Abstract
The PCAF and GCN5 acetyltransferases, but not p300 or CBP, stimulate DNA replication when tethered near the polyomavirus origin. Replication stimulation by PCAF and GCN5 is blocked by mutational inactivation of their acetyltransferase domains but not by deletion of sequences that bind p300 or CBP. Acetylation of histones near the polyomavirus origin assembled into chromatin in vivo is not detectably altered by expression of these acetyltransferases. PCAF and GCN5 interact with polyomavirus large T antigen in vivo, PCAF acetylates large T antigen in vitro, and large T-antigen acetylation in vivo is dependent upon the integrity of the PCAF acetyltransferase domain. These data suggest replication stimulation occurs through recruitment of large T antigen to the origin and acetylation by PCAF or GCN5.
Stimulation of DNA replication by auxiliary sequence elements located in cis to origins that function in eukaryotic cells was first observed with murine polyomavirus (Py) (28) and has since been documented for many other viruses. These auxiliary sequence elements bind proteins postulated to recruit and/or activate additional proteins involved directly in replication or to modify origin sequences and chromatin structures or their intranuclear localization (9, 19, 21, 22, 38-40, 48, 60, 63, 64, 72, 99, 100, 106, 108, 113; for reviews, see references 26 and 74). Much can be learned about the control of DNA replication from studying these proteins and their functions.
The Py enhancer PEA1 and PEA3 sites are particularly important for stimulating Py DNA replication (18, 20, 39, 48, 69, 71, 75, 83, 90, 91, 105). Jun, a member of the AP1 (PEA1) complex, recruits Py large T antigen (PyLT) to the origin to stimulate DNA unwinding, particularly at early times after infection when PyLT is limiting (39, 48, 69, 71, 75, 91). The AP1 complex and ets family proteins (that bind the PEA3 site) as well as Gal4VP16, NF-κB, E1a, Sp1, and p53, which also can stimulate Py DNA replication (8, 10, 11, 37, 46, 53, 74, 77, 111; reviewed in references 26 and 74), interact with p300/CBP (2, 4-7, 31, 36, 50, 57, 61, 65, 68, 82, 101, 116), PCAF, and GCN5 (15, 67, 107, 110) and other coactivators that acetylate histones and nonhistone proteins involved in transcription, including HMG17, HMGI(Y), E2Fs, p53, c-Jun (109), MyoD, YY1, Tat, TFIIE, TFIIF, and TFI68 (17, 92, 96). Acetylation regulates these proteins' functions and interactions with other proteins (17, 55, 92, 96, 97).
Proteins directly involved in DNA replication also interact with acetyltransferases, including PyLT, which interacts with p300/CBP (23, 76), MCM2 and ORC1 (which interact with acetyltransferase HBO1 [14, 44]), and MCM3, whose acetylation affects DNA replication (103). Also, acetyltransferases are recruited to double-stranded DNA breaks to facilitate DNA repair (13, 45, 70; for a review see reference 17). However, neither the specific roles for histone acetylation in these processes nor the proteins that catalyze them have been established.
Here we demonstrate that the PCAF and GCN5 acetyltransferases, when tethered near the Py origin via Gal4 DNA binding, stimulate DNA replication in vivo. Our initial hypothesis was that these acetyltransferases modify chromatin structures near the origin, but this was not supported by our experimental evidence. Instead, we observed that PCAF and GCN5 bound PyLT in vivo and that PCAF acetylated PyLT. We propose that PCAF and GCN5 activate replication at the Py origin by helping to recruit and to modify PyLT function by acetylation.
MATERIALS AND METHODS
Plasmids.
pBSPyΔE (Fig. 1A) contains Py sequences between nucleotides (nt) 4999 (AccI) and 372 (DraI) at the XhoI site of pBluescript (Stratagene), encompassing the Py origin with the viral enhancer (nt 5047 to 5291) replaced by an XhoI site. pBSPyGal (Fig. 1A) is a derivative of pBSPyΔE with five Gal4-binding sites from pG5-E4T (16) cloned at the XhoI site of pPyXhoI, which contains the polyomavirus A3 genome cloned at the EcoRI site of pBR322 with a viral enhancer (nt 5047 to 5291) replaced by an XhoI site. pBM129 was described by Hermansen et al. (42). pMKSO11 contains the entire Py genome (with a defective ori) cloned in pMK16 (104).
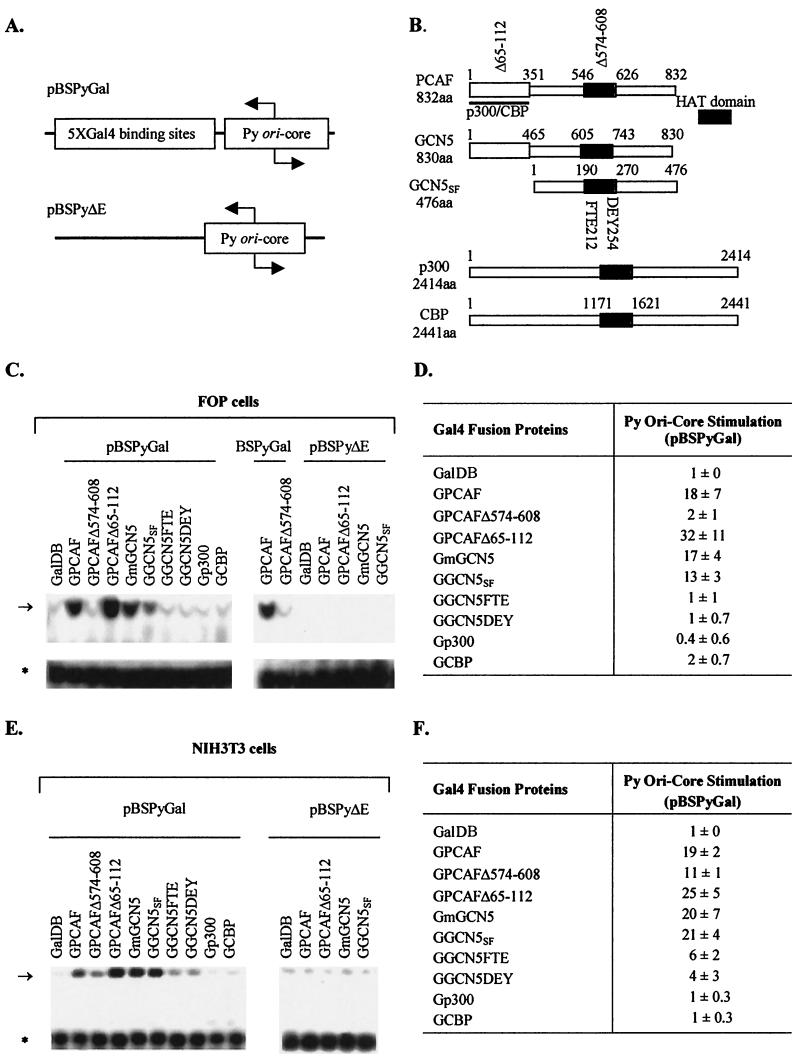
FIG.1.
Replication of Py ori core DNA by acetyltransferases. (A) Structures of test plasmids used to measure replication stimulation. (B) Structures of acetyltransferases fused to GalDBs. Acetyltransferase (HAT) domains are indicated by dark bars. Two HAT substitution mutants of human GCN5 (FTE and DEY), one HAT deletion mutant of PCAF (Δ574-608), and one deletion mutant of PCAF in the p300/CBP interacting domain (Δ65-112) also are indicated. aa, amino acid. (C) Examples of replication assays performed with the test plasmids pBSPyGal and pBSPyΔE in FOP cells. Test plasmids and cotransfected expression vectors are indicated above lanes. The arrow and the asterisk indicate the positions of replicated test DNA and the nonreplicated test DNA (input), respectively. (D) Averages from three independent replication assays (means ± standard deviations). (E) Examples of replication assays performed with the test plasmids pBSPyGal and pBSPyΔE in NIH 3T3 cells. Test plasmids and cotransfected expression vectors are indicated above lanes. The arrow and the asterisk indicate the positions of replicated test DNA (top band) and the nonreplicated test DNA as a reference (lower band), respectively. (F) Averages from three independent transfection assays (means ± standard deviations).
Expression vectors for Gal4 fusion acetyltransferase proteins (Fig. 1B) include the following: pcDNA3GalDB for the Gal4 DNA-binding domain (GalDB) (15); pcDNA3GalhGCN5SF for Gal4 human GCN5SF (GGCN5SF) (15); pcDNA3GalhGCN5FTE for an FTE212-214AAA substitution histone acetyltransferase (HAT) mutant of Gal4 human GCN5SF (GGCN5FTE) (15); pcDNA3hGCN5DEY for a DEY254-256AAA substitution HAT mutant of Gal4 human GCN5SF (GGCN5DEY) (15); pcDNA3GGmGCN5FL-flag for Gal4 (2×) mouse GCN5 (GmGCN5) (114); pcDNA3Galp300 for Gal4 p300 (Gp300) (119); pRc/RSVGalCBPFL-flag for Gal4 CBP (GCBP) (102); pCXflag-GalPCAF for Gal4 human PCAF (GPCAF) (56); pCXflag-GalPCAFΔ574-608 for a HAT-truncated mutant of Gal4PCAF (GPCAFΔ574-608) (56); and pCXflag-GalPCAFΔ65-112 for an N-terminal-truncated mutant of Gal4PCAF (GPCAFΔ65-112) (56). PcDNA3GalDB, pcDNA3GalhGCN5SF, pcDNA3GalhGCN5FTE, and pcDNA3GalDEY were constructed by transferring Gal4 chimeras from pM2 vectors (15) into pcDNA3 (Invitrogen) by using PCR. pcDNA3GGmGCN5FL-flag containing duplicate GalDBs was constructed by inserting EcoRI-digested PCR products of the GalDB from pCXflag-GalPCAF by inserting primers 5′-CCGGAATTCATGAAGCTACTGTCTTCTATC-3′ and 5′-CTGTATCGCCGGAAGAATTCGCCAC-3′ into the EcoRI site of pCMVSPORT2mGCN5fl (114). Expression vectors for Gal4PCAF and its deletion mutants were described by Krumm et al. (56).
Cell cultures.
NIH 3T3 (mouse fibroblast cell line), FOP (polyomavirus transformed mouse mammary carcinoma cell line), and Cos7 (simian virus 40 [SV40]-transformed Africa green monkey kidney cell line) were cultured in Dulbecco's modified Eagle's medium (DMEM; low glucose) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 4 mM l-glutamine.
Replication assays.
NIH 3T3 cells were seeded in 12-well plates (1.5 × 105 cells/well) and were incubated overnight at 37°C. Cells were transiently transfected by using LipofectAMINE PLUS (Invitrogen) with expression plasmids for Gal4 fusion proteins (0.2 μg) and a test plasmid (0.2 μg) pBSPyGal4 containing the Py ori core flanked by 5 Gal4-binding sites or pBSPyΔE containing only the Py ori core (Fig. 1A). PyLT, which was required for replication, was provided by cotransfection of pMKSO11 (0.01 μg), and the total amount of DNA (0.41 μg) was kept constant by adding vector DNA. After incubating cells with a DNA:LipofectAMINE PLUS mixture for 4 to 5 h in 400 μl of serum-free DMEM, the transfection solution was replaced with 1 ml of DMEM containing 10% FBS.
Similarly, FOP cells (1.5 × 106 cells per well), a cell line that constitutively expresses PyLT (11), were transfected with expression plasmids (0.8 μg) and test plasmid pBSPyGal or pBSPyΔE (0.4 μg) by using LipofectAMINE. After incubating cells with DNA:LipofectAMINE mixture for 6 h in 350 μl of serum-free DMEM, 800 μl of DMEM containing 10% FBS was added. For both types of cells, the medium was changed at 12 h and was incubated for another 24 h. DNAs were isolated by the Hirt procedure (43), digested with RNase A (0.2 μg/μl) and with EcoRI and HindIII to linearize plasmids, and were digested with DpnI in the presence of 200 mM NaCl to distinguish input (methylated) DNA from DNA replicated in animal cells (80, 93). The replicated DpnI-resistant DNA was resolved from DpnI-digested DNA by electrophoresis in agarose gels (0.8%), transferred to a nylon membrane, detected by Southern Blotting with probes generated from the test plasmid, labeled, and visualized with the North2South detection kit (Pierce). Transfection efficiencies were normalized by using input DNA digested by DpnI. For analysis of protein expression, an identical set of transfections was performed, except test plasmids were not included. Cells were lysed with 1% Triton X-100 lysis buffer 36 h after transfection. Proteins in lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and expression of Gal4 fusion proteins was detected by Western blotting by using the anti-GalDB monoclonal antibody sc-510 (Santa Cruz, Inc.).
Transcription assays.
NIH 3T3 and FOP cells were transiently transfected as described above with expression plasmids for Gal4 fusion proteins, reporter plasmid pFR-Luc (Stratagene; Fig. 2A), and expression plasmid pRL-CMV (Promega; Fig. 2A) with Renilla luciferase as an internal control. At 36 to 48 h after transfection, extracts were prepared by passive lysis and luciferase activities were assayed according to the protocol of the Dual-Luciferase Reporter Assay System (Promega). Luciferase activities were normalized with Renilla luciferase activities. All transfections were repeated at least three times. The relative luciferase activities are represented as the means relative to the basal activity of a transfected GalDB (pcDNA3Gal4DB). For analysis of protein expression, a sample of whole-cell extracts was resolved by SDS-PAGE, and expression of Gal4 fusion proteins was detected by Western blotting by using the anti-GalDB monoclonal antibody sc-510 (Santa Cruz, Inc.).
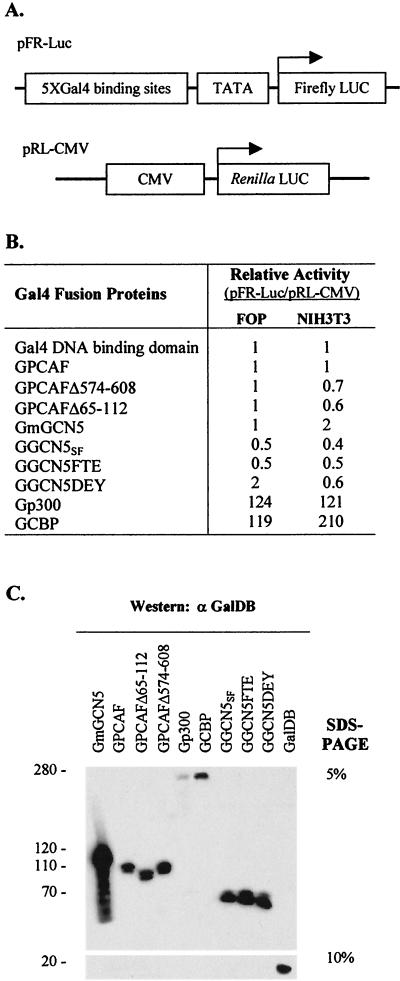
FIG. 2.
Transcriptional activation and expression of acetyltransferases. (A) Structure of the test plasmids used to measure transcription stimulation. LUC, luciferase; CMV, cytomegalovirus. (B) Summary of luciferase assays with different acetyltransferases in FOP and NIH 3T3 cells. Each value represents the mean of three independent assays. (C) Protein expression of Gal4 fusion acetyltransferases in NIH 3T3 cells. Molecular sizes (in kilobase pairs) of the proteins are indicated on the left.
Immunoprecipitation and Western blot analyses.
NIH 3T3 cells (approximately 2.5 × 106) were transfected with expression vectors for Gal4 fusion proteins (2.5 μg) and pMKSO11 (2.5 μg) by using LipofectAMINE PLUS in a 100-mm-diameter plate. After 36 h, the cells were extracted with 500 μl of single-detergent lysis buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.02% sodium azide, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× Complete protease inhibitors [Roche Diagnostics], 1% Triton X-100) for 1 h at 4°C, and lysates were cleared by centrifugation twice at 13,000 × g for 10 min. A sample (10 μl) was taken for a protein input control, and the remaining supernatants were incubated with 10 μg of rabbit anti-Gal4 polyclonal antibody sc-577 (Santa Cruz, Inc.) or mouse anti-PyLT monoclonal antibody KF4 (78) overnight at 4°C, and the immune complexes were collected by adding 80 μl of protein A-Sepharose beads for sc-577 or protein G-Sepharose beads for KF4. Samples were incubated for 2 h at 4°C, followed by five washes with wash buffer (50 mM Tris-Cl [pH 8.0], 1 mM EDTA, 150 mM NaCl, 1% NP-40, 4 mM NaF, 2 mM Na-Orthovanadate). Following the fifth wash, pelleted beads were suspended in 40 μl of 1× SDS sample buffer (50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 4% SDS, 0.1% bromophenol blue, 10% [vol/vol] glycerol), boiled for 10 min, and centrifuged at 13,000 × g for 10 min. Samples of 20 μl of the supernatant were removed for SDS-PAGE. After SDS-PAGE and transfer to polyvinylidene difluoride membrane, the bound T antigen or GalDB fusion proteins were detected with mouse KF4 antibody or rabbit anti-GalDB antibody sc-577 by Western blot analysis. For estimation of Gal4 fusion protein and PyLT expression, 20 μl of the whole-cell extract was resolved by SDS-PAGE and detected by Western blotting by using mouse anti-Gal4DB monoclonal antibodies sc-510 and KF4, respectively, and subsequently horseradish peroxidase-conjugated antibody followed by chemiluminescence measurement.
In vitro acetylation assays.
Acetylation reactions were performed with a mixture of 2.8 μg of baculovirus-expressed histidine-tagged PyLT purified by nickel chelate chromatography (62) (or bovine serum albumin [BSA] as a negative control), 0.8 μg of PCAF (Upstate), and 5.0 nmol of [3H]acetyl-coenzyme A (200 mCi/mmol; DuPont-NEN) in 30 μl of HAT assay buffer (50 mM Tris-HCl [pH 8.0], 0.1 mM EDTA, 10 mM sodium butyrate, 1.0 mM dithiothreitol, 10% glycerol) at 30°C for 1 h. Reaction mixtures were stopped by addition of 1× SDS sampling buffer, and a 25-μl volume was resolved by SDS-PAGE. The gel was stained with Coomassie blue to estimate the protein quantities, soaked in EN3HANCE solution (DuPont) for 30 min and vacuum dried, and subsequently subjected to autoradiography for 3 weeks. The acetylation reaction was also carried out in 25 μl of HAT assay buffer with unlabeled acetyl-coenzyme A (0.2 mM; Sigma) and PyLT (2.5 μg) as substrates and active PCAF (0.7 μg) or p300 acetyltransferase (0.9 μg or 7 U; Upstate) as a catalyst, and acetylation in half a reaction was detected by Western blotting by using rabbit anti-acetyl lysine polyclonal antibody (Upstate). For filter-binding assays, acetylation reactions were performed with a mixture of 2.5 μg of PyLT (or chicken histones as a positive control), 0.25 μg of active PCAF (Upstate), and 1.25 nmol of [3H]acetyl-coenzyme A (200 mCi/mmol; DuPont-NEN) in 25 μl of HAT assay buffer at 30°C for 1 h. Five microliters of each reaction sample was transferred onto P81 paper (Whatman), washed five times for 5 min per wash with 50 ml of 50 mM Na2HPO4 (pH 9.0) and once with 50 ml of acetone, and dried. Each paper was then equilibrated overnight in a scintillation vial containing scintillation fluid and was counted.
Analysis of PyLT acetylation in vivo.
NIH 3T3 cells (approximately 5.0 × 106) were transfected with expression vectors for Gal4 fusion proteins (6.0 μg) and pMKSO11 (4.0 μg) by using LipofectAMINE PLUS. After 36 h, acetylation of PyLT was assessed by immunoprecipitation of total cell extracts with rabbit anti-acetyl lysine polyclonal antibody (Upstate) followed by Western blot analysis with mouse KF4 antibody. For estimation of Gal4 fusion protein and PyLT expression, 10 μl of the whole-cell extract was resolved by SDS-PAGE and detected by Western blot analysis by using rabbit anti-GalDB polyclonal antibodies sc-577 and KF4, respectively.
Preparation and analysis of SV40 virus with Py origin.
A PCR product with the Py ori core flanked by 5× Gal4 binding sites was prepared by using pBSPyGal4 as a template with the reverse M13 primer and Ori94 primer (5′-GTGCAAGGCGCCAGTCCTG). The PCR product was digested with EcoRV and inserted into the PmlI site of pBM129 (42) to create construct TSPy5 (see Fig. 6A). The C-to-T point mutation at nt 2151 (i.e., P to S substitution in the viral capsid protein VP1) present in the SV40 tsC219 mutant (24, 25) was introduced into TSPy5 by site-directed mutagenesis. The orientation of the insert was determined by double digestion with MluI and SalI, and the insert sequence was confirmed by DNA sequencing. To prepare TSPy5 virus, the pBR322 sequences were excised by EcoRI digestion and the linearized SV40-TSPy5 DNA was transfected into Cos7 cells by using LipofectAMINE, and the supernatant was harvested after transfected cells were incubated at 37°C for 7 days. Viruses were amplified by further infection of Cos7 cells, and the virus titers were estimated by determining the quantity of purified viral DNA in samples of the infected cells (34). Nuclear extracts containing TSPy5 chromatin were prepared from Cos7 cells infected with the TSPy5 virus, and characterization of TSPy5 chromatin by microccocal nuclease (MNase) was performed as described by Kingston (54). To ascertain whether nucleosomes occupied the Py ori core, TSPy5 minichromosomes were prepared in hypotonic buffers as previously described (34) and were digested with excess restriction endonucleases as described by Hermansen et al. (42) and were analyzed by Southern blotting.
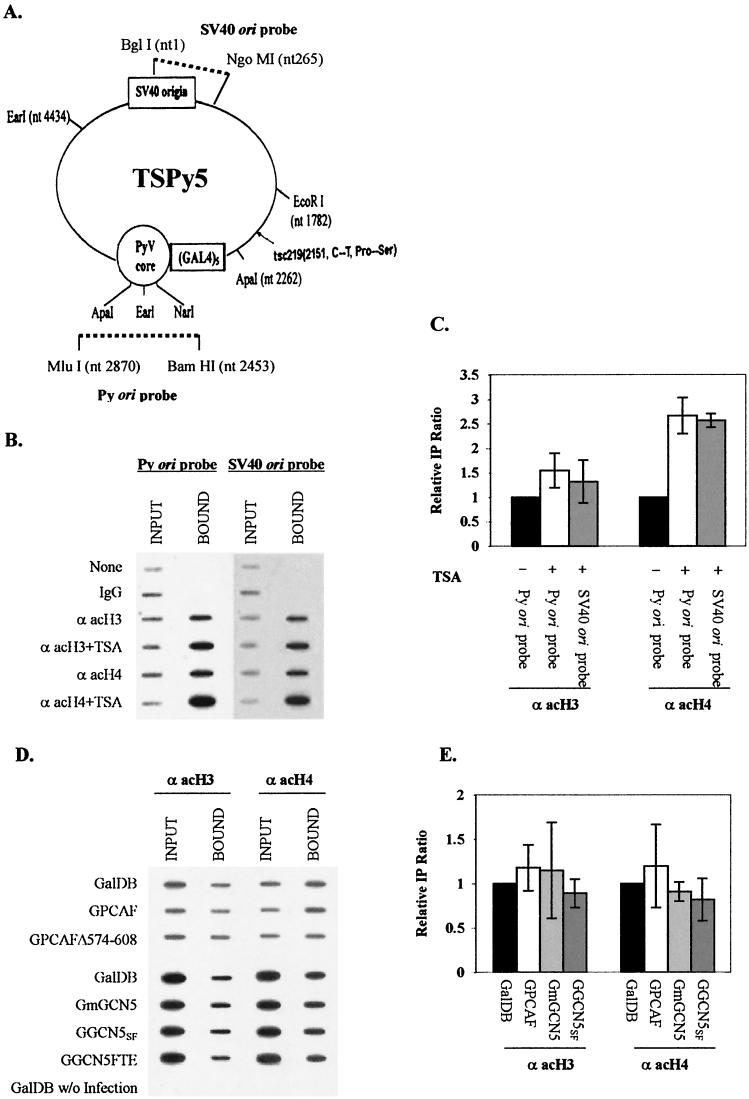
FIG.6.
Analysis of acetylation of histones around Py ori core. (A) Schematic representation of TSPy5 with location and orientation of the Py ori core and adjacent Gal4-binding sites. Fragments used to generate the Py ori probe and SV40 ori probe for ChIP assays are indicated by broken lines. (B) Example of ChIP assays from TSA-treated or untreated cells. TSPy5 chromatin fragments immunoprecipitated with anti-acetylated H3 and H4, and DNA input (INPUT) and DNA immunoprecipitated (BOUND) analyzed by Py ori and SV40 ori probes as indicated above the lanes. Antibodies used for ChIP assays and TSA treatment are indicated to the left to the corresponding rows. (C) Average from three independent experiments represented in panel B. Immunoprecipitation ratio for each Gal4 chimera protein was represented as the ratio of bound DNA intensity to input DNA intensity quantified from slot blots. Relative immunoprecipitation ratio was measured by comparing each immunoprecipitation ratio relative to that (set as 1) for cells without TSA treatment. (D) Example of ChIP assays with acetyltransferases using Py ori probe. TSPy5 minichromosome fragments for ChIP assays from Cos7 cells infected with TSPy5 virus and transfected with expression vectors of Gal4 fusion acetyltransferases. DNA input (INPUT) and coprecipitated chromatin DNA with anti-acetyl H3 or H4 antibody (BOUND) analyzed by slot blot using Py ori probe are indicated above the lanes, and Gal4 fusion proteins are indicated at the left. (E) Average from three independent experiments represented in panel D. Relative immunoprecipitation ratio was measured by comparing each immunoprecipitation ratio relative to that (set as 1) for GalDB. IgG, immunoglobulin G.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described by Boyd et al. (12) with the following modifications. Cos7 cells were seeded into 100-mm-diameter plates (1.5 × 106 to 2.4 × 106 cells/plate), and after 12 to 16 h they were infected with TSPy5 virus for 1 h and then transfected with expression plasmids (8 μg) for Gal4 fusion proteins by using LipofectAMINE 2000 (Invitrogen). Aphidicolin (10 μg/ml, prepared as 1-mg/ml concentration of stock solution in dimethylsulfoxide [DMSO] and stored at −20°C; Sigma) was added to the medium at 16 h after transfection to block DNA replication and virion assembly (32). After 24 h of additional incubation, cross-linking solution (11% formaldehyde diluted from 37% stock, 0.1 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 50 mM HEPES, pH 8.0) was added to the culture medium to a final concentration of 1% and was incubated for 30 min at 37°C and then for 24 h at 4°C. As a positive control for the ChIP assays, Cos7 cells were infected with virus and were treated at 24 h after infection with or without trichostatin A (TSA), a histone deacetylase (HDAC) inhibitor (117), at 100 ng/ml (TSA [Sigma] was prepared as a 1-mg/ml concentration of stock solution in DMSO). After 12 h of additional incubation cells were cross-linked as described above, and cross-linking was stopped by the addition 0.55 ml of 2.5 M glycine to a final concentration of 0.125 M for 5 min at room temperature. The cells were scraped and collected by centrifugation at 7,400 × g for 5 min after two washes with ice-cold phosphate-buffered saline. The cells were washed twice with ice-cold phosphate-buffered saline plus 0.5 mM PMSF and resuspended in 120 μl of a solution containing 5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 8.0), 85 mM KCl, 0.5% NP-40, 0.5 mM PMSF, and 1× complete protease inhibitors for 20 min on ice. The lysate was sonicated four times for 30 s on ice with a Vibra Sonicator and then was digested for 10 min by MNase (0.5 U/μl) at room temperature to reduce the DNA length to between 200 and 1,000 bp. Debris was removed by centrifugation for 10 min at 16,000 × g at 4°C. The supernatant was diluted in 1.2 ml of immunoprecipitation buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 16.7 mM NaCl, 1 mM PMSF, and 1× Complete protease inhibitors). The chromatin solution was precleared with 50 to 80 μl of a 50% protein A-Sepharose (Sigma) slurry containing 0.2 μg of sonicated salmon sperm DNA/μl and 0.5 μg of BSA in TE (10 mM Tris, 1 mM EDTA [pH 8.0], 0.05% sodium azide)/μl for 30 min at 4°C with agitation. Sepharose beads were pelleted at 12,000 × g for 5 min, and 500 μl of supernatant was immunoprecipitated with 5 μg of anti-acetyl histone H3 and H4 polyclonal antibodies (Upstate) each for 12 h at 4°C with rotation. A portion of the supernatant (50 μl) was kept as an input chromatin control. Immune complexes were collected with 60 μl of 50% protein A-Sepharose slurry containing 0.2 μg of sonicated salmon sperm DNA/μl and 0.5 μg of BSA in TE/μl for 1 h at 4°C with rotation. Beads were pelleted by centrifugation and were washed a total of five times with 500 μl of each of following buffers: 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), 150 mM NaCl (one washing); 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), 500 mM NaCl (one washing); 0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl (pH 8.1) (one washing); and TE (pH 8.0) (two washings). Immune complexes were eluted by two washes with 250 μl of elution buffer (1% SDS, 0.1 M NaHCO3). NaCl (5 M; 20 μl) was added to the combined eluates, and cross-links, including the input chromatin control, were reversed by incubation at 65°C for at least 4 h. Proteinase K (2 μl of a 10-mg/ml concentration) was added to the eluate and was incubated for 1 h at 45°C. DNA was recovered by phenol-chloroform extraction and ethanol precipitation by using 20 μg of glycogen as a carrier, denatured in 100 μl of denaturation solution (1.5 M NaCl, 0.5 M NaOH), and transferred onto nylon membranes by using a slot blot apparatus. Sequences from immunoprecipitated (bound) and input control (input) DNAs were detected by using a North2South detection kit with a Py ori probe generated from the BamHI-MluI small fragment of TSPy5 and an SV40 ori probe generated from the BglI-NgoMI small fragment of TSPy5 (see Fig. 6A).
RESULTS
PCAF and GCN5 acetyltransferases tethered near the Py origin stimulate DNA replication.
Plasmid pBSPyGal containing the Py ori core flanked by Gal4-binding sites in place of the natural enhancer (Fig. 1A) was transfected into FOP or NIH 3T3 cells together with plasmids that express the GalDB, either alone or fused to cellular acetyltransferases, and the replicated pBSPyGal DNA was measured after 48 h. The absence of the natural viral enhancer caused replication of pBSPyGal plasmid to be dependent upon the expression of Gal4 fusion proteins that stimulate DNA replication (Fig. 1C to F) (8, 11, 37). PCAF and GCN5 and their various mutationally altered forms stimulated replication of pBSPyGal to differing degrees (Fig. 1C to F). Compared to GalDB alone, GPCAF and GGCN5SF stimulated replication 18- and 13-fold, respectively, in FOP cells (Fig. 1D) and 19- and 21-fold in NIH 3T3 cells (Fig. 1F). Mutationally altered forms of PCAF and GCN5SF lacking intrinsic acetyltransferase activities (GPCAFΔ574-608, GGCN5FTE, and GGCN5DEY) did not stimulate replication in FOP cells (Fig. 1C and D) and stimulated replication at reduced levels in NIH 3T3 cells (Fig. 1E and F), indicating a requirement for their intrinsic acetyltransferase activities. GPCAFΔ65-112, a mutant containing a deletion within the p300/CBP-interacting domain, activated replication even better than its parent, and GGCN5SF, which does not associate with p300/CBP due to the lack of an N-terminal sequence (114, 115), also activated replication as well as full-length GmGCN5 (Fig. 1C to F), suggesting that the p300 and CBP accessory acetyltransferases are not required to act in concert with PCAF and GCN5 to stimulate replication. This was further supported by the observation that p300 and CBP tethered to the Py origin did not stimulate replication (Fig. 1C to F). A parallel assay with plasmid pBSPyΔE, which lacks Gal4-binding sites, revealed that replication stimulation required tethering the PCAF and GCN5 fusion proteins to the test plasmid (Fig. 1C and E).
Differential stimulation of transcription by PCAF, GCN5, p300, and CBP acetyltransferases.
Stimulation of DNA replication by PCAF and GCN5 did not correlate with their capacity to stimulate transcription. For example, Gal4 fusion GCN5 (GmGCN5 and GGCN5SF) and PCAF (GPCAF) proteins did not enhance expression of the luciferase gene placed downstream of Gal4 sites (Fig. 2A and B), but Gal4 fusion p300 and CBP proteins significantly stimulated gene expression (Fig. 2B), the latter agreeing with results previously reported by others (102). Western analyses of extracts from transiently transfected NIH 3T3 cells demonstrated that the Gal4 PCAF and GCN5 fusion proteins were expressed equivalently (Fig. 2C). Expression of these proteins was not detected in FOP cells, possibly due to inefficient transfection. These data suggest that activation of transcription and replication by these acetyltransferases differs mechanistically, perhaps as a result of their being present in different multiprotein complexes.
Interaction of acetyltransferases with PyLT.
Work by others has indicated that p300 and CBP interact with PyLT (23, 76). Were PCAF and GCN5 also to interact with PyLT, they might facilitate recruitment of PyLT to the origin (as does Jun when bound to the Py enhancer [39, 48, 69, 75]). To examine this possibility, NIH 3T3 cells were cotransfected with expression plasmids for PyLT and Gal4 fusion proteins and cell lysates were immunoprecipitated with either anti-GalDB antibody or anti-PyLT antibody, separated by SDS-PAGE, and immunoblotted correspondingly with anti-T antisera and anti-GalDB antibody. Anti-GalDB antibody brought down PyLT from cell extracts containing GPCAF, GmGCN5, or GGCN5SF as well as Gp300 and GCBP, but not GalDB alone (Fig. 3A and B). Also, anti-PyLT antibody, but not normal serum immunoglobulin G, coprecipitated GPCAF, GmGCN5, or GGCN5SF but not GalDB (Fig. 3C and D). These results show PyLT stably bound PCAF and GCN5, with the bound fraction of PyLT estimated to be roughly 1 to 4% by comparing band densities of PyLT in internal and coimmunoprecipitate lanes.
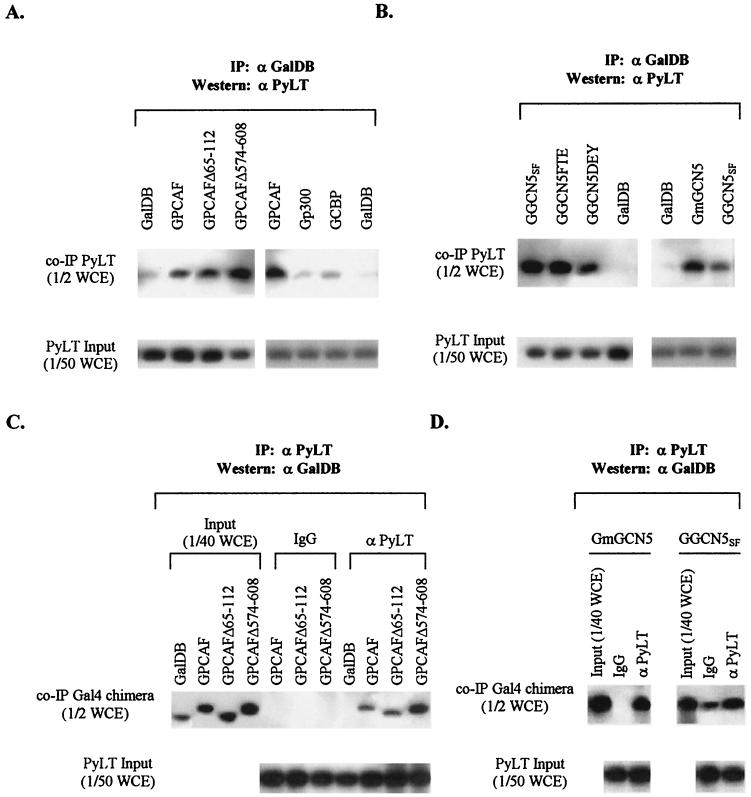
FIG. 3.
Coimmunoprecipitation of acetyltransferases and PyLT. (A) Analysis of PyLT coimmunoprecipitated with Gal4 fusion PCAF. Cell extracts from NIH 3T3 cells transiently cotransfected with expression vector for PyLT and an expression vector for Gal4 fusion acetyltransferase were precipitated with rabbit polyclonal anti-GalDB antibody. One half of the immune complexes (top band), equivalent to one half of whole-cell extracts (WCE), and one fiftieth of WCE for measurement of input PyLT (lower band) were analyzed by Western blots. (B) Analysis of PyLT coimmunoprecipitated with Gal4 fusion GCN5 as described for panel A. (C and D) Analysis of acetyltransferases coimmunoprecipitated with PyLT. Cell extracts from NIH 3T3 cells transiently cotransfected with expression vector for PyLT and an expression vector for Gal4 fusion acetyltransferase were incubated overnight at 4°C with mouse monoclonal anti-PyLT antibody and mouse normal serum immunoglobulin G (IgG). One half of the immune complexes, equivalent to one half of WCE, and one fortieth of WCE for measurement of input Gal4 fusion acetyltransferases were analyzed by Western blotting by using rabbit polyclonal anti-GalDB antibody (top lane) and one fiftieth of WCE for measurement of input PyLT analyzed by Western blotting by using anti-PyLT antisera (lower lane). Gal4 fusion acetyltransferases transfected and antibodies (anti-PyLT and normal IgG) used for immunoprecipitation are indicated above each lane. α PyLT, anti-PyLT antibody; α GalDB, anti-GalDB antibody; IP, immunoprecipitation.
Nearly equivalent amounts of PyLT were coprecipitated with mutants of PCAF (GPCAFΔ574-608) or GCN5 (GGCN5FTE and GGCN5DEY) lacking acetyltransferase activities whose expression levels were comparable to those of their parents (Fig. 3A and B). The mutant of PCAF with a deletion in the p300/CBP-interacting domain (GPCAFΔ65-112) also complexed with a similar amount of PyLT. Similarly, GGCN5SF lacking the p300/CBP-interacting domain was associated with PyLT to the same extent as full-length GCN5 (GmGCN5) (Fig. 3B). When anti-PyLT antisera were used for immunoprecipitation, mutants of PCAF with expression levels comparable to those of the parent (GPCAF) were coprecipitated with PyLT at nearly the same level, and expression of PyLT was comparable for these assays (Fig. 3). These results suggest that the mutations in PCAF and GCN5 affect neither their association with PyLT nor, likely, its recruitment to the origin. As mutations abrogating their intrinsic acetyltransferase activities reduce the ability of PCAF or GCN5 to stimulate Py DNA replication, these data indicate that both recruitment of PyLT as well as acetylation are required to stimulate DNA replication.
Acetylation of PyLT by PCAF.
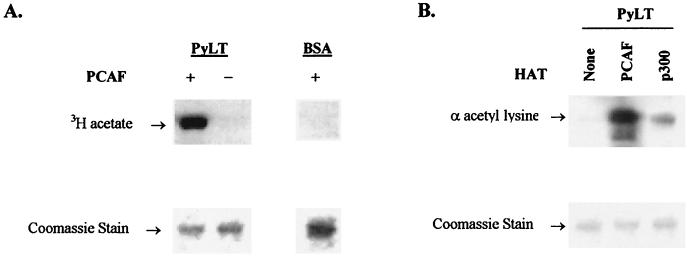
Incubation of PCAF with purified PyLT and acetyl-coenzyme A revealed PyLT was a substrate for acetylation (Fig. 4A and B). Based on the data from filter binding assays, acetylation of PyLT was approximately 12% that of purified histones without considering the difference in mass. Approximately 2% of PyLT was acetylated if one [3H]acetyl group was introduced per PyLT molecule (data not shown). Since GCN5 is highly related to PCAF in structure and function (114), it is likely that GCN5 also acetylates PyLT. PyLT appeared to be acetylated very weakly, if at all, by p300 (Fig. 4B).
FIG. 4.
In vitro acetylation of PyLT by PCAF. (A) PyLT incubated with (+) or without (−) PCAF with [3H]acetyl-coenzyme A as described in Materials and Methods. The right-hand panel shows BSA incubated with PCAF as a control. Reaction products were separated by SDS-PAGE and stained with Coomassie blue, and [3H]acetate reaction products were visualized by fluorography. (B) PyLT incubated with acetyl-coenzyme A and PCAF or p300 detected by Western blotting by using rabbit anti-acetyl-lysine polyclonal antibody (α acetyl lysine). Upper panel, Western blot with anti-α-acetyl-lysine antibody; lower panel, Coomassie-stained gel.
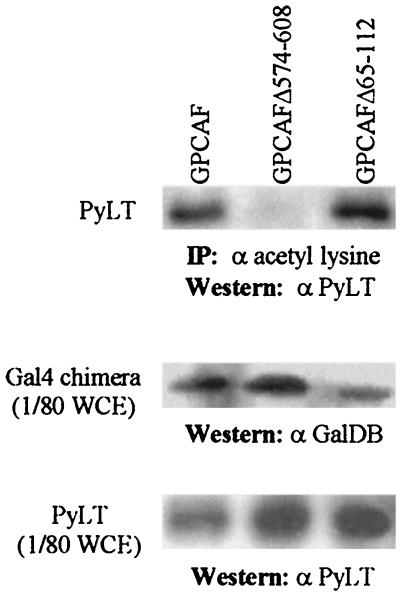
To assess acetylation of PyLT in vivo, extracts from NIH 3T3 cells expressing PyLT and Gal4 fusion PCAF were immunoprecipitated by using rabbit anti-acetyl lysine antibody and were analyzed by Western blotting by using anti-PyLT antibody KF4. PyLT was immunoprecipitated by the anti-acetyl lysine antibody from extracts of cells expressing GalPCAF and GPCAFΔ65-112 (Fig. 5), both of which stimulate DNA replication. By contrast, no PyLT was detected in the immunoprecipitates of cells expressing GPCAFΔ574-608, the mutant of GPCAF lacking HAT activity. This result suggests that PyLT is acetylated in vivo and that the acetyltransferase activity of PCAF is required.
FIG. 5.
Analysis of PyLT acetylation in vivo. Cell extracts from NIH 3T3 cells transiently cotransfected with expression vector for PyLT and an expression vector for Gal4 fusion PCAF were immunoprecipitated with rabbit polyclonal anti-acetyl-lysine antibody (α acetyl lysine). The immunoprecipitates (upper panel) were analyzed for PyLT by Western blotting by using anti-PyLT antibody (α PyLT). Expression of Gal4 fusion PCAF and its mutants (middle panel) and expression of PyLT transfected (bottom panel) also were analyzed in one eightieth of whole-cell extract (WCE) by Western blotting by using anti-GalDB (α GalDB) and anti-PyLT (α PyLT) antibodies. Gal4 fusion PCAF and its mutants transfected are indicated above the lanes. IP, immunoprecipitation.
Py origin chromatin acetylation is not detectably changed by PCAF and GCN5.
While plasmids with bacterial DNA back-bones transfected into eukaryotic cells can be assembled into chromatin, a significant portion is assembled into nonnucleosomal structures or has an atypical nucleosomal structure (3, 51, 52, 88) and is unlikely to be representative of chromatin structures in vivo. In contrast, the structure of the SV40 minichromosome is nearly indistinguishable from that of cellular chromatin (27, 73). Consequently, to study the chromatin structures assembled near the Py origin in vivo we utilized an SV40 virus whose genome contained the Py origin sequences (TSPy5; Fig. 6A). MNase digestion of extracts of TSPy5 virus-infected Cos7 cells indicated that the TSPy5 genome was assembled into chromatin, and restriction enzyme analysis of TSPy5 minichromosomes revealed that the Py ori core region was occupied by nucleosomes (data not shown). To inhibit extensive DNA replication and SV40 virion assembly that might disrupt histone modifications caused by Gal4 fusion proteins expressed in the Cos7 cells (49), aphidicolin was added to the cells at 16 h after infection.
To validate ChIP assays as a means to study TSPy5 chromatin acetylation, we analyzed whether TSA treatment of Cos7 cells infected with TSPy5 viruses changed the acetylation status of TSPy5 chromatin. HDAC inhibitors, such as TSA, cause hyperacetylation of core histones (especially H4) in cellular chromatin (98, 118) and SV40 minichromosomes (1). ChIP assays with probes for Py ori core and SV40 ori demonstrated that the acetylation of H4 was increased by 2.6-fold, and acetylation of H3 slightly increased in TSPy5 minichromosomes extracted from TSA-treated cells compared to that of minichromosomes from untreated cells (Fig. 6B and C).
By contrast, introduction of Gal4 fusion PCAF or GCN5 (and also GGCN5SF and GmGCN5) into the Cos7 cells infected by TSPy5 virus caused no detectable changes of acetylation relative to GalDB protein alone, as detected by a probe for Py ori core sequences (Fig. 6D and E). Neither PCAF nor GCN5 had any effect on the chromatin structure near the SV40 origin (data not shown). Expression of Gal4 fusion proteins in Cos7 cells was detected by Western analysis (data not shown). Thus, it appears that stimulation of DNA replication by PCAF and GCN5 is not accompanied by detectable changes in histone acetylation around the Py ori core.
DISCUSSION
PCAF and GCN5 acetyltransferases interact, either directly or through cofactors, with a large variety of cellular proteins, including nuclear steroid receptors, c-Jun, c-Fos, c-Myb, p53, YY1, NF-κB, Stat1, Stat2, Ets-1, HIF1, GATA1, MyoD, Smad proteins, E2F-1, and members of RNA polymerase II holoenzyme (33, 92, 96), some of which stimulate Py origin-dependent DNA replication when tethered in cis. Evidence presented here indicates the PCAF and GCN5 acetyltransferases bind to and acetylate PyLT. Inactivation of their intrinsic acetylation activities greatly reduces the stimulation of DNA replication. By contrast, p300 and CBP acetyltransferases bind PyLT but acetylate it to a much lower extent, if at all, and do not detectably stimulate DNA replication. These data implicate acetylation of PyLT as being important for replication in vivo.
As tethering of PCAF and GCN5 to the origin by using Gal4-binding sites is required to stimulate replication, structural features of PyLT associated with prereplication complex at the origin are likely to be important. There may be parallels between these events and those occurring at origins in cellular chromosomes: the recent finding that MCM2 and ORC1 interact with the HAT HBO1 (14, 44) and that acetylation of MCM3 by a novel acetyltransferase affects DNA replication (103) suggest that acetyltransferases also have direct roles in DNA replication of higher eukaryotes.
Mammalian PCAF and GCN5 are elements of several distinct multiprotein HAT complexes (17, 92, 96), including those recruited to double-stranded DNA breaks that facilitate DNA repair (13, 45, 70). PCAF and GCN5, in the NIH 3T3 and FOP cells used here, did not activate expression of Luc genes when tethered upstream; equivocal transcriptional activation by PCAF and GCN5 also has been reported in other studies (56, 58, 86, 95). Perhaps, as with transcription activation, distinct complexes are involved in replication stimulation.
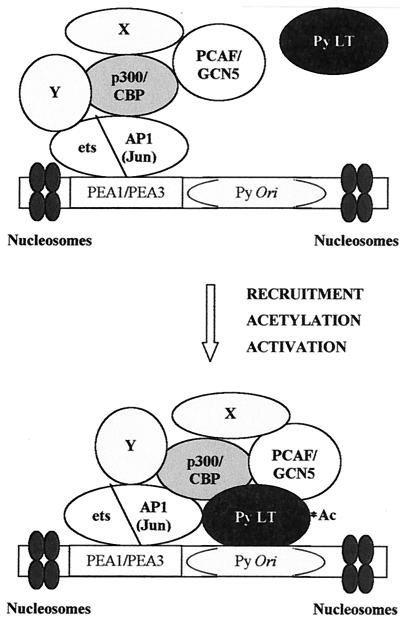
For the Py natural enhancer, AP-1 complex and ets family protein interactions with p300/CBP (4, 6, 7, 50, 116) might recruit PCAF and GCN5 to the origin region (Fig. 7). Although p300/CBP is required for certain CREB-dependent transcriptional responses (33, 79, 89, 94) and thereby assists in the activation of viral transcription by the enhancer, others have noted that CREB did not stimulate DNA replication when bound near the Py ori core (75). Our data indicating p300/CBP is not directly involved in replication stimulation can be explained by the lack of acetylation of PyLT by p300/CBP, or alternatively by their being inhibited by PyLT, as has been suggested for activation of transcription (23). Viruses with mutationally altered PyLT incapable of binding p300/CBP are restricted in forming tumors in newborn mice, leading to the suggestion these interactions are essential for virus replication and spread (23). Dissecting the roles of these complexes should provide new information about the control of eukaryotic replication and transcription.
FIG. 7.
Model for Py enhancer-origin complexes containing accessory proteins PCAF, GCN5, and PyLT. See the text for details.
Phosphorylation of proteins involved in DNA replication provides a very important means to regulate initiation, and this has been extensively dissected for SV40 and Py DNA replication (85, 112). Analogously, acetylation of PyLT might affect its DNA binding and association/disassociation with itself or other proteins in the replisome. Low pH enhances the binding of PyLT to the origin in vitro (81), and substitution of Asn-286 for the negatively charged Asp-286 within a positively charged region of the PyLT DNA-binding domain affects association with the origin and DNA replication (104). Were acetylation to neutralize the positively charged Lys residues in the DNA-binding domain of PyLT, this might affect its binding to the origin.
As PyLT helicase activity is required for movement of the replisome, acetyltransferases associated with PyLT might also mediate chromatin remodeling to facilitate elongation. Hyperacetylation of histones, or removal of histone tails, the primary targets for acetylation, facilitate elongation of SV40 DNA replication (1, 87). Acetylation of PyLT also could affect its association with cellular proteins, such as pRB or p53, that are important for inducing G1-to-S cell cycle progression and DNA replication (29, 30, 59, 84). These and other possibilities can be better understood once the extent to which PCAF (and GCN5) acetylate PyLT in vivo and the sites that are acetylated are known.
In Saccharomyces cerevisiae, nucleosome positioning mediated by ORC (origin recognition complex) affects replication initiation (66) and SV40 large T antigen promotes the formation of an origin free of nucleosomes and competent for replication (35, 47). Normal SV40 minichromosomes are more highly acetylated than cellular chromatin, with H4 occurring in mono-, di-, and triacetylated forms and H3 occurring in a diacetylated form (1). The sites for these particular hyperacetylations remain undetermined, although acetylation sites of core histones in vertebrate chromatin include K5 and K9 of H2A, K5, K12, K15, and K20 of H2B, K9, K14, K18, and K23 of H3, and K5, K8, K12, and K16 of H4 (92). Our not being able to detect changes in the acetylation of histones near the Py origin sequences in SV40 minichromosomes might be due to high levels of normal acetylation. It is still possible that specific sites in these histones are targeted by PCAF and GCN5 and thus could be detected by ChIP assays more refined than were conducted here.
Acknowledgments
We thank the members of the Folk laboratory for helpful suggestions and Sarah Scanlon for her invaluable assistance. We also thank S. L. Berger for GCN5SF expression vectors, J. C. Chrivia for pRc/RSVGalCBPFL-flag, S. Y. Dent for pCMVSPORT2mGCN5fl, A. J. Giordano for pcDNA3Galp300, B. Milavetz for pBM129, and X. J. Yang for PCAF expression vectors. We are greatly indebted to S. L. Berger and S. Y. Dent for insightful discussions during the course of these studies and for their suggestions about the manuscript.
This work was supported in part by National Institutes of Health grant R01 CA38538, U.S. Army grant DAMD17-98-1-8321, and by the University of Missouri—Columbia.
REFERENCES
- 1.Alexiadis, V., L. Halmer, and C. Gruss. 1997. Influence of core histone acetylation on SV40 minichromosome replication in vitro. Chromosoma 105:324-331. [DOI] [PubMed] [Google Scholar]
- 2.Arany, Z., W. R. Sellers, D. M. Livingston, and R. Eckner. 1994. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell 77:799-800. [DOI] [PubMed] [Google Scholar]
- 3.Archer, T. K., P. Lefebvre, R. G. Wolford, and G. L. Hager. 1992. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science 255:1573-1576. [DOI] [PubMed] [Google Scholar]
- 4.Arias, J., A. S. Alberts, P. Brindle, F. X. Claret, T. Smeal, M. Karin, J. Feramisco, and M. Montminy. 1994. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature 370:226-229. [DOI] [PubMed] [Google Scholar]
- 5.Avantaggiati, M. L., V. Ogryzko, K. Gardner, A. Giordano, A. S. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89:1175-1184. [DOI] [PubMed] [Google Scholar]
- 6.Bannister, A. J., T. Oehler, D. Wilhelm, P. Angel, and T. Kouzarides. 1995. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene 11:2509-2514. [PubMed] [Google Scholar]
- 7.Bannister, A. J., and T. Kouzarides. 1995. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 14:4758-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baru, M., M. Shlissel, and H. Manor. 1991. The yeast GAL4 protein transactivates the polyomavirus origin of DNA replication in mouse cells. J. Virol. 65:3496-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baumann, M., R. Feederle, E. Kremmer, and W. Hammerschmidt. 1999. Cellular transcription factors recruit viral replication proteins to activate the Epstein-Barr virus origin of lytic DNA replication, oriLyt. EMBO J. 18:6095-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett, E. R., M. Naujokas, and J. A. Hassell. 1989. Requirements for species-specific papovavirus DNA replication. J. Virol. 63:5371-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett-Cook, E. R., and J. A. Hassell. 1991. Activation of polyomavirus DNA replication by yeast GAL4 is dependent on its transcriptional activation domains. EMBO J. 10:959-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyd, K. E., J. Wells, J. Gutman, S. M. Bartley, and P. J. Farnham. 1998. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc. Natl. Acad. Sci. USA 95:13887-13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brand, M., J. G. Moggs, M. Oulad-Abdelghani, F. Lejeune, F. J. Dilworth, J. Stevenin, G. Almouzni, and L. Tora. 2001. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 20:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke, T. W., J. G. Cook, M. Asano, and J. R. Nevins. 2001. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem. 276:15397-15408. [DOI] [PubMed] [Google Scholar]
- 15.Candau, R., P. A. Moore, L. Wang, N. Barlev, C. Y. Ying, C. A. Rosen, and S. L. Berger. 1996. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol. Cell. Biol. 16:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carey, M. Y., Y. S. Lin, M. R. Green, and M. Ptashne. 1990. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature 345:361-363. [DOI] [PubMed] [Google Scholar]
- 17.Chen, H., M. Tini, and R. M. Evans. 2001. HATs on and beyond chromatin. Curr. Opin. Cell Biol. 13:218-224. [DOI] [PubMed] [Google Scholar]
- 18.Chen, L., and M. Fluck. 2001. Kinetic analysis of the steps of the polyomavirus lytic cycle. J. Virol. 75:8368-8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, L. F., K. Ito, Y. Murakami, and Y. Ito. 1998. The capacity of polyomavirus enhancer binding protein 2αB (AML1/Cbfa2) to stimulate polyomavirus DNA replication is related to its affinity for the nuclear matrix. Mol. Cell. Biol. 18:4165-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, M. C., D. Redenius, F. Osati-Ashtiani, and M. M. Fluck. 1995. Enhancer-mediated role for polyomavirus middle T/small T in DNA replication. J. Virol. 69:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng, L., and T. J. Kelly. 1989. Transcriptional activator nuclear factor I stimulates the replication of SV40 minichromosomes in vivo and in vitro. Cell 59:541-551. [DOI] [PubMed] [Google Scholar]
- 22.Cheng, L., J. L. Workman, R. Kinston, and T. J. Kelly. 1992. Regulation of DNA replication in vitro by the transcriptional activation domain of GAL4-VP16. Proc. Natl. Acad. Sci. USA 89:589-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho, S. Y., Y. Tian, and T. L. Benjamin. 2001. Binding of p300/CBP co-activators by polyoma large T antigen. J. Biol. Chem. 276:33533-33539. [DOI] [PubMed] [Google Scholar]
- 24.Chou, J. Y., and R. G. Martin. 1974. Complementation analysis of simian virus 40 mutants. J. Virol. 13:1101-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Contreras, R., C. Cole, P. Berg, and W. Fiers. 1979. Nucleotide sequence analysis of two simian virus 40 mutants with deletions in the late region of the genome. J. Virol. 29:789-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DePamphilis, M. L. 1993. How transcription factors regulate origins of DNA replication in eukaryotic cells. Trends Cell Biol. 3:161-167. [DOI] [PubMed] [Google Scholar]
- 27.DePamphilis, M. L., S. Anderson, R. Bar-Shavit, E. Collins, H. Edenberg, T. Herman, B. Karas, G. Kaufmann, H. Krokan, E. Shelton, R. Su, D. Tapper, and P. M. Wassarman. 1979. Replication and structure of simian virus 40 chromosomes. Cold Spring Harb. Symp. Quant. Biol. 43(Pt 2):679-692. [DOI] [PubMed] [Google Scholar]
- 28.de Villiers, J., W. Schaffner, C. Tyndall, S. Lupton, and R. Kamen. 1984. Polyoma virus DNA replication requires an enhancer. Nature 312:242-246. [DOI] [PubMed] [Google Scholar]
- 29.Freund, R., P. H. Bauer, H. A. Crissman, E. M. Bradbury, and T. L. Benjamin. 1994. Host range and cell cycle activation properties of polyomavirus large T-antigen mutants defective in pRB binding. J. Virol. 68:7227-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freund, R., R. T. Bronson, and T. L. Benjamin. 1992. Separation of immortalization from tumor induction with polyoma large T mutants that fail to bind the retinoblastoma gene product. Oncogene 7:1979-1987. [PubMed] [Google Scholar]
- 31.Gerritsen, M. E., A. J. Williams, A. S. Neish, S. Moore, Y. Shi, and T. Collins. 1997. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. USA 94:2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilbert, D. M., A. Neilson, H. Miyazawa, M. L. DePamphilis, and W. C. Burhans. 1995. Mimosine arrests DNA synthesis at replication forks by inhibiting deoxyribonucleotide metabolism. J. Biol. Chem. 270:9597-9606. [DOI] [PubMed] [Google Scholar]
- 33.Giordano, A., and M. L. Avantaggiati. 1999. P300 and CBP: partners for life and death. J. Cell. Physiol. 181:218-230. [DOI] [PubMed] [Google Scholar]
- 34.Gruss, C., and R. Knippers. 1995. The SV40 minichromosome, p. 101-113. In K. W. Adolph (ed.), Viral gene techniques. Methods in molecular genetics, vol. 7. Academic Press, San Diego, Calif.
- 35.Gruss, C., J. Wu, T. Koller, and J. M. Sogo. 1993. Disruption of the nucleosomes at the replication fork. EMBO J. 12:4533-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu, W., X. L. Shi, and R. G. Roeder. 1997. Synergistic activation of transcription by CBP and p53. Nature 387:819-823. [DOI] [PubMed] [Google Scholar]
- 37.Guo, Z. S., and M. L. DePamphilis. 1992. Specific transcription factors stimulate simian virus 40 and polyomavirus origins of DNA replication. Mol. Cell. Biol. 12:2514-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo, Z. S., C. Gutierrez, U. Heine, J. M. Sogo, and M. L. DePamphilis. 1989. Origin auxiliary sequences can facilitate initiation of simian virus 40 DNA replication in vitro as they do in vivo. Mol. Cell. Biol. 9:3593-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo, W., W. J. Tang, X. Bu, V. Bermudez, M. Martin, and W. R. Folk. 1996. AP1 enhances polyomavirus DNA replication by promoting T-antigen-mediated unwinding of DNA. J. Virol. 70:4914-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gutierrez, C., Z. S. Guo, J. Roberts, and M. L. DePamphilis. 1990. Simian virus 40 origin auxiliary sequences weakly facilitate T-antigen binding but strongly facilitate DNA unwinding. Mol. Cell. Biol. 10:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He, Z., B. T. Brinton, J. Greenblatt, J. A. Hassell, and C. J. Ingles. 1993. The transactivator proteins VP16 and GAL4 bind replication factor A. Cell 73:1223-1232. [DOI] [PubMed] [Google Scholar]
- 42.Hermansen, R., M. A. Sierra, J. Johnson, M. Friez, and B. Milavetz. 1996. Identification of simian virus 40 promoter DNA sequences capable of conferring restriction endonuclease hypersensitivity. J. Virol. 70:3416-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 44.Iizuka, M., and B. Stillman. 1999. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 274:23027-23034. [DOI] [PubMed] [Google Scholar]
- 45.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 46.Ishikawa, H., M. Asano, T. Kanda, S. Kumar, C. Gelinas, and Y. Ito. 1993. Two novel functions associated with the Rel oncoproteins: DNA replication and cell-specific transcriptional activation. Oncogene 8:2889-2896. [PubMed] [Google Scholar]
- 47.Ishimi, Y. 1992. Preincubation of T antigen with DNA overcomes repression of SV40 DNA replication by nucleosome assembly. J. Biol. Chem. 267:10910-10913. [PubMed] [Google Scholar]
- 48.Ito, K., M. Asano, P. Hughes, H. Kohzaki, C. Masutani, F. Hanaoka, T. Kerppola, T. Curran, Y. Murakami, and Y. Ito. 1996. C-Jun stimulates origin-dependent DNA unwinding by polyomavirus large T antigen. EMBO J. 15:5636-5646. [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson, V., D. Granner, and R. Chalkley. 1976. Deposition of histone onto the replicating chromosome: newly synthesized histone is not found near the replication fork. Proc. Natl. Acad. Sci. USA 73:2266-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jayaraman, G., R. Srinivas, C. Duggan, E. Ferreira, S. Swaminathan, K. Somasundaram, J. Williams, C. Hauser, M. Kurkinen, R. Dhar, S. Weitzman, G. Buttice, and B. Thimmapaya. 1999. p300/cAMP-responsive element-binding protein interactions with ets-1 and ets-2 in the transcriptional activation of the human stromelysin promoter. J. Biol. Chem. 274:17342-17352. [DOI] [PubMed] [Google Scholar]
- 51.Jeong, S., and A. Stein. 1994. Micrococcal nuclease digestion of nuclei reveals extended nucleosome ladders having anomalous DNA lengths for chromatin assembled on non-replicating plasmids in transfected cells. Nucleic Acids Res. 22:370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeong, S. W., and A. Stein. 1994. DNA sequence affects nucleosome ordering on replicating plasmids in transfected COS-1 cells and in vitro. J. Biol. Chem. 269:2197-2205. [PubMed] [Google Scholar]
- 53.Kanda, T., K. Segawa, N. Ohuchi, S. Mori, and Y. Ito. 1994. Stimulation of polyomavirus DNA replication by wild-type p53 through the DNA-binding site. Mol. Cell. Biol. 14:2651-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kingston, R. E. 1999. Chromatin assembly and analysis, p. 21.0.1-21.1.15. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc. New York, N.Y.
- 55.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krumm, A., L. Madisen, X. J. Yang, R. Goodman, Y. Nakatani, and M. Groudine. 1998. Long-distance transcriptional enhancement by the histone acetyltransferase PCAF. Proc. Natl. Acad. Sci. USA 95:13501-13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kundu, T. K., V. B. Palhan, Z. Wang, W. An, P. A. Cole, and R. G. Roeder. 2000. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6:551-561. [DOI] [PubMed] [Google Scholar]
- 58.Kurooka, H., and T. Honjo. 2000. Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 275:17211-17220. [DOI] [PubMed] [Google Scholar]
- 59.Larose, A., N. Dyson, M. Sullivan, E. Harlow, and M. Bastin. 1991. Polyomavirus large T mutants affected in retinoblastoma protein binding are defective in immortalization. J. Virol. 65:2308-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lednicky, J., and W. R. Folk. 1992. Two synthetic Sp1-binding sites functionally substitute for the 21-base-pair repeat region to activate simian virus 40 growth in CV-1 cells. J. Virol. 66:6379-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee, J. S., R. H. See, T. Deng, and Y. Shi. 1996. Adenovirus E1A downregulates cJun- and JunB-mediated transcription by targeting their coactivator p300. Mol. Cell. Biol. 16:4312-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li, L., B. L. Li, M. Hock, E. Wang, and W. R. Folk. 1995. Sequences flanking the pentanucleotide T-antigen binding sites in the polyomavirus core origin help determine selectivity of DNA replication. J. Virol. 69:7570-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li, R. L., and M. R. Botchan. 1993. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell 73:1207-1221. [DOI] [PubMed] [Google Scholar]
- 64.Li, R., and M. R. Botchan. 1994. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc. Natl. Acad. Sci. USA 91:7051-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lill, N. L., S. R. Grossman, D. Ginsberg, J. DeCaprio, and D. M. Livingston. 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature 387:823-827. [DOI] [PubMed] [Google Scholar]
- 66.Lipford, J. R., and S. P. Bell. 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell 7:21-30. [DOI] [PubMed] [Google Scholar]
- 67.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lundblad, J. R., R. P. Kwok, M. E. Laurance, M. L. Harter, and R. H. Goodman. 1995. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature 374:85-88. [DOI] [PubMed] [Google Scholar]
- 69.Martin, M. E., J. Piette, M. Yaniv, W. J. Tang, and W. R. Folk. 1988. Activation of the polyomavirus enhancer by a murine activator protein 1 (AP1) homolog and two contiguous proteins. Proc. Natl. Acad. Sci. USA 85:5839-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Müeller, C. R., W. J. Muller, and J. A. Hassell. 1988. The polyomavirus enhancer comprises multiple functional elements. J. Virol. 62:1667-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Müller, K., and N. Mermod. 2000. The histone-interacting domain of nuclear factor I activates simian virus 40 DNA replication in vivo. J. Biol. Chem. 275:1645-1650. [DOI] [PubMed] [Google Scholar]
- 73.Müller, U., H. Zentgraf, I. Eicken, and W. Keller. 1978. Higher order structure of simian virus 40 chromatin. Science 201:406-415. [DOI] [PubMed] [Google Scholar]
- 74.Murakami, Y., and Y. Ito. 1999. Transcription factors in DNA replication. Front. Biosci. 4:D824-D833. [DOI] [PubMed] [Google Scholar]
- 75.Murakami, Y., M. Satake, Y. Yamaguchi-Iwai, M. Sakai, M. Muramatsu, and Y. Ito. 1991. The nuclear protooncogenes c-jun and c-fos as regulators of DNA replication. Proc. Natl. Acad. Sci. USA 88:3947-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nemethova, M., and E. Wintersberger. 1999. Polyomavirus large T antigen binds the transcriptional coactivator protein p300. J. Virol. 73:1734-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nilsson, M., M. Osterlund, and G. Magnusson. 1991. Analysis of polyomavirus enhancer-effect on DNA replication and early gene expression. J. Mol. Biol. 218:479-483. [DOI] [PubMed] [Google Scholar]
- 78.Pallas, D. C., C. Schley, M. Mahoney, E. Harlow, B. S. Schaffhausen, and T. M. Roberts. 1986. Polyomavirus small T antigen: overproduction in bacteria, purification, and utilization for monoclonal and polyclonal antibody production. J. Virol. 60:1075-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parker, D., K. Ferreri, T. Nakajima, V. J. Lamorte, R. Evans, S. C. Koerber, C. Hoeger, and M. R. Montminy. 1996. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol. Cell. Biol. 16:694-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peden, K. W., J. M. Pipas, S. Pearson-White, and D. Nathans. 1980. Isolation of mutants of an animal virus in bacteria. Science 209:1392-1396. [DOI] [PubMed] [Google Scholar]
- 81.Peng, Y. C., and N. H. Acheson. 1998. Polyomavirus large T antigen binds cooperatively to its multiple binding sites in the viral origin of DNA replication. J. Virol. 72:7330-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Perkins, N. D., L. K. Felzien, J. C. Betts, K. Leung, D. H. Beach, and G. J. Nabel. 1997. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science 275:523-527. [DOI] [PubMed] [Google Scholar]
- 83.Piette, J., and M. Yaniv. 1987. Two different factors bind to the alpha-domain of the polyoma virus enhancer, one of which also interacts with the SV40 and c-fos enhancers. EMBO J. 6:1331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pilon, A. A., P. Desjardins, J. A. Hassell, and A. M. Mes-Masson. 1996. Functional implications of mutations within polyomavirus large T antigen Rb-binding domain: effects on pRb and p107 binding in vitro and immortalization activity in vivo. J. Virol. 70:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prives, C. 1990. The replication functions of SV40 T antigen are regulated by phosphorylation. Cell 61:735-738. [DOI] [PubMed] [Google Scholar]
- 86.Puigserver, P., G. Adelmant, Z. Wu, M. Fan, J. Xu, B. O'Malley, and B. M. Spiegelman. 1999. Activation of PPARγ coactivator-1 through transcription factor docking. Science 286:1368-1371. [DOI] [PubMed] [Google Scholar]
- 87.Quintini, G., K. Treuner, C. Gruss, R. Knippers. 1996. Role of amino-terminal histone domains in chromatin replication. Mol. Cell. Biol. 16:2888-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reeves, R., C. M. Gorman, and B. Howard. 1985. Minichromosome assembly of non-integrated plasmid DNA transfected into mammalian cells. Nucleic Acids Res. 13:3599-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Richards, J. P., H. P. Bächinger, R. H. Goodman, and R. G. Brennan. 1996. Analysis of the structural properties of cAMP-responsive element-binding protein (CREB) and phosphorylated CREB. J. Biol. Chem. 271:13716-13723. [DOI] [PubMed] [Google Scholar]
- 90.Rochford, R., C. T. Davis, K. K. Yoshimoto, and L. P. Villarreal. 1990. Minimal subenhancer requirements for high-level polyomavirus DNA replication: a cell-specific synergy of PEA3 and PEA1 sites. Mol. Cell. Biol. 10:4996-5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rochford, R., J. P. Moreno, M. L. Peake, and L. P. Villarreal. 1992. Enhancer dependence of polyomavirus persistence in mouse kidneys. J. Virol. 66:3287-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 93.Sanchez, J. A., D. Marek, and L. J. Wangh. 1992. The efficiency and timing of plasmid DNA replication in Xenopus eggs: correlations to the extent of prior chromatin assembly. J. Cell Sci. 103:907-918. [DOI] [PubMed] [Google Scholar]
- 94.Shaywitz, A. J., S. L. Dove, J. M. Kornhauser, A. Hochschild, and M. E. Greenberg. 2000. Magnitude of the CREB-dependent transcriptional response is determined by the strength of the interaction between the kinase-inducible domain of CREB and the KIX domain of CREB-binding protein. Mol. Cell. Biol. 20:9409-9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spencer, T. E., G. Jenster, M. M. Burcin, C. D. Allis, J. Zhou, C. A. Mizzen, N. J. McKenna, S. A. Onate, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194-198. [DOI] [PubMed] [Google Scholar]
- 96.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modification. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 98.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 99.Stuiver, M. H., W. G. Bergsma, A. C. Arnberg, H. van Amerongen, R. van Grondelle, and P. C. van der Vliet. 1992. Structural alterations of double-stranded DNA in complex with the adenovirus DNA-binding protein. Implications for its function in DNA replication. J. Mol. Biol. 225:999-1011. [DOI] [PubMed] [Google Scholar]
- 100.Sun, D., and L. H. Hurley. 1994. Cooperative bending of the 21-base-pair repeats of the SV40 viral early promoter by human Sp1. Biochemistry 33:9578-9587. [DOI] [PubMed] [Google Scholar]
- 101.Suzuki, T., A. Kimura, R. Nagai, and M. Horikoshi. 2000. Regulation of interaction of the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA binding. Genes Cells 5:29-41. [DOI] [PubMed] [Google Scholar]
- 102.Swope, D. L., C. L. Mueller, and J. C. Chrivia. 1996. CREB-binding protein activates transcription through multiple domains. J. Biol. Chem. 271:28138-28145. [DOI] [PubMed] [Google Scholar]
- 103.Takei, Y., M. Swietlik, A. Tanoue, G. Tsujimoto, T. Kouzarides, and R. Laskey. 2001. MCM3AP, a novel acetyltransferase that acetylates replication protein MCM3. EMBO Rep. 2:119-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tang, W. J., and W. R. Folk. 1989. Asp-286 224 Asn-286 in polyomavirus large T antigen relaxes the specificity of binding to the polyomavirus origin. J. Virol. 63:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tang, W. J., S. L. Berger, S. J. Triezenberg, and W. R. Folk. 1987. Nucleotides in the polyomavirus enhancer that control viral transcription and DNA replication. Mol. Cell. Biol. 7:1681-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Turner, W. J., and M. E. Woodworth. 2001. DNA replication efficiency depends on transcription factor-binding sites. J. Virol. 75:5638-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 108.Verrijzer, C. P., J. A. van Oosterhout, W. W. van Weperen, and P. C. van der Vliet. 1991. POU proteins bend DNA via the POU-specific domain. EMBO J. 10:3007-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vries, R. G., M. Prudenziati, C. Zwartjes, M. Verlaan, E. Kalkhoven, and A. Zantema. 2001. A specific lysine in c-Jun is required for transcriptional repression by E1A and is acetylated by p300. EMBO J. 20:6095-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang, L., S. R. Grossman, and E. Kieff. 2000. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl. Acad. Sci. USA 97:430-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wasylyk, C., J. Schneikert, and B. Wasylyk. 1990. Oncogene v-jun modulates DNA replication. Oncogene 5:1055-1058. [PubMed] [Google Scholar]
- 112.Weisshart, K., and E. Fanning. 1996. Roles of phosphorylation in DNA replication, p. 295-330. In M. L. Depamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 113.Wilderman, P. J., B. Hu, and M. E. Woodworth. 1999. Conformational changes in simian virus 40 rearranged regulatory regions: effects of the 21-base-pair promoters and their location. J. Virol. 73:10254-10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu, W., D. G. Edmondson, and S. Y. Roth. 1998. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol. Cell. Biol. 18:5659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 116.Yang, C., L. H. Shapiro, M. Rivera, A. Kumar, and P. K. Brindle. 1998. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol. Cell. Biol. 18:2218-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 118.Yoshida, M., S. Horinouchi, and T. Beppu. 1995. Trichostatin A and trapoxin-novel chemical probes for the role of histone acetylation in chromatin structure and function. BioEssays 17:423-430. [DOI] [PubMed] [Google Scholar]
- 119.Yuan, W., G. Condorelli, M. Caruso, A. Felsani, and A. J. Giordano. 1996. Human p300 protein is a coactivator for the transcription factor MyoD. J. Biol. Chem. 271:9009-9013. [DOI] [PubMed] [Google Scholar]