. J. Biotechnol. 103:51-65. [DOI] [PubMed] [Google Scholar]
- 28.Rückert, C., A. Pühler, and J. Kalinowski. 2003. Genome-wide analysis of the l-methionine biosynthetic pathway in Corynebacterium glutamicum by targeted gene deletion and homologous complementation. J. Biotechnol. 104:213-228. [DOI] [PubMed] [Google Scholar]
- 29.Tallan, H. H., T. A. Pascal, K. Schneidman, B. M. Gillam, and G. E. Gaull. 1971. Homolanthionine synthesis by human liver cystathionase. Biochem. Biophys. Res. Commun. 43:303-310. [DOI] [PubMed] [Google Scholar]
- 30.Wittmann, C., and A. de Graaf. 2005. Metabolic flux analysis in Corynebacterium glutamicum, p. 277-304. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, Fla.
- 31.Wittmann, C., M. Hans, and E. Heinzle. 2002. In vivo analysis of intracellular amino acid labelings by GC/MS. Anal. Biochem. 307:379-382. [DOI] [PubMed] [Google Scholar]