Abstract
Standard biochemical tests have revealed that hemin and menadione auxotrophic Staphylococcus aureus small-colony variants (SCVs) exhibit multiple phenotypic changes. To provide a more complete analysis of the SCV phenotype, two genetically defined mutants with a stable SCV phenotype were comprehensively tested. These mutants, generated via mutations in menD or hemB that yielded menadione and hemin auxotrophs, were subjected to phenotype microarray (PM) analysis of over 1,500 phenotypes (including utilization of different carbon, nitrogen, phosphate, and sulfur sources; growth stimulation or inhibition by amino acids and other nutrients, osmolytes, and metabolic inhibitors; and susceptibility to antibiotics). Compared to parent strain COL, the hemB mutant was defective in utilization of a variety of carbon sources, including Krebs cycle intermediates and compounds that ultimately generate ATP via electron transport. The phenotype of the menD mutant was similar to that of the hemB mutant, but the defects in carbon metabolism were more pronounced than those seen with the hemB mutant. In both mutant strains, hexose phosphates and other carbohydrates that provide ATP in the absence of electron transport stimulated growth. Other phenotypes of SCV mutants, such as hypersensitivity to sodium selenite, sodium tellurite, and sodium nitrite, were also uncovered by the PM analysis. Key results of the PM analysis were confirmed in independent growth studies and by using Etest strips for susceptibility testing. PM technology is a new and efficient technology for assessing cellular phenotypes in S. aureus.
The discovery and characterization of S. aureus small-colony variants (SCVs) have provided new insight into the pathogenesis associated with staphylococcal diseases (20, 21). These variants are increasingly found in antibiotic-refractory, recurrent, and persistent infections, such as chronic osteomyelitis, chronic skin and soft-tissue infections, and chronic airway infections in cystic fibrosis patients (10, 21, 24, 26-28). SCVs gain a survival advantage by their ability to hide within host cells, which protects these naturally occurring variants of S. aureus from host defenses and decreases exposure to antibiotics. Indeed, intracellular persistence assays revealed that >200-fold more SCV cells than cells of the clonal identical parent strain with the normal phenotype persisted intracellularly (9, 15, 27, 29). Many studies have shown that SCVs can be phagocytosed by nonprofessional phagocytes, such as fibroblasts and endothelial cells, but do not lyse these cells, probably because the SCVs produce very little or no alpha-toxin (11, 25, 29).
Clinical S. aureus SCVs are frequently auxotrophic for menadione or hemin, two compounds that are involved in the biosynthesis of the electron transport chain components menaquinone and cytochromes, respectively (15, 26). The following phenotypic characteristics of SCVs can be tied together by a common thread, alterations in electron transport, and are very likely linked to disruption of the electron transport chain: (i) slow growth, because cell wall synthesis requires large quantities of ATP, (ii) decreased pigment formation, because carotenoid biosynthesis requires electron transport, (iii) resistance to gentamicin and other aminoglycosides, because uptake of these compounds requires a large membrane potential generated by electron transport, and (iv) partial or complete loss of mannitol fermentation, because utilization of sugar alcohols, such as mannitol, is decreased when electron transport cannot oxidize the NADH produced as mannitol is metabolized to fructose-6-phosphate (20, 29).
To date, there has been only limited phenotypic analysis of genetically undefined, clinical SCV isolates. Complicating the analysis was the high rate of reversion of the SCV isolates to a large-colony form (9, 10, 27, 28). Therefore, two genetically defined, stable SCVs were generated by (i) interrupting one of the heme biosynthetic genes by inserting a ermB cassette into hemB (29) and (ii) interrupting one of the genes required for menadione biosynthesis by inserting a ermC cassette into menD (2). Both mutants showed phenotypic characteristics typical of clinical SCVs, including slow growth, decreased pigment formation, resistance to aminoglycosides, low coagulase activity, and reduced hemolytic activity. These SCV phenotypes were nearly restored to normal by growth with appropriate medium supplements (hemin or menadione) or by complementation with a wild-type allele of the mutant gene (2, 29).
To provide a more complete analysis of SCV phenotypes and to obtain deeper insight into the physiological changes that lead to in vivo antibiotic resistance in S. aureus SCVs, mutants that reproduce the SCV phenotype were compared to their parental strain using phenotype microarray (PM) technology (4, 5, 31). This technology is an integrated system of cellular assays, instrumentation, and bioinformatic software for high-throughput screening of cellular phenotypes (5, 31). With this approach, changes in a large number of cellular pathways were identified in both SCV mutants. Some differences between the two mutants were also revealed.
MATERIALS AND METHODS
Bacterial strains.
A hemB mutant of S. aureus strain COL was constructed by allelic replacement with a ermB cassette-inactivated hemB gene, as previously described (25, 29). A menD mutant containing a chromosomal insertion of ermC within menD was constructed by allelic exchange using S. aureus strain 8325-4 as the parent strain (2), and for this study, the mutation was transduced into strain COL and the insertion site was confirmed by Southern analysis.
PM analysis.
Sixteen 96-well PMs (metabolic panels PM1 to PM8; sensitivity panels PM9, PM10, and PM31 to PM36) were used in this study. To monitor cell phenotypes, PMs use a patented redox chemistry. If bacterial growth is supported by the medium in an assay well, the actively metabolizing cells reduce a tetrazolium dye. Reduction of the dye results in color formation in the well, and the phenotype is considered “positive.” If metabolism is hampered or growth is poor or not supported, the phenotype is “weakly positive” or “negative,” and little or no color is formed in the well. This colorimetric redox assay allows both amplification and precise quantitation of bacterial growth. An alternative option is to omit the redox dye and measure turbidity, which directly reflects cell growth. In our experiments, the redox dye chemistry was used for PM1 to PM8, and direct turbidity readouts were used for the remaining PMs because the turbidity signal did not need amplification with dye. Incubation and recording of phenotypic data were performed with an OmniLog instrument, which captured a digital image of the microarray and stored quantitative color change or turbidity values in a computer file. The computer files could be displayed in the form of kinetic graphs. For each strain, 1,536 phenotypes were recorded simultaneously four times each hour by the OmniLog.
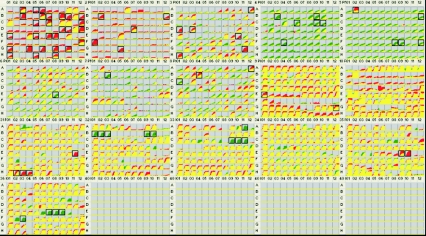
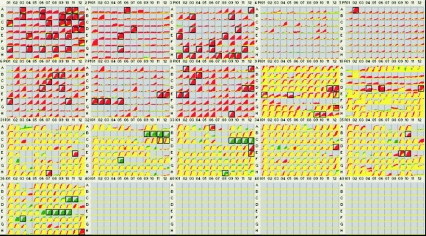
To assess the altered phenotypes of the hemB and menD mutants, each mutant was compared to its parent strain (S. aureus COL). The parent strain was recorded as a red tracing, and the mutant was recorded as a green tracing. Color-coded kinetic graphs could then be overlaid by the OmniLog PM bioinformatic software, and differences were visualized and quantified. When the data for two strains revealed equivalent metabolism or growth responses in a well, the red and green kinetic graphs overlapped and were yellow. When one strain gave a stronger response in a well, either the red or green kinetic graph superseded the other graph and was visually highlighted as a patch of red or green. Standard PM testing protocols were used (http://www.biolog.com/PM_FAQ.html) with conditions similar to those used previously (5, 22, 31). A carbon source consisting of sodium pyruvate and sodium α-ketoglutarate was used in PM3 to PM8. The inoculating cell densities used in this study were 85% transmittance in metabolic panels and a 1:100 dilution of 85% transmittance in sensitivity panels (31). Both mutants and parent strain COL were tested twice, and a consensus result was obtained. The phenotypic changes listed in Table 1 were the changes detected in both PM runs (for a complete analysis, see Tables S1 and S2 in the supplemental material). The data shown in Fig. 1 and 2 were obtained after 24 h of incubation in the OmniLog.
TABLE 1.
Results of comparing the hemB and menD mutants with their wild-type parental strain, S. aureus COL
| Mode of action | Substrate(s) | Phenotype of:
|
|
|---|---|---|---|
| hemB mutant | menD mutant | ||
| Nutrient stimulation | d-(+)-glucose, N-acetyl d-glucosamine, 2′-deoxyguanosine, thymidine | Gained | No difference |
| N source, amine | Ethylenediamine | No difference | Lost |
| N source, amino acid | Lys-Thr, Gly-Lys, Lys-Gly | Gained | No difference |
| His-Glu, Gly-Asn, δ-Amino-N-Valeric Acid, Ala-Asp, l-pyroglutamic acid, l-glutamic acid | Lost | Lost | |
| N-Acetyl-l-glutamic acid | Lost | No difference | |
| Thr-Gln, Thr-Leu, Thr-Glu, Thr-Arg, Thr-Phe, Met-Thr, Leu-Gly-Gly, Thr-Gly, Thr-Asp, Thr-Pro, Asn-Glu, Arg-Glu, Thr-Ser, Thr-Ala, Gly-Thr, His-Pro, Gln-Gly, Glu-Gly, Arg-Gln, Gly-Glu, histamine, l-histidine, Ala-Thr, Ala-Glu, Gly-Gln, l-proline | No difference | Lost | |
| N source, carbohydrate | d-Mannosamine | Lost | No difference |
| d-Glucosamine | No difference | Lost | |
| N source, nucleic acid | Guanine, alloxan | No difference | Lost |
| N source, nucleoside | Xanthosine | No difference | Lost |
| N source, urea | Parabanic acid | No difference | Lost |
| P source, carbohydrate | dl-α-Glycerol phosphate, d-glucosamine-6-phosphate, guanosine-3′-monophosphate, cytidine-3′-monophosphate, cytidine-5′-monophosphate | Gained | No difference |
| S source | Tetrathionate | Gained | No difference |
| C source, fatty acid | Tween 80, Tween 20 | Lost | No difference |
| C source, amino acid | l-Threonine, Ala-Gly, glycine, l-glutamic acid, glycyl-l-glutamic acid | Lost | Lost |
| C source, amino acid | Glycyl-l-proline, l-glutamine, | Lost | No difference |
| C source, carbohydrate | Glycerol, N-acetyl-d-mannosamine, β-methyl-d-glucoside, d-galactose, d-fructose, N-acetyl-d-glucosamine, d-gluconic acid, turanose | Lost | Lost |
| N-Acetyl-neuraminic acid, dl-α-glycerol phosphate, d-psicose, α-d-lactose, 3-0-β-d-galacto-pyranosyl-d-arabinose, lactulose | Lost | No difference | |
| d-Glucosamine, dextrin, d-mannitol, maltose, d-(+)-glucose, maltotriose, sucrose, d-fructose-6-phosphate, d-trehalose, d-glucose-6-phosphate, d-mannose | No difference | Lost | |
| C source, carboxylate | dl-α-Hydroxybutyric acid, l-lactic acid, methyl pyruvate, formic acid | Lost | Lost |
| d-Lactic acid methyl ester, pyruvic acid, succinic acid | Lost | No difference | |
| C source, nucleoside | Uridine, thymidine, inosine | Lost | Lost |
| DNA intercalator | Acriflavine | Gained | No difference |
| DNA polymerase | Phleomycin | No difference | Gained |
| DNA gyrase (GN), DNA topoisomerase (GP) | Norfloxacin | No difference | Lost |
| DNA synthesis, nitro compound | Nitrofurantoin, furaltadone | No difference | Lost |
| Folate antagonist, dihyldrofolate reductase | Trimethoprim | Gained | No difference |
| Folate antagonist | Salicylanilide | No difference | Lost |
| Protein synthesis, aminoglycoside | Kanamycin, gentamicin, amikacin, neomycin, sisomicin | Gained | Gained |
| Protein synthesis, lincosamide | Lincomycin | Gained | No difference |
| Protein synthesis, macrolide | Spiramycin | No difference | Lost |
| Cell wall, β-lactam | Penicillin G | No difference | Gained |
| Cell wall, cephalosporin | Cefmetazole | No difference | Gained |
| Inhibitor of calmodulin stimulation of cyclic nucleotide phosphodiesterase | Chlorpromazine | Lost | Lost |
| Membrane detergent | Triton X-100 | Lost | No difference |
| Osmotic sensitivity | 1% sodium formate | Lost | Lost |
| 20% ethylene glycol, 15% ethylene glycol, 7% urea, 5% sodium formate | No difference | Lost | |
| pH, decarboxylase | pH 4.5 + β-hydroxyglutamate, pH 4.5 + urea, pH 4.5 + α-amino-N-butyric acid | No difference | Lost |
| Toxicity | 100 mM ammonium sulfate (pH 8) | Lost | Lost |
| 100 mM sodium nitrite | No difference | Lost | |
| Transport, toxic anion | Sodium selenite | Lost | Lost |
| Sodium thiosulfate, potassium tellurite, potassium chromate | No difference | Lost | |
FIG. 1.
hemB and menD mutants were subjected to PM analysis of 1,536 phenotypes (16 PMs). Incubation and recording of phenotypic data were performed by using an instrument which captures a digital image of the microarray and stores quantitative color change or turbidity values in a computer file displayed in the form of kinetic graphs. The PM kinetic results show consensus data comparing the hemB mutant (green) and its wild-type parental strain, S. aureus COL (red). Red indicates a stronger response by the parent, and green indicates a stronger response by the hemB mutant; when the two strains have equivalent metabolism or equivalent growth responses in a well, the red and green kinetic graphs overlap and are yellow. A box around a growth curves indicates a significant difference in response. The phenotypic changes are listed in Table 1; for a complete analysis, see Tables S1 and S2 in the supplemental material.
FIG. 2.
PM kinetic results showing the consensus data for a comparison of the menD mutant (green) and its wild-type parental strain, S. aureus COL (red). The color codes and boxes are explained in the legend to Fig. 1.
Confirmation studies.
To confirm the PM analysis results, some of the key findings were subsequently investigated by performing independent studies. Thus, growth studies were performed in chemically defined medium (3) to test (i) the utilization of selected carbon sources (compared to PM1 and PM2), (ii) growth stimulation or inhibition by selected osmolytes (compared to PM9), and (iii) inhibition by metabolic inhibitors (compared to PM35) (Table 2). To study growth with and without selected substances, defined concentrations of these compounds were added to overnight cultures (in which the optical density at 578 nm was adjusted to 0.05). Experiments were repeated at least two times. Numbers of CFU and optical densities were determined in duplicate after growth for 24 h at 37°C with shaking at 180 rpm. In addition, susceptibilities to selected antimicrobial agents, particularly aminoglycosides, were tested using Etest strips (AB-Biodisk, Solna, Sweden) (compared to PM32, PM33, and PM36).
TABLE 2.
Results of independent growth studies with selected substances
| Strain | Growth without supplementation (CFU/ml)
|
Growth with supplementationa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Start of growth | After 24 h | 0.2% Mannitol
|
0.2% Galactose
|
5% Ethylene glycol
|
0.2% Sodium selenite
|
|||||
| CFU/ml | Differenceb | CFU/ml | Differenceb | CFU/ml | Differenceb | CFU/ml | Differenceb | |||
| COL | 1.83 × 107 | 1.41 × 1010 | 1.32 × 1011 | 9.38 | 9.37 × 1010 | 6.66 | 2.83 × 109 | 2.01 × 10−1 | 2.80 × 106 | 1.99 × 10−4 |
| hemB mutant | 9.80 × 106 | 1.18 × 109 | 1.37 × 109 | 1.16 | 5.33 × 108 | 4.51 × 10−1 | 4.74 × 108 | 4.02 × 10−1 | 5.00 × 102 | 4.24 × 10−7 |
| menD mutant | 3.92 × 107 | 4.49 × 108 | 1.60 × 106 | 3.57 × 10−3 | 5.98 × 108 | 1.33 | 3.70 × 106 | 8.24 × 10−3 | 1.70 × 102 | 3.79 × 10−7 |
Growth was determined after 24 h.
Difference between growth after 24 h without supplementation and growth after 24 h with supplementation.
RESULTS
The consensus PM results for the hemB mutant tested without hemin supplementation and for the menD mutant tested without menadione supplementation are shown in the Fig. 1 and 2, respectively.
The PM assay of the hemB mutant (Fig. 1, Table 1) verified the previously observed resistance to the aminoglycoside antibiotics kanamycin, gentamicin, amikacin, neomycin, and sisomicin (green wells in PM32, PM33, and PM36). Resistance to lincomycin was also detected (PM32 wells B-1 to B-3), but this was presumably due to the lincosamide resistance on the erm cassette used to disrupt the hemB gene. Slight resistance to trimethoprim (in PM33 well A-11, better growth of the mutant compared to the growth of COL with 3 μg/ml trimethoprim) and slight resistance to the intercalating agent acriflavine (PM31 well A-3) were also detected. The mutant was notably hypersensitive to the detergent Triton X-100 (PM31 wells H-2, H-3, and H-4) and to sodium selenite (PM35 wells E-9 and E-10), as indicted by the pattern of growth. Compared to COL, the mutant grew less well in the rich medium control (PM5 well A-2) but slightly better in the minimal medium (all other wells in PM5).
Another salient difference between the hemB mutant and its parental strain was found in the metabolism of carbon sources. The mutant strain was unable to utilize carbon sources that are metabolized by wild-type S. aureus strain COL (red wells in PM1 and PM2). This defect was very pronounced for carboxylic acids and amino acids. However, the metabolism of carbohydrate carbon sources by the mutant was quantitatively but not qualitatively different, as shown by the fact that the carbohydrate metabolism wells were yellow rather than red. Specifically, there was little or no loss of metabolism of N-acetyl-d-glucosamine, d-trehalose, d-mannose, d-mannitol, d-glucose-6-phosphate, d-glucose, maltose, sucrose, d-fructose-6-phosphate, maltotriose, inosine, dextrin, turanose, and d-glucosamine (PM1 wells A-3, A-10, A-11, B-11, C-1, C-9, C-10, D-11, E-4, E-10, and F-12 and PM2 wells A-6, D-7, and E-5, respectively).
The menD mutant was metabolically more restricted than the hemB mutant (Fig. 2 and Table 1). While the menD mutant has many of the same phenotypes as the hemB mutant, the defects in growth with the carbon sources were quantitatively more pronounced. Again, resistance to aminoglycoside antibiotics was observed (PM32, PM33, and PM36), and the menD mutant grew in the presence of higher concentrations of the aminoglycosides. As observed with the hemB mutant, the menD mutant was notably hypersensitive to sodium selenite (PM35 wells E-9 and E-10) and slightly hypersensitive to sodium tellurite (PM32 wells C-11 and C-12) and sodium nitrite (PM9 wells H-9 and H-10). Unlike the hemB mutant, it was also hypersensitive to ethylene glycol (PM9 wells D-11 and D-12).
The carbon metabolism defects were even more pronounced with the menD strain; however, again at least minimal growth was observed with carbohydrate carbon sources, including N-acetyl-d-glucosamine, d-trehalose, d-mannose, d-glucose-6-phosphate, d-glucose, maltose, sucrose, d-fructose-6-phosphate, and inosine (wells A-3, A-10, A-11, C-1, C-9, C-10, D-11, E-4, F-12, respectively). Growth of the menD mutant was restricted in media with d-mannitol, maltotriose, and dextrin as carbon sources. Growth and redox activity were very poor in PM3 to PM8; however, this was probably a reflection of the defective metabolism of pyruvate and α-ketoglutarate used as the carbon sources in these arrays.
It is possible that in vivo, SCV strains can scavenge low levels of hemin or menadione at their infection site. Testing of the hemB and menD mutants supplemented with hemin or menadione was complicated by the fact that these nutrients, which are needed for growth, are also toxic to the cells. To titrate the level of nutrient supplementation, we found that optimal growth and maximal restoration of normal phenotypes occurred with 1.0 μM hemin or 0.375 μM menadione. We repeated the PM analysis by supplementing the medium with the optimal levels of either hemin or menadione. However, although these compounds increased the growth rates of SCV strains, they were also toxic and simultaneously decreased the growth rate of the parental COL strain. This tended to diminish and obscure the phenotypic differences (data not shown).
Some of the key findings were subsequently confirmed by independent studies to verify the PM analysis (Table 2). In growth studies, we independently demonstrated that both mutants were in fact defective for growth using (for example) 0.2% galactose as a carbon source. Also, differences between the two mutants could be demonstrated, e.g., by adding mannitol, which had a major effect on the menD mutant (resulting in distinctly reduced growth). Furthermore, hypersensitivity of both mutants to sodium selenite and hypersensitivity of the menD mutant to ethylene glycol (in contrast to the hemB mutant) were demonstrated by simple growth studies. Finally, by testing the susceptibilities to antimicrobial agents (e.g., penicillin, gentamicin, and amikacin), we were also able to confirm the results observed with the Biolog system. By determining MICs by Etests, both mutants were shown to be less susceptible to aminoglycosides than the parent strain, and some of the MICs were above the CLSI/NCCLS breakpoints (gentamicin MIC for COL, 0.125 μg/ml; gentamicin MIC for the hemB mutant, 2 μg/ml; gentamicin MIC for the menD mutant, 4 μg/ml; amikacin MIC for COL, 2 μg/ml; amikacin MIC for the hemB mutant, 24 μg/ml; amikacin MIC for the menD mutant, 96 μg/ml).
DISCUSSION
This is the first detailed and comprehensive report on the biochemical profile of S. aureus menD and hemB mutants, both of which display the SCV phenotype. While previously detected single phenotypic traits of SCVs were confirmed by PM technology, this approach also offered a more complete phenotypic analysis of SCVs and provided insight into the physiological changes that lead to in vivo antibiotic resistance in S. aureus SCVs.
Heme and menaquinone are used both by aerobic electron transport and by the anaerobic nitrate reductase complex. Because the electron transport chain of the mutants is interrupted due to the deficiency of heme or menadione biosynthesis, these mutants should be unable to use oxygen or nitrate as a terminal electron acceptor. Consistent with this hypothesis, a previous study using a proteomic approach showed that proteins involved in the glycolytic and related pathways and in fermentation pathways were overexpressed in exponentially growing cells of the hemB mutant compared to the parent strain (11). Thus, this previous study strongly indicated that the hemB mutant generates ATP from glucose or fructose only by substrate phosphorylation.
Both the hemB mutant and the menD mutant are defective in utilizing carboxylic acids or amino acids because of limited Krebs cycle activity (11). They become primarily or completely dependent on carbohydrates as carbon and energy sources. The menD mutant's carbon metabolism defect is even more severe, as observed for metabolism of mannitol, maltotriose, and dextrin. Notably, it affects mannitol metabolism more strongly than it affects mannose metabolism. The principal difference between mannose and mannitol is the reduction of NAD+ as mannitol is metabolized to fructose-6-phoshate (16). Because a loss of electron transport limits the reoxidation of NADH, this means that the quantity of NAD+ is insufficient to allow the large number of dehydrogenase reactions to function efficiently (18). As mannitol salt agar is often used in clinical laboratories to recover and identify S. aureus (17), menadione-auxotrophic SCVs from clinical specimens would not be efficiently recovered on this medium.
As to why the hemB mutant is less profoundly affected than the menD mutant, this finding can be related to the larger number of metabolic pathways that use menadione than use hemin. While menaquinone and heme are both required for aerobic and anaerobic electron transport, menadione is also found in quinoproteins (SA0665) (6), malate quinol oxidoreductase (Mqo2), dihydroorotate dehydrogenase (PyrD, thymidine biosynthesis), and NAD(P)H:quinone oxidoreductase (SA1989). An example of how menaquinone deficiency affects metabolism can be drawn from Mqo2, an NADH minaquinol oxidoreducate. Mqo2 is important for the “horseshoe” metabolism of the Krebs cycle, in which oxaloacetate is metabolized to malate (with concomitant oxidation of NADH). Hence, the menD mutant should be unable to metabolize oxaloacetate to malate to fumarate, but the hemB mutant should be able to do this. This allows pyruvate to enter the fumarate reductase system in the hemB mutant. Fumarate forms the link between the urea cycle and the Krebs cycle, thereby allowing more extensive metabolism of amino acids. Hence, the absence of Mqo2 in the menD mutant should result in widespread metabolic alterations. While heme is also used in specific places, such as catalase, the biosynthesis of heme and the biosynthesis of menaquinone seem to be coordinately regulated since a defect in menadione biosynthesis results in decreased heme biosynthesis, but the reverse does not occur when heme biosynthesis is interrupted (5, 18). Therefore, the metabolic defects found in the menD mutant are expected to be greater than those found in the hemB mutant.
In a previous study analyzing the physiology and antibiotic susceptibility of S. aureus SCVs, it was shown that SCVs can generate a ΔΨ that is large enough that the SCVs are susceptible to gentamicin only in the presence of glucose (3). This is consistent with having the F0F1 ATPase run “backwards,” when ATP is produced via glycolysis and consumed to produce a ΔΨ. However, glucose is not used to its full potential because the Krebs cycle is not active in electron transport-deficient cells (3).
In contrast to the parent strain, both mutants were clearly susceptible to selenite, and the more severely restricted menD mutant also appeared to be susceptible to tellurite and nitrite. These results are consistent with previous observations which revealed that S. aureus cells in biofilms were fivefold more susceptible to tellurite than planktonic cultured bacteria (8), probably due to the decreased electron transport activity of biofilm S. aureus cells (19). These chemicals are toxic to many microbial species because they result in oxidant damage to membranes. SCVs have limited abilities to generate a ΔΨ, and any damage to the membrane presents a major challenge to SCVs as disrupted membranes are more permeable to proton fluxes, thereby dissipating the ΔΨ. In addition, heme is the prosthetic group for catalase, making the hemB mutant more susceptible to oxidant damage by hydrogen peroxide.
There is a similar situation in the menD mutant because interruption of menadione biosynthesis is accompanied by a coordinate decrease in heme biosynthesis in gram-positive bacteria (7, 23). Because the menD mutant is more metabolically restricted, it may be less able to maintain its ΔΨ, making it more susceptible to these compounds. Since media containing potassium tellurite are often used to isolate S. aureus, susceptibility of SCVs to tellurite should hinder recovery of this subpopulation in clinical specimens. Other genera also show increased susceptibility to these compounds when their energy metabolism is compromised (1). Production of reduced sulfur compounds requires energy from ATP. Recently, it was shown that mutation of cysM encoding cysteine synthase caused increased sensitivity of S. aureus to tellurite, hydrogen peroxide, acid, and diamide and also significantly reduced the ability of S. aureus to recover from starvation in amino acid- or phosphate-limiting conditions, indicating that cysteine has a role in the S. aureus oxidant stress response and survival mechanisms (12). Also of interest, mutation of a membrane-bound, ATP- and Zn2+-dependent “AAA-type” protease gene in S. aureus led to pleiotropic defects, including slower growth, sensitivity to salt, acid, methyl viologen, and potassium tellurite stresses, and reduced survival in amino acid- or phosphate-limiting conditions (13). Thus, these data provide further support for the hypothesis that the reduced ΔΨ and ATP of the SCVs play a role in the susceptibility of these variants to selenite, tellurite, and nitrite.
Similarly, the observed aminoglycoside resistance of the mutants that potentially results in therapeutic failures could also be due to defective uptake of these positively charged chemicals by strains with reduced ΔΨ (3, 14). Acriflavine resistance, phleomycin resistance, and trimethoprim resistance were also observed, and these compounds are also positively charged compounds. In contrast, the observed increased utilization of three positively charged lysine peptides in the hemB mutant is consistent with active transport of amino acids in S. aureus that is not charge dependent (30). Therefore, the explanation related to transport of cationic compounds is consistent for many compounds, but for other compounds the explanation may be more complex and could involve energy-driven influx pumps.
In conclusion, PM analyses have made it possible on a metabolic level to quantitatively measure over 1,000 cellular phenotypes at once and thus to provide biochemical profiles whose capacities are comparable to those of DNA microarrays and proteomic technologies. In this study, this technology proved to be appropriate for defining the metabolic differences between genetically defined SCVs and wild-type S. aureus. The hemB mutant and even more so the menD mutant were defective in utilizing a variety of carbon sources, including Krebs cycle intermediates and compounds that generate ATP via electron transport. Notably, carbohydrates that provided ATP in the absence of electron transport supported growth of the SCV mutants. Overall, the observed phenotypic changes documented in SCVs can best be viewed as consequences of alterations in multiple metabolic and energy-dependent pathways.
Supplementary Material
Acknowledgments
This work was supported by a grant from BMBF, Germany (Pathogenomic Network), to C.V.E., K.B., and G.P., by grant AI42072 from the National Institutes of Health to R.A.P., and by NIH grant AI053235-01 to P.J.M.
We sincerely thank S. Weber for excellent technical assistance.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Avazeri, C., R. J. Turner, J. Pommier, J. H. Weiner, G. Giordano, and A. Vermeglio. 1997. Tellurite reductase activity of nitrate reductase is responsible for the basal resistance of Escherichia coli to tellurite. Microbiology 143:1181-1189. [DOI] [PubMed] [Google Scholar]
- 2.Bates, D. M., C. von Eiff, P. J. McNamara, G. Peters, M. R. Yeaman, A. S. Bayer, and R. A. Proctor. 2003. Staphylococcus aureus menD and hemB mutants are as infective as the parent strains, but the menadione biosynthetic mutant persists within the kidney. J. Infect. Dis. 187:1654-1661. [DOI] [PubMed] [Google Scholar]
- 3.Baumert, N., C. von Eiff, F. Schaaff, G. Peters, R. A. Proctor, and H. G. Sahl. 2002. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 8:253-260. [DOI] [PubMed] [Google Scholar]
- 4.Bochner, B. R. 2003. New technologies to assess genotype-phenotype relationships. Nat. Rev. Genet. 4:309-314. [DOI] [PubMed] [Google Scholar]
- 5.Bochner, B. R., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson, V. L. 2004. Electron transfer in quinoproteins. Arch. Biochem. Biophys. 428:32-40. [DOI] [PubMed] [Google Scholar]
- 7.Goldenbaum, P. E., P. D. Keyser, and D. C. White. 1975. Role of vitamin K2 in the organization and function of Staphylococcus aureus membranes. J. Bacteriol. 121:442-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison, J. J., H. Ceri, C. Stremick, and R. J. Turner. 2004. Differences in biofilm and planktonic cell mediated reduction of metalloid oxyanions. FEMS Microbiol. Lett. 235:357-362. [DOI] [PubMed] [Google Scholar]
- 9.Kahl, B., M. Herrmann, A. S. Everding, H. G. Koch, K. Becker, E. Harms, R. A. Proctor, and G. Peters. 1998. Persistent infection with small colony variant strains of Staphylococcus aureus in patients with cystic fibrosis. J. Infect. Dis. 177:1023-1029. [DOI] [PubMed] [Google Scholar]
- 10.Kahl, B. C., A. Duebbers, G. Lubritz, J. Haeberle, H. G. Koch, B. Ritzerfeld, M. Reilly, E. Harms, R. A. Proctor, M. Herrmann, and G. Peters. 2003. Population dynamics of persistent Staphylococcus aureus isolated from the airways of cystic fibrosis patients during a 6-year prospective study. J. Clin. Microbiol. 41:4424-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohler, C., C. von Eiff, G. Peters, R. A. Proctor, M. Hecker, and S. Engelmann. 2003. Physiological characterization of a heme-deficient mutant of Staphylococcus aureus by a proteomic approach. J. Bacteriol. 185:6928-6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lithgow, J. K., E. J. Hayhurst, G. Cohen, Y. Aharonowitz, and S. J. Foster. 2004. Role of a cysteine synthase in Staphylococcus aureus. J. Bacteriol. 186:1579-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lithgow, J. K., E. Ingham, and S. J. Foster. 2004. Role of the hprT-ftsH locus in Staphylococcus aureus. Microbiology 150:373-381. [DOI] [PubMed] [Google Scholar]
- 14.Mates, S. M., E. S. Eisenberg, L. J. Mandel, L. Patel, H. R. Kaback, and M. H. Miller. 1982. Membrane potential and gentamicin uptake in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 79:6693-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNamara, P. J., and R. A. Proctor. 2000. Staphylococcus aureus small colony variants, electron transport and persistent infections. Int. J. Antimicrob. Agents 14:117-122. [DOI] [PubMed] [Google Scholar]
- 16.Murphey, W. H., and E. D. Rosenblum. 1964. Mannitol catabolism by Staphylococcus aureus. Arch. Biochem. Biophys. 107:292-297. [DOI] [PubMed] [Google Scholar]
- 17.Murray, P. R., E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken. 2003. Manual of clinical microbiology, vol. 1 and 2. ASM Press, Washington, D.C.
- 18.Nelson, D. L., and M. M. Cox. 2004. Lehninger principles of biochemistry. W. H. Freeman, New York, N.Y.
- 19.Proctor, R. A. 2000. Microbial pathogenic factors, including small colony variants, p. 41-54. In F. A. Waldvogel and A. L. Bisno (ed.), Infections associated with indwelling medical devices. ASM Press, Washington, D.C.
- 20.Proctor, R. A., and G. Peters. 1998. Small colony variants in staphylococcal infections: diagnostic and therapeutic implications. Clin. Infect. Dis. 27:419-422. [DOI] [PubMed] [Google Scholar]
- 21.Proctor, R. A., P. van Langevelde, M. Kristjansson, J. N. Maslow, and R. D. Arbeit. 1995. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 20:95-102. [DOI] [PubMed] [Google Scholar]
- 22.Prüß, B. M., J. W. Campbell, T. K. Van Dyk, C. Zhu, Y. Kogan, and P. Matsumura. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer 1. J. Bacteriol. 185:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin, X., and H. W. Taber. 1996. Transcriptional regulation of the Bacillus subtilis menp1 promoter. J. Bacteriol. 178:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seifert, H., H. Wisplinghoff, P. Schnabel, and C. von Eiff. 2003. Small colony variants of Staphylococcus aureus and pacemaker-related infection. Emerg. Infect. Dis. 9:1316-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaudaux, P., P. Francois, C. Bisognano, W. L. Kelley, D. P. Lew, J. Schrenzel, R. A. Proctor, P. J. McNamara, G. Peters, and C. von Eiff. 2002. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect. Immun. 70:5428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Eiff, C., and K. Becker. 2003. Small colony variants: another mechanism by which Staphylococcus aureus can evade the immune response and antimicrobial therapy, p. 253-273. In A. C. Fluit and F.-J. Schmitz (ed.), MRSA: current perspectives. Caister Academic Press, Wymondham, United Kingdom.
- 27.von Eiff, C., K. Becker, D. Metze, G. Lubritz, J. Hockmann, T. Schwarz, and G. Peters. 2001. Intracellular persistence of Staphylococcus aureus small-colony variants within keratinocytes: a cause for antibiotic treatment failure in a patient with Darier's disease. Clin. Infect. Dis. 32:1643-1647. [DOI] [PubMed] [Google Scholar]
- 28.von Eiff, C., D. Bettin, R. A. Proctor, B. Rolauffs, N. Lindner, W. Winkelmann, and G. Peters. 1997. Recovery of small colony variants of Staphylococcus aureus following gentamicin bead placement for osteomyelitis. Clin. Infect. Dis. 25:1250-1251. [DOI] [PubMed] [Google Scholar]
- 29.von Eiff, C., C. Heilmann, R. A. Proctor, C. Woltz, G. Peters, and F. Götz. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179:4706-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams, W. A., R. G. Zhang, M. Zhou, G. Joachimiak, P. Gornicki, D. Missiakas, and A. Joachimiak. 2004. The membrane-associated lipoprotein-9 GmpC from Staphylococcus aureus binds the dipeptide GlyMet via side chain interactions. Biochemistry 43:16193-16202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou, L., X. H. Lei, B. R. Bochner, and B. L. Wanner. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 185:4956-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.