Abstract
Bradyrhizobium japonicum, the nitrogen-fixing soybean symbiont, possesses a heme uptake system encoded by the gene cluster hmuVUT-hmuR-exbBD-tonB. Transcription of the divergently oriented hmuT and hmuR genes was previously found to be induced by iron limitation and to depend on a 21-bp promoter-upstream iron control element (ICE). Here, we show by deletion analysis that the full-length ICE is needed for this type of positive control. Additional genes associated with ICE-like motifs were identified in the B. japonicum genome, of which bll6680 and blr7895 code for bacterioferritin and rubrerythrin homologs, respectively. Transcription start site mapping revealed that their ICEs directly overlap with either the −10 promoter region or the transcription initiation site, suggesting an involvement of the ICE in negative control of both genes. Consistent with this inference was the observed down-regulation of both genes under iron limitation, which in the case of bll6680 was shown to require an intact ICE motif. Using a yeast one-hybrid system, we demonstrated in vivo interaction of the iron response regulator (Irr) with all three ICEs. Moreover, specific in vitro binding of purified Irr protein to the ICE motifs of bll6680 and blr7895 was shown in electrophoretic mobility shift experiments. A genome-wide survey for iron-regulated genes with a custom-made Affymetrix gene chip revealed 17 genes to be induced and 68 to be repressed under iron-replete conditions. Remarkably, ICE-like motifs are associated with a large subset of those B. japonicum genes. We propose the ICE as an important cis-acting element in B. japonicum which represents the DNA-binding site for the Irr protein and, depending on its location within promoter regions, is involved in positive or negative control of the associated iron-regulated genes.
Genes involved in iron acquisition or metabolism are not only conserved within and between many bacterial taxa but are also under tight control in response to the external iron supply. Iron availability is usually restricted because the element—even when abundantly present in the environment—occurs mostly in its insoluble, ferric form (2, 4, 21). To get access to this metal as a nutrient, microorganisms have evolved different strategies. Iron-chelating molecules (siderophores), synthesized under iron-limiting conditions, are secreted to bind to ferric iron (Fe3+), and the Fe-siderophore complexes are then imported via active transport mechanisms into the cell (11, 60). Alternative iron sources are also used, including heme (both as a free molecule or protein bound, as in hemoglobins), Fe-containing proteins, ferric citrate, or inorganic iron (ideally in the ferrous form). Once the intracellular iron concentration is sufficiently high, transcription of genes that are involved in Fe uptake (e.g., those for siderophore synthesis and transport) is shut off.
After its first identification in Escherichia coli (27), the ferric uptake regulator Fur was thought to be the sole “global” regulator for all processes concerned with iron acquisition and metabolism. In its repressive, Fe2+-liganded form, the Fur protein binds to the so-called Fur boxes on DNA, thus preventing transcription of the iron-regulated genes. Although Fur homologs are now known to exist in a vast range of different bacterial species (26), most studies on Fur have been carried out with γ-proteobacteria such as E. coli, Vibrio cholerae, or Pseudomonas aeruginosa (47, 48). Recently, E. coli Fur was demonstrated to act also as an indirect activator in that it represses the transcription of a small regulatory RNA, ryhB, which in turn inhibits expression of several target genes (38). Unexpectedly, Neisseria meningitidis Fur appears to activate RNA transcription directly by binding to upstream sequences of the pan1, norB, and nuoA genes in response to iron accumulation in the cell (9).
Further Fe-dependent regulators have been discovered. One is DtxR of Corynebacterium diphtheriae, a repressor of diphtheria toxin production (6), which in gram-positive bacteria such as Bacillus subtilis assumes some of the roles that Fur plays in other bacterial species (26). RirA is an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Null mutation of rirA causes high-level expression of at least eight operons whose products are involved in synthesis and uptake of siderophores, uptake of heme, and iron acquisition from other sources (57). The zinc uptake regulator Zur of Bacillus subtilis (15) and the manganese uptake regulator Mur of R. leguminosarum, which represses transcription of the sitABCD operon (10, 15), differ from other members of the Fur family, as they do not respond solely to Fe2+ but rather to Zn2+ or Mn2+.
Another member of the Fur family of transcriptional regulators is the iron-response regulator Irr, first identified in the nitrogen-fixing soybean symbiont, Bradyrhizobium japonicum (23). At present, Irr occurrence appears to be restricted to α-proteobacteria, including rhizobia, Agrobacterium tumefaciens, the animal pathogen Brucella abortus, and Rhodopseudomonas palustris (62). It has been argued particularly for the rhizobia that they have a high demand for iron due to the intracellular abundance of iron-sulfur proteins and hemoproteins (nitrogenase complex and cytochromes) during symbiotic nitrogen fixation (42, 20). The Irr protein was shown to repress transcription of hemB, the gene for δ-aminolevulinic acid dehydratase in the heme biosynthesis pathway (23). Interestingly, Irr itself is modified at the posttranslational level: under iron-replete conditions, Irr interacts directly with ferrochelatase and becomes degraded in a process that depends on heme being delivered by the ferrochelatase enzyme (49, 50). In addition, transcription of the irr gene is repressed in Fe-replete conditions by Fur, even though the irr promoter region shares no sequence similarity with canonical Fur boxes (25). Since other roles of Irr, apart from regulating hemB, were so far unknown, a DNA sequence motif for Irr binding had yet to be defined.
B. japonicum and R. leguminosarum can grow on heme as the sole iron source (43, 46, 63), a property that until recently was thought to be unique to pathogenic bacteria (16). The heme uptake system (Hmu) of B. japonicum consists of a TonB-dependent heme receptor (HmuR), a periplasmic heme-binding protein (HmuT), an ABC transporter (HmuUV), and a Ton system (ExbBD and TonB). Transcription of hmuT and hmuR was shown to be up-regulated by iron limitation via a mechanism that involves a 21-bp A/T-rich incomplete inverted repeat named the iron control element (ICE) (43). This motif is located in between the divergently oriented hmuT and hmuR genes and might represent the binding site for a regulatory protein that, presumably, acts positively on the expression of both genes (43).
Here, we have followed up on this previous work and have addressed several open questions. Are both of the half-sites of the symmetric ICE needed for control? Does one of the known B. japonicum iron control proteins, Irr or Fur, bind to the ICE? Can we solidify the postulated positive control at this ICE? The experiments reported here provide answers to these questions. Moreover, we describe the use of a microarray for the identification of iron-regulated B. japonicum genes on a genome-wide scale. Many of those genes are associated with ICE-like motifs which are involved in positive or negative control, depending on their location within promoters.
MATERIALS AND METHODS
Strains and plasmids.
Strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Bacterial and yeast strains, cloning vectors, and recombinant plasmids used in this study
| Strain or plasmid | Relevant genotype or properties | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories Inc., Gaithersburg, MD |
| S17-1 | Smr SprhsdR (RP4-2 kan::Tn7 tet::Mu, integrated in the chromosome) | 54 |
| BL21(DE3) | hsdS gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) | 55 |
| B. japonicum | ||
| 110spc4 | Spr wild type | 51 |
| GEM4 | Sprfur::Ω (parental strain: USDA I110) | 24 |
| S. cerevisiae | ||
| YM4271 | MATaura3-52 his3-200 ade2-101 lys2-801 leu2-3,112 trp1-901 tyr1-501 gal4-Δ512 gal80-Δ538 ade5::hisG | 36 |
| 8730 | ICEhmu chromosomally integrated in YM4271 | This work |
| 8732 | ICEhmumut chromosomally integrated in YM4271 | This work |
| 8733 | ICE6680 chromosomally integrated in YM4271 | This work |
| 8734 | ICE7895 chromosomally integrated in YM4271 | This work |
| 8765 | Fur box chromosomally integrated in YM4271 | This work |
| Plasmids | ||
| pGEM-T Easy | Apr TA cloning vector | Promega, Madison, WI |
| pME3535XhoI | Tcr pME3535 derivative with XhoI site (Klenow fill-in of StuI site, addition of XhoI linker) | 64 |
| pASK-IBA3+ | Apr expression vector, Tet repressor, tetA-promoter/operator, C-terminal Strep-tag II | IBA, Göttingen, Germany |
| pRK290X | Tcr pRK290 derivative with XhoI site | 1 |
| pRKPolScaI | Tcr pRK290 derivative with ScaI linker | Laboratory collection |
| pNM482 | Apr pUC8 ′lacZ | 41 |
| pSUP482 | Tcr ′lacZ part from pNM482 in pSUP202pol4 | Laboratory collection |
| pUC18/19 | Apr high-copy-number cloning vectors | 45 |
| pGAD424 | Apr expressing the GAL4 activation domain (AD) | Clontech, Palo Alto, CA |
| pLacZi | Apr for integration at the URA3 locus; carries the lacZ reporter gene | Clontech, Palo Alto, CA |
| pRJ2562 | Apr (pGEM-T Easy) 1,301-bp PCR fragment comprising bll6680 and upstream region | This work |
| pRJ2563 | Apr (pGEM-T Easy) 1,306-bp PCR fragment comprising blr7895 and upstream region | This work |
| pRJ8710 | Apr (pUC18) with 1,852-bp EcoRI/BamHI fragment comprising the hmuT-hmuR intergenic region and hmuR | This work |
| pRJ8722 | Tcr (pRK290X) with 5,538-bp hmuR-lacZ fusion | This work |
| pRJ8723 | Tcr (pRK290X) with 5,521-bp hmuR-lacZ fusion | This work |
| pRJ8724 | Tcr (pRK290X) with 5,505-bp hmuR-lacZ fusion | This work |
| pRJ8730 | Apr (pLacZi) triple tandem copy of ICEhmu | This work |
| pRJ8732 | Apr (pLacZi) triple tandem copy of ICEhmumut | This work |
| pRJ8733 | Apr (pLacZi) triple tandem copy of ICE6680 | This work |
| pRJ8734 | Apr (pLacZi) triple tandem copy of ICE7895 | This work |
| pRJ8735 | Apr (pGAD424) 493-bp EcoRI fragment comprising the fur gene | This work |
| pRJ8737 | Apr (pGAD424) 504-bp SmaI fragment comprising the irr gene | This work |
| pRJ8750 | Apr (pASK-IBA3+) 491-bp Eco31I fragment comprising the irr gene | This work |
| pRJ8765 | Apr (pLacZi) triple tandem copy of Fur box | This work |
| pRJ8766 | Apr (pGEM-T Easy) 1,017-bp PCR fragment comprising bll6680 and upstream region with &12frac;-ICE6680 | This work |
| pRJ8767 | Apr (pNM482) 726-bp EcoRI/ScaI fragment of pRJ2562 comprising bll6680′-′lacZ and upstream region | This work |
| pRJ8768 | Apr (pNM482) 730-bp EcoRI/ScaI fragment of pR8766 comprising bll6680′-′lacZ and upstream region with &12frac;-ICE6680 | This work |
| pRJ8769 | Tcr (pRKPolScaI) 5,829-bp EcoRI/StuI fragment of pR8767 | This work |
| pRJ8770 | Tcr (pRKPolScaI) 5,829-bp EcoRI/StuI fragment of pR8768 | This work |
Media and growth conditions.
E. coli was grown in Luria-Bertani (LB) medium (40) containing the following concentrations of antibiotics for plasmid selection (μg ml−1): ampicillin, 200; kanamycin, 30; and tetracycline, 10. B. japonicum was cultivated in peptone-salts-yeast extract (PSY) medium (51) containing 0.1% l-arabinose and 1.2 μM FeCl3 for routine use. Concentrations of antibiotics were as follows (μg ml−1): spectinomycin, 100; kanamycin, 100; streptomycin, 50; and tetracycline, 50 (solid media) and 25 (liquid media). For growth under “low-iron” conditions, PSY medium lacked the iron supplement. PSY medium representing “high-iron” conditions contained 50 μM FeSO4. Yeast strains were grown at 30°C in synthetic dropout medium or yeast extract-peptone-dextrose medium (yeast protocols handbook; Clontech, Palo Alto, CA).
DNA work and sequence analysis.
Recombinant DNA work was performed according to standard protocols (53). B. japonicum chromosomal DNA was isolated as described previously (22). Programs DNA Star MegAlign 5.06, Clone Manager 7.02, GeneDoc version 2.6.002 (http://www.psc.edu/biomed/genedoc), and WebLogo version 2.8 (http://weblogo.berkeley.edu/) were used for computer-assisted analyses of DNA and protein sequences. Similarity searches were done on the National Center for Biotechnology Information BLAST network server (http://www.ncbi.nlm.nih.gov/BLAST/) and RhizoBase, the database for rhizobial genomes (http://www.kazusa.or.jp/rhizobase/). Designations for B. japonicum genes and open reading frames were also taken from RhizoBase (blr, bll, bsr, and bsl plus 4-digit numbers).
RNA isolation, cDNA synthesis, and microarray analysis.
Cultures of B. japonicum 110spc4 were grown to an optical density (at 600 nm) of 0.35 to 0.4 in PSY medium either lacking or containing 50 μM FeSO4 iron. Grown cultures (200 ml) were immediately transferred into cold tubes containing 1/10 of the culture volume of stop solution (10% Tris-HCl-buffered phenol [pH 8] in ethanol). After centrifugation for 5 min (10,800 × g; 4°C) the supernatant was decanted, and cells were frozen in liquid nitrogen and stored at −80°C. Isolation of total RNA, cDNA synthesis, and conditions for microarray hybridization were described elsewhere (28). Based on the B. japonicum genome sequence (31), a custom Affymetrix-type gene chip representing all 8,317 annotated open reading frames plus 51 RNA genes and intergenic regions was designed (Affymetrix Inc., Santa Clara, CA). Further details of the BJAPETHa520090 microarray will be described elsewhere, and they are also available on request from the authors. Primary data analysis was done with the Affymetrix GeneChip operating software (GCOS). GeneSpring 4.2 software (Silicon Genetics) was used for comparison analysis.
In silico search for ICE motifs.
Initial searches for ICE-like motifs in intergenic regions of the B. japonicum genome were performed with the program “FUZZNUC” (http://emboss.ch.embnet.org/Pise) using TTTA-N9-TAAA as a query sequence. In subsequent searches, the Darwin software (19) was used. The ICE motif was represented as a position-specific frequency matrix (PSFM), which provides a good approximation of protein-DNA interactions (5). The PSFM is a matrix consisting of the frequency of each nucleotide at each motif position, based on a collection of known motifs.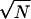 pseudocounts (where N is the number of motifs in the PSFM) were used to enable finding new motifs that were similar, but not identical, to the known motifs (32). The genomic sequence 500 bp upstream of each annotated gene was searched for the motif PSFM. Because transcriptional start sites were not known for most genes, the translational start codon was used as a substitute. Each (overlapping) sequence of the same length as the profile was scored by multiplying the frequencies from the PSFM: ∏i=1n PSFM[i,s(i)], where n is the length of the PSFM and s is the sequence (of length n) to be scored.
pseudocounts (where N is the number of motifs in the PSFM) were used to enable finding new motifs that were similar, but not identical, to the known motifs (32). The genomic sequence 500 bp upstream of each annotated gene was searched for the motif PSFM. Because transcriptional start sites were not known for most genes, the translational start codon was used as a substitute. Each (overlapping) sequence of the same length as the profile was scored by multiplying the frequencies from the PSFM: ∏i=1n PSFM[i,s(i)], where n is the length of the PSFM and s is the sequence (of length n) to be scored.
Construction of transcriptional hmuR-lacZ fusions.
Starting from a 1.8-kb DNA fragment comprising the entire hmuT-hmuR intergenic region (plasmid pRJ8710), a set of nested deletion derivatives with variable 5′ ends relative to hmuR was generated by PCR amplification using Taq DNA polymerase and primer pairs GR-3/GR-7, GR-4/GR-7, and GR-5/GR-7 (Table 2). Amplification products of 408 bp, 391 bp, and 375 bp were digested with EcoRI and BamHI, cloned into the pUC19 vector, and verified by sequencing. Plasmid-derived EcoRI-BamHI fragments of 394 bp, 377 bp, and 361 bp were then inserted between the EcoRI and BamHI sites of the transcriptional lacZ fusion vector pME3535XhoI, from which the entire transcriptional lacZ fusions, including the variable upstream promoter fragments, were transferred into the broad-host-range vector pRK290X by cloning in between the EcoRI and XhoI restriction sites. The final constructs pRJ8722, pRJ8723, and pRJ8724 (Fig. 1) were mobilized by conjugation into B. japonicum strains 110spc4 and GEM4. The correct plasmid contents of these strains were verified by PCR.
TABLE 2.
Oligonucleotides used in this work
| Designation | Nucleotide sequence (5′ to 3′)a,b |
|---|---|
| GR-3 | CCGGAATTCGGCGGAAATTTACAATCGATATAAACTGC7,793,570 |
| GR-4 | CCGGAATTCGATATAAACTGCAACCAGACGTC7,793,559 |
| GR-5 | CCGGAATTCAGACGTCATTGACCGTCAG7,793,547 |
| GR-7 | GCGCGGGGATCCTAGGCGTCAGGCGTTGCCTGCTG7,793,233 |
| GR-14 | CGGAATTCATGACCGCACTGAAACCTT856,706 |
| GR-15 | CGGAATTCCGCAGTCGAAGATGATGAGA856,257 |
| GR-21 | TCCCCCGGGAATGAGCGAGAATACCG823,348 |
| GR-22 | TCCCCCGGGTAGATCTCAGCGCTTCTT822,883 |
| GR-26 | ATGGTAGGTCTCAAATGAGCGAGAATACCGCGCCCCAT823,340 |
| GR-27 | ATGGTAGGTCTCAGCGCTGCGCTTCTTGCGCAGGCGCA822,894 |
| fwICE-2 | AATTC-(AATTTACAATCGATATAAACT)3x-G |
| revICE-2 | TCGAC-(AGTTTATATCGATTGTAAATT)3x-G |
| 5′ ICE E/Smut | AATTC-(AATTTACAATCGCGATATAAACT)3x-G |
| 3′ ICE E/Smut | TCGAC-(AGTTTATATCGCGATTGTAAATT)3x-G |
| furbox fw | AATTC-(GATAATGATAATCATTATC)3x-G |
| furbox rev | TCGAC-(GATAATGATTATCATTATC)3x-G |
| 6680-1 | GGGAATTCAGCTGTTCCAGCAGCGATAG7,350,980 |
| 6680-2 | GGGAATTCTACAAGCAGCAGCTGTAGGC7,349,730 |
| 6680-7 | GTGCCTTGTTGAGGTAGTCGATGA7,350,328 |
| 6680-8 | GTACTGGTTGATCGCAGTCAGCTC7,350,296 |
| 6680-9 | AATTC-(ATTTAGAAGCGTTCTAAAT)3x-G |
| 6680-10 | TCGAC-(ATTTAGAACGCTTCTAAAT)3x-G |
| 6680-11c | aaactcATTTAGAAGCGTTCTAAATtcggtt7,350,384 |
| 6680-mut1c,d | aaactcATTTAGAAGCGCGTTCTAAATtcggtt |
| 6680-16c | aaactcATTTAGAAGCCAAGATTAAtcggtt |
| 6680-18 | ATTTAGAAGCCAAGATTAATCGGTTTAGGATCGAGTTGG |
| 6680-19 | CCTAAACCGATTAATCTTGGCTTCTAAATGAGTTTACGG |
| F2 | ATGATCTCCTTGACGTTCT |
| SP6 | ATTTAGGTGACACTATAG |
| hmu-1c | ggcggaAATTTACAATCGATATAAACTgcaacc7,793,566 |
| hmu-mut1c,d | ggcggaAATTTACAATCGCGATATAAACTgcaacc |
| 7895-1 | GGGAATTCGTTCGAGGCGATGCAGAAAAG8,656,690 |
| 7895-2 | GGGAATTCGATCGCGATTGATATGTGGATG8,657,942 |
| 7895-8 | GAAGGTCATGTAGATGCGGCTGT8,656,963 |
| 7895-9 | AATTC-(AATTTAGAATCATTCTAAACT)3x-G |
| 7895-10 | TCGAC-(AGTTTAGAATGATTCTAAATT)3x-G |
| 7895-11 | GCTCTTCCTCGGCCATCTGTT8,657,032 |
| GS-7895-11c | ctatatAATTTAGAATCATTCTAAACTagtggg8,656,884 |
| 7895-mut1c,d | ctatatAATTTAGAATCGCATTCTAAACTagtggg |
Nucleotide positions in the B. japonicum genome are indicated by subscript numbers at the 3′ ends of oligonucleotides. 3× indicates that the sequence in parentheses occurs three times in a row.
Nucleotides shown in italics belong to restriction sites.
Capital letters refer to the 19-to-21-bp ICE motifs of the respective promoter regions; lowercase letters represent adjacent nucleotides (Fig. 2).
The spacing between the half-sites of the mutated ICE motifs is increased by the two underlined nucleotides.
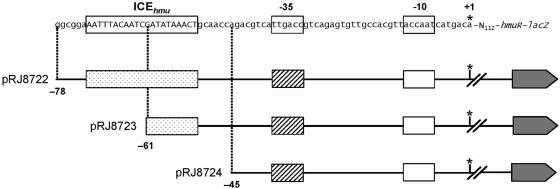
FIG. 1.
Promoter-upstream deletions of transcriptional hmuR-lacZ fusions. The top line shows the nucleotide sequence of the hmuR promoter region comprising the −10/−35 region and the iron regulatory element (ICEhmu). The three lower lines represent different 5′ deletion constructs. Plasmid names and nucleotide positions of the deletion end points are indicated to the left of each construct. An asterisk marks the transcription start at genome position 7,793,520. The arrow on the far right symbolizes lacZ.
Cloning of genes bll6680 and blr7895.
B. japonicum chromosomal regions comprising bll6680 and blr7895 were amplified by PCR using Taq DNA polymerase and primer pairs 6680-1/6680-2 and 7895-1/7895-2 (Table 2). The resulting PCR products of 1,301 bp (bll6680) and 1,306 bp (blr7895) were cloned into vector pGEM-T Easy, yielding plasmids pRJ2562 and pRJ2563. Cloned DNA regions were verified by sequencing.
Construction of translational bll6680′-′lacZ fusions.
A translational bll6680′-′lacZ fusion was obtained by isolating a 726-bp EcoRI-ScaI fragment from plasmid pRJ2562 and cloning it between the EcoRI and SmaI sites of pNM482. The resulting plasmid, pRJ8767, carried the 5′ end of bll6680 and its promoter region, including ICE6680. In parallel, a mutated version of the translational bll6680′-′lacZ fusion was constructed in which the nucleotides of the 3′ portion of ICE6680 were systematically replaced. This resulted in the alteration of the wild-type ICE6680 (ATTTAGAAGCGTTCTAAAT) to the mutated 1/2-ICE6680 (ATTTAGAAGCCAAGATTAA; nucleotides in italics are part of an AseI site introduced in the course of the mutagenesis). Using plasmid pRJ2562 as a template, two overlapping fragments comprising the desired mutations were generated in separate amplification reactions with Pfu DNA polymerase and primer pairs SP6/6680-19 and 6680-18/F2 (Table 2). Products of these reactions were mixed, annealed via their overlapping ends, and subjected to a third PCR step using the external primers SP6 and F2 according to the method of Higuchi et al. (29). After attachment of 3′ A overhangs in an additional reaction with Taq polymerase and dATP, the final amplification product of 1,017 bp, including 1/2-ICE6680 and the 5′ portion of bll6680, was cloned into vector pGEM-T Easy, yielding plasmid pRJ8766. The cloned DNA region was verified by sequencing. A translational bll6680′-′lacZ fusion containing the mutated 1/2-ICE6680 was obtained by cloning a 726-bp EcoRI-ScaI fragment from pRJ8766 between the EcoRI and SmaI sites of pNM482, resulting in plasmid pRJ8768. Finally, 5,829-bp EcoRI-StuI fragments of pRJ8767 and pRJ8768, comprising the desired lacZ fusions, were cloned between the EcoRI and ScaI restriction sites of the broad-host-range vector pRKPolScaI to yield plasmids pRJ8769 (wild-type ICE6680) and pRJ8770 (1/2-ICE6680). These plasmids were mobilized by conjugation into B. japonicum strain 110spc4.
Transcript mapping.
The transcription start sites of bll6680 and blr7895 were determined by primer extension. B. japonicum wild-type cells were grown aerobically to mid-exponential phase in PSY medium either lacking iron or supplemented with 50 μM FeSO4. RNA was isolated and used for reverse transcription as described previously (3, 44), using primers 6680-7 and 6680-8 (for bll6680) and 7895-8 and 7895-11 (for blr7895). Extension products were analyzed on 6% denaturing polyacrylamide gels adjacent to sequencing ladders generated with the same oligonucleotides and plasmids pRJ2562 and pRJ2563.
Construction of an Irr-Strep-tag expression plasmid.
The B. japonicum 110spc4 chromosomal region comprising the irr gene (bll0768) was amplified by PCR using Pfu DNA polymerase and primer pair GR-26/GR-27 (Table 2) containing Eco31I restriction sites (520-bp PCR product). 3′ A overhangs were attached through an additional treatment with Taq polymerase, which then enabled cloning into the pGEM-T Easy vector. The construct was verified by sequencing. Cloning of the pGEM-T Easy vector-derived 491-bp Eco31I irr fragment into the Eco31I-digested expression vector pASK-IBA3+ (encoding the C-terminal Strep-tag II; IBA, Göttingen, Germany) resulted in the irr expression plasmid pRJ8750.
Overproduction and purification of recombinant Irr protein.
E. coli strain BL21(DE3) harboring plasmid pRJ8750 was used for overproduction of the Irr-Strep-tag II fusion protein. One-liter LB medium cultures in 5,000-ml Erlenmeyer flasks were grown on a rotary shaker (160 rpm) at 37°C until an optical density (at 550 nm) of 0.4 to 0.6 was reached. Expression of irr was induced by adding 0.2 μg ml−1 anhydrotetracycline, and the cultures were further incubated for 2 to 3 h. Cells were harvested by centrifugation (5 min, 8,000 × g, 4°C), resuspended in 3 ml of buffer W (100 mM Tris-HCl [pH 8.0], 150 mM NaCl), and lysed by three passages through a French pressure cell (11,000 lb in−2). The extract was centrifuged for 30 min (16,000 × g, 4°C), and the supernatant with soluble recombinant Irr protein was applied to a 1-ml Strep-Tactin column (IBA, Göttingen, Germany) and affinity purified according to the instructions in the manufacturer's protocol. A further purification step was added by anion ion-exchange chromatography using a 10-ml Mono Q column (4.6 by 100 mm; Amersham Biosciences). The column was developed with 10 mM Tris-HCl (pH 8), 1 mM MgCl2, 1 mM dithiothreitol, and 5% glycerol using a salt gradient from 0.04 M to 1 M KCl. For desalting, the fraction containing the Irr-Strep-tag II fusion protein was gel filtrated over a PD-10 Sephadex G-25 column (Amersham Biosciences) with DNA-binding buffer (10 mM Tris-HCl (pH 8), 40 mM KCl, 1 mM MgCl2, 1 mM dithiothreitol, 5% glycerol). Protein concentration was determined with the Bio-Rad assay (Bio-Rad Laboratories, Richmond, CA) and the degree of purity was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Electrophoretic mobility shift assay.
Electrophoretic mobility shift assays were used to monitor Irr binding to the ICE. Irr protein and double-stranded, end-labeled ICE oligonucleotides were incubated for 10 to 15 min at room temperature in a 20-μl volume of DNA-binding buffer to which 1 μg poly(dI-dC) (Amersham Biosciences) was added. Double-stranded ICE DNA probes were produced by annealing the single-stranded oligonucleotides representing the ICE motifs with their corresponding complementary oligonucleotides (oligonucleotide hmu-1 for ICEhmu; 6680-11 for ICE6680; 6680-16 for 1/2-ICE6680; GS-7895-11 for ICE7895; hmu-mut1, 6680-mut1, and 7895-mut1 for 2-bp insertions in ICEhmu, ICE6680, and ICE7895, respectively) (Table 2). End labeling and annealing of oligonucleotides was performed as follows: oligonucleotides (30 pmol), T4 polynucleotide kinase (10 units; MBI Fermentas, Vilnius, Lithuania) and [γ-32P]ATP (10 μCi, 3,000 Ci/mmol) were incubated in polynucleotide kinase-buffer A (MBI Fermentas) for 30 min at 37°C in a total volume of 50 μl. After addition of 2 μl stop solution (4 M Na-acetate, pH 7.5, 25 mM EDTA), 60 pmol of the unlabeled complementary oligonucleotide was added. After 10 min at 95°C, the reaction mixture was slowly cooled down to room temperature. Annealed, labeled oligonucleotides were purified over a NAP-10 Sephadex G-25 column (Amersham Biosciences). Following incubation of protein and DNA (see above), 5 μl of loading dye solution (30% glycerol, 0.02% bromophenol blue) was added and reaction mixtures were analyzed on 6% nondenaturing polyacrylamide gels in electrophoresis buffer (90 mM Tris base, 90 mM boric acid, pH 8) that were prerun for 10 to 20 min at 180 V. After electrophoresis for 25 to 30 min at 180 V, gels were dried and exposed on a phosphorimager screen.
Construction of yeast one-hybrid plasmids and reporter strains.
Target DNAs were created by first annealing complementary oligonucleotides (Table 2) representing three tandem copies of the respective ICE (primer pair fwICE-2/revICE-2 for ICEhmu; primer pair 6680-9/6680-10 for ICE6680; primer pair 7895-9/7895-10 for ICE7895; and, as control, primer pair 5′ICE E/S−mut/3′ICE E/Smut for ICEhmumut or the consensus Fur box (primer pair furbox fw/furbox rev). Annealed oligonucleotides carrying overhanging EcoRI and SalI ends were inserted into vector pLacZi (Clontech) between the EcoRI and SalI sites upstream of the GAL4-dependent lacZ reporter gene, resulting in constructs pRJ8730 (ICEhmu), pRJ8732 (ICEhmumut), pRJ8733 (ICE6680), pRJ8734 (ICE7895), and pRJ8765 (Fur box). Correct orientation and sequence of the ICEs or Fur box were confirmed by DNA sequencing. ICE- or Fur box-containing plasmids were linearized by digestion with NcoI and integrated into the genome of yeast strain YM4271 upon transformation according to the lithium acetate method (17). The five resulting yeast strains are listed in Table 1. For the construction of translational fusions between the activation domain of yeast GAL4 and B. japonicum prey proteins, B. japonicum fur (bll0797) and irr (bll0768) were PCR amplified with primer pairs GR-14/GR-15 and GR-21/GR-22, respectively. Genes were cloned in frame with the GAL4 activation domain-encoding region on the EcoRI- or SmaI-linearized cloning vector pGAD424 (Matchmaker one-hybrid system; Clontech), yielding plasmids pRJ8735 and pRJ8737 that were confirmed by sequencing. These plasmids were transferred to all five reporter yeast strains (Table 1) by lithium acetate transformation (17).
β-Galactosidase assays.
Assays for β-galactosidase activity in B. japonicum were carried out as described previously (12). β-Galactosidase activity assays with Saccharomyces cerevisiae were performed as described in the user manual for the Matchmaker one-hybrid system (Clontech).
RESULTS
Maximal hmuR expression depends on full-length ICEhmu.
The ICE is located in the promoter regions between the divergently oriented hmuT and hmuR genes and is necessary for low-iron-inducible expression of these genes. In order to determine the length of the ICE needed for control of hmuR, it was functionally characterized by 5′ deletion analysis. A set of transcriptional hmuR-lacZ fusions with variable 5′ ends relative to hmuR was generated (see Materials and Methods and Fig. 1). Expression from the plasmid-borne hmuR-lacZ fusions was monitored as β-galactosidase activity measured in B. japonicum wild-type (110spc4) and fur mutant (GEM4) cells grown under high- and low-iron conditions (Table 3). The latter strain was included to investigate a potential role of the Fur protein in iron regulation of hmuR.
TABLE 3.
ICE-dependent Fe regulation of plasmid-borne hmuR-lacZ fusions in a B. japonicum wild-type and fur background
| Plasmid | ICEhmu genotype |
hmuR-lacZ expressiona
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 110spc4 (wt)
|
GEM4 (fur mutant)
|
Ratio (fur mutant:wt)
|
|||||||
| Miller units
|
Ratio (low:high Fe) | Miller units
|
Ratio (low:high Fe) | High Fe | Low Fe | ||||
| High Fe | Low Fe | High Fe | Low Fe | ||||||
| pRJ8722 | Wild type | 51 ± 5 | 521 ± 67 | 10.2 | 26 ± 11 | 1,045 ± 93 | 40.2 | 0.5 | 2.0 |
| pRJ8723 | Deleted 5′ half-site | 27 ± 1 | 89 ± 10 | 3.3 | 15 ± 8 | 182 ± 7 | 12.1 | 0.6 | 2.0 |
| pRJ8724 | Complete deletion | 23 ± 2 | 69 ± 9 | 3.0 | 11 ± 2 | 83 ± 9 | 7.5 | 0.5 | 1.2 |
Two aerobic cultures per strain and condition were inoculated to an optical density (at 600 nm) of 0.02 and grown for 3 days at 30°C in PSY medium without supplemented FeSO4 (low Fe) or supplemented with 50 μM FeSO4 (high Fe) and assayed in duplicate. Mean values ± standard errors of β-galactosidase activities (Miller units) are shown for one out of three independent experiments, which all gave similar results. wt, wild type.
Maximal expression was observed under iron-depleted conditions in both B. japonicum backgrounds harboring pRJ8722 with full-length ICEhmu. Deletion of the 5′ ICE half-site (pRJ8723) resulted in a sixfold decrease of expression in both backgrounds. Complete elimination of the ICE (pRJ8724) further decreased hmuR-lacZ expression, maximally twofold. Thus, maximal hmuR activation under iron-depleted conditions depended on full-length ICEhmu. Under iron-replete conditions, expression of hmuR was repressed in both backgrounds, with slightly higher residual expression levels when full-length ICE was present (pRJ8722).
Because the regulatory pattern of hmuR expression was largely the same in wild-type and fur mutant cells, the Fur protein is unlikely to be directly involved. Yet, Fur may contribute indirectly (via control of irr; see Discussion), because gene activation factors in the fur background were about fourfold higher than in the wild type when assayed with pRJ8722 and pRJ8723.
The fact that significant iron regulation was retained in both backgrounds, even when ICEhmu was completely deleted, may point to the existence of an (unknown) additional regulatory mechanism which is independent of the ICE and Fur.
B. japonicum possesses several ICE-like sequences.
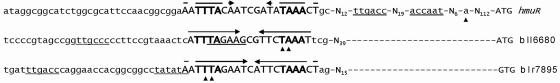
A systematic search was performed to look for the possible presence of additional ICEs in the B. japonicum strain 110 genome. Indeed, using the TTTA-N9-TAAA motif as a probe with the “FUZZNUC” program, six additional ICE-like motifs were identified, four of them located in presumed intergenic regions upstream of blr0586 (ICE0586), blr4157 (ICE4157), bll6680 (ICE6680), and blr7895 (ICE7895) (Fig. 2). The open reading frames blr0586 and blr4157 code for conserved hypothetical proteins without significant similarities to functionally characterized proteins. They were not further analyzed in this study. By contrast, ICE6680 and ICE7895 are associated with genes that code for proteins likely to contain iron. The predicted product of bll6680 (162 amino acids) shares a 50% identity (68% similarity) with bacterioferritin from E. coli. The N-terminal half of the predicted product from blr7895 (∼170 of 323 amino acids) exhibits a 24% identity (36% similarity) to rubrerythrin of Moorella thermoacetica (191 amino acids). A detailed regulatory analysis of bll6680 and blr7895 is presented below.
FIG. 2.
DNA sequence comparison and location of ICE-like motifs within the promoter regions of three iron-regulated genes investigated in this work (ICEhmu, ICE6680, and ICE7895). The alignment shows the three ICEs, all in capital letters, with highly conserved nucleotides printed in bold. Putative promoter core elements (−35/−10 regions) are underlined. Inverse complementary ICE sequences are marked by convergent arrows. Experimentally determined transcription start sites are specified by solid triangles. Each sequence line ends on the right with the start codon of the ICE-associated open reading frame as annotated in RhizoBase.
ICEhmu and the newly identified putative ICE motifs ICE6680 and ICE7895 were used to create the initial profile for an advanced search of ICE motifs using the Darwin software (see Materials and Methods). In total, 172 putative ICE motifs were found (see Table S1 in the supplemental material). ICEhmu, ICE6680, and ICE7895 showed up as the top scoring hits, which reflects their pronounced similarity and the high specificity of the matrix used for the search.
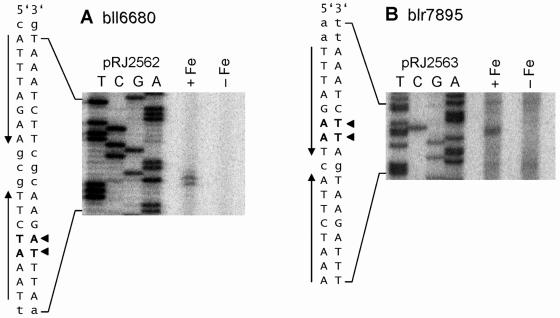
Transcription start sites of bll6680 and blr7895.
To identify the positions of the ICE-like sequences associated with bll6680 and blr7895 relative to the respective promoters, the 5′ ends of the transcripts were mapped by primer extension (Fig. 3A and B). Transcripts of both genes were detected with RNA isolated from cells grown in the presence of iron but not in iron-depleted cells. Moreover, the 5′ ends of both transcripts corresponded to nucleotides located within the predicted ICE motifs (TA7,350,393 [bll6680] and AA8,656,866 [blr7895] in the B. japonicum genome sequence). Possible −35/−10-type promoters could be identified upstream of both transcription start sites, which are underlined in Fig. 2. The positioning of the transcription start sites within the ICE suggests that the ICE acts negatively on transcription of bll6680 and blr7895, in clear contrast to hmuR (and hmuT) where the promoter-upstream ICE (Fig. 1 and 2) is involved in the positive control of gene expression. This difference is accentuated by the fact that low iron concentrations induce hmuR (Table 3) but repress bll6680 and blr7895 (Fig. 3).
FIG. 3.
Transcription start site mapping of genes bll6680 and blr7895. RNA used for primer extension reactions was isolated from exponentially growing B. japonicum cells cultivated in PSY medium that either contained 50 μM FeSO4 supplement (+Fe) or lacked iron supplement (−Fe). Shown are the extension products obtained with primers 6680-7 (A) and 7895-11 (B). The same primers were used in combination with plasmid pRJ2562 (A) and pRJ2563 (B) to generate the sequence ladders included in both panels. The relevant sequence is shown at the left of each panel, with the transcript starts detected in iron-replete cells marked by solid arrowheads. ICE half-sites associated with bll6680 and blr7895 are emphasized with capital letters and vertical arrows (Fig. 2 shows a more detailed representation of the bll6680 and blr7895 promoter regions). The same transcription start sites were detected with at least one disparate primer for each gene (not shown).
Irr, not Fur, interacts with the ICE in a yeast one-hybrid system.
Given their known role in iron regulation, Irr and Fur were candidate regulators for ICE-dependent iron control. Based on the genetic evidence presented above (Table 3), Fur is unlikely to interact with the ICE. Unfortunately, we were unable to apply the same strategy for testing the potential involvement of Irr, because repeated attempts to construct a null mutation in the respective gene (bll0768) of our B. japonicum wild-type strain 110spc4 were not successful. Therefore, a yeast one-hybrid system, employing Irr- and Fur-expressing yeast strains, was used to examine whether any of these regulatory proteins might be able to interact with the ICE directly. Yeast reporter strains were constructed as described in Material and Methods. Reproducible one-hybrid interaction was observed between Irr and all three of the ICEs associated with hmuR, bll6680, and blr7895 (Table 4). To validate that interaction between Irr and the three ICEs was specific, a negative control strain carrying a mutated ICEhmu with a 2-bp insertion between the ICE half-sites was used (ICEhmumut). No interaction between Irr and the mutant ICE was detected (Table 4). Similarly, no interaction was observed when Gal4′-′Fur (pRJ8735) or Gal4′ alone (pGAD424) was expressed in the ICE-containing reporter strains. When pRJ8735 was present in the yeast strain 8765 which contains an E. coli Fur box upstream of the lacZ reporter gene, β-galactosidase activity was detected, indicating that the Gal4′-′Fur hybrid protein was functional.
TABLE 4.
Expression of ICE-lacZ and Fur box-lacZ reporter fusions in S. cerevisiae strains synthesizing Gal4′-′Irr or Gal4′-′Fur hybrid prey proteinsa
| Yeast strain | Target | β-Galactosidase activity (Miller units)
|
|
|---|---|---|---|
| Gal4′-′Irr | Gal4′-′Fur | ||
| 8730 | ICEhmu | 36.1 ± 9.7 | 0.0 |
| 8732 | ICEhmumut | 0.0 | ND |
| 8733 | ICE6680 | 10.1 ± 1.3 | 0.0 |
| 8734 | ICE7895 | 93.5 ± 8.4 | 0.0 |
| 8765 | Fur box | ND | 5.3 ± 0.3 |
Cells of yeast reporter strains were transfected with prey plasmid pRJ8737 (Gal4′-′Irr) or pRJ8735 (Gal4′-′Fur). Cultures grew in synthetic dropout medium (−URA, −LEU) for 3 to 5 h until they reached mid-exponential phase (optical density at 600 nm of 0.5 to 0.8), and one-hybrid interactions were examined as β-galactosidase activity. Activity was measured from two cultures per strain, each assayed in duplicate. Control yeast strains 8730, 8733, 8734 and 8765 harboring the vector pGAD424 (encoding Gal4′ only) showed no β-galactosidase activity. Data are shown for one out of four independent experiments, all yielding similar results. ND, not done.
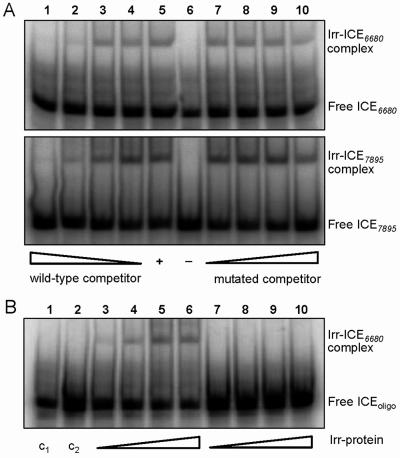
Irr protein binds to ICE6680 and ICE7895 in an electrophoretic mobility shift assay.
Strep-tagged Irr protein was overproduced, purified (see Materials and Methods), and tested for possible binding to the ICE in gel retardation experiments. While Irr binding to ICEhmu was very weak and not reproducibly detected (data not shown), we observed clear Irr-dependent retardation in the cases of ICE6680 and ICE7895 (Fig. 4A). To test whether Irr binds specifically to these two ICE motifs, competitive gel retardation experiments were done. Nonlabeled, wild-type ICE6680 (31 bp) and ICE7895 (33 bp) were used as competing DNAs, whereas two variants, termed ICE6680* (33 bp) and ICE7895* (35 bp), were assumed to be noncompetitive. ICE6680* and ICE7895* are derivatives of ICE6680 and ICE7895, respectively, which contain two additional nucleotides (GC) inserted between the ICE half-sites. The same mutation inserted in ICEhmu was shown previously to abolish activation of hmuR and hmuT under iron-depleted conditions. Indeed, the mutated oligonucleotides ICE6680* and ICE7895* showed only little competition even when added at a 300-fold molar excess, whereas competition with the wild-type oligonucleotides ICE6680 and ICE7895 became detectable already at a 3-fold molar excess (compare lanes 7 to 10 and 1 to 4 in Fig. 4A). This suggested a specific binding of Irr to ICE6680 and ICE7895.
FIG. 4.
Analysis of Irr binding to ICE targets by electrophoretic mobility shift assays. (A) Constant amounts of purified, recombinant Irr protein (1.5 μM) and double-stranded 32P-labeled oligonucleotides ICE6680 (upper) and ICE7895 (lower), each at approximately 2 nM, were incubated with increasing amounts of the corresponding nonlabeled wild-type (ICE6680 and ICE7895) or mutated (ICE6680* and ICE7895*) competitor oligonucleotides. Molar ratios between labeled and nonlabeled oligonucleotides were 1:1 (lanes 4 and 7), 1:3 (lanes 3 and 8), 1:30 (lanes 2 and 9), and 1:300 (lanes 1 and 10). No competitor oligonucleotide and no Irr protein was added to the control reactions in lanes 5 and 6, respectively. (B) Increasing amounts of purified, recombinant Irr protein were incubated with constant amounts of double-stranded 32P-labeled oligonucleotide ICE6680 (lanes 3 through 6) or 1/2-ICE6680 (lanes 7 through 10), each at approximately 2 nM. Concentrations of Irr protein were 0.26 μM (lanes 3 and 7), 0.52 μM (lanes 4 and 8), 1.0 μM (lanes 5 and 9), and 2.1 μM (lanes 6 and 10). No Irr protein was added to the control reactions C1 and C2 with oligonucleotides ICE6680 (lane 1) and 1/2-ICE6680 (2), respectively. Samples were run on 6% nondenaturing polyacrylamide gels and visualized by exposing the dried gels to a phosphorimager screen. ICEoligo, free digonucleotide ICE6680 or 1/2-ICE6680.
It was obvious to test whether iron or heme has an effect on Irr binding to its target DNA sequences. Addition of iron (FeSO4), heme, or hemin (all at 0.3 to 30 μM) to the in vitro binding reactions had no effect on specific binding of Irr to ICE6680 or ICE7895 (data not shown).
ICE6680 mediates negative control of bll6680, and its function depends on both half-sites.
One of the two newly identified Irr binders, ICE6680, was chosen for further analysis of its role in iron regulation of the associated bll6680 gene. To do so, a variant of ICE6680, termed 1/2-ICE6680, was designed in which eight nucleotides at the 3′ end of ICE6680 were simultaneously exchanged (Table 2). Based on our transcript mapping results (Fig. 2 and 3), we anticipated that this mutation would not interfere with the function of the −10 box of the bll6680 promoter.
Plasmids pRJ8769 and pRJ8770, harboring a bll6680′-′lacZ translational fusion with full-length ICE6680 or 1/2-ICE6680, respectively, were transferred to the B. japonicum 110spc4 wild-type strain, and the resulting reporter strains were grown at low-iron (no iron supplement) or high-iron (50 μM FeSO4) conditions. Expression of bll6680′-′lacZ with full-length ICE6680 (pRJ8769) was reproducibly about 1.5-fold higher under iron-replete conditions than under iron limitation (Fig. 5). Under the latter conditions, the strain with plasmid pRJ8770 showed more than threefold higher β-galactosidase activity than that containing pRJ8769. As expected, both fusions yielded comparable β-galactosidase activities under iron-replete conditions; however, for reasons that are not obvious, they did not reach the same expression level as did the strain with plasmid pRJ8770 grown under iron limitation. Iron regulation of bll6680 was further substantiated by measuring expression from a chromosomally integrated bll6680′-′lacZ reporter fusion. β-Galactosidase activity was consistently 1.7-fold higher in cells grown under iron-replete conditions than in iron-depleted cells, and the same result was obtained with the fusion integrated in the chromosome of the fur mutant strain GEM4 (data not shown). Taken together, these results indicated that bll6680 is under negative control and that intact ICE6680, but not Fur, is crucial for repression of the promoter under iron-limiting conditions.
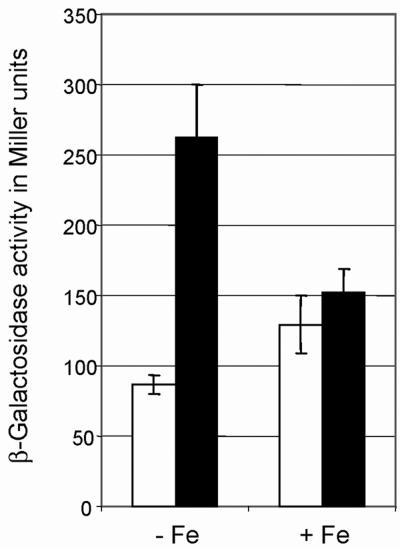
FIG. 5.
β-Galactosidase activity derived from expression of plasmid-borne bll6680′-′lacZ fusions with wild-type ICE6680 (pRJ8769, open columns) or mutant 1/2-ICE6680 (pRJ8770, filled columns) in B. japonicum 110spc4 cells. Cultures were inoculated to an optical density (at 600 nm) of 0.02 and grown aerobically for about 36 h at 30°C in PSY medium lacking (−Fe) or supplemented (+Fe) with iron (50 μM FeSO4) and assayed in duplicate. Shown are mean values ± standard errors of β-galactosidase activities (Miller units) derived from two independent experiments, each comprising two cultures per strain and condition.
Finally, Irr binding to 1/2-ICE6680 was also tested in vitro in the mobility shift assay. In contrast to ICE6680, no interaction was observed between 1/2-ICE6680 and the Irr protein (0.26 to 2.1 μM) (Fig. 4B). Thus, both half-sites of ICE6680 are critical for productive Irr binding.
Identification of iron-regulated, ICE-associated genes by microarray analysis.
Microarray analysis was used to identify iron-regulated B. japonicum genes on a genome-wide scale. All genes whose transcription changed reproducibly at least twofold (P ≤ 0.01) in the presence of iron (50 μM FeSO4) than in iron-depleted conditions are listed in Table S2 in the supplemental material. For each condition, four biological replicas were analyzed. We identified 17 genes to be induced and 68 genes to be repressed under iron-replete conditions. Remarkably, almost 40% of the genes are predicted to code for iron-containing proteins or for proteins involved in iron or energy metabolism.
The overlap of iron-regulated genes and genes associated with putative ICE motifs consisted of 6 genes whose expression was increased and 11 genes whose expression was decreased in the presence of iron (Table 5). Among the genes whose expression was increased were those encoding fumarate hydratase and aconitase, which both contain an iron-sulfur cluster. While the change value of blr7895 (2.7-fold) was above the cutoff, bll6680 was excluded from the selection because its change value was too low (see Discussion). Members of the hmuVUT-hmuR-exbBD-tonB gene cluster were found in the group of genes whose expression was decreased under high-iron conditions. The hmuR and hmuT genes which share the ICEhmu motif showed very high change values of 51-fold and 18-fold, respectively.
TABLE 5.
Fe-regulated B. japonicum genes or open reading frames associated with an ICE-like motifa
| Gene no. | Gene name | Description | Fold change | Putative ICE-like motifb | Fe-regulated genes possibly organized in an operon with the gene listed in column 1c |
|---|---|---|---|---|---|
| Increased expression under iron repletion | |||||
| bll5796 | Fumarate hydratase | 4.5 | TTTGAAAATCATGATAAA−260 | ||
| bll0466 | acnA | Aconitase | 3.6 | GGTTAGAAAGCTTCTATA−117 | |
| blr2219 | Dehydrogenase | 3.5 | CTTTTGACGAATTCCAAA−152 | blr2217d | |
| blr6742 | Putative glutamate synthase, small subunit | 3.4 | CTTCGGAAGAGTTCTAAT−107 | blr6743 | |
| blr0488 | leuC | 3-Isopropylmalate dehydratase, large subunit | 3.3 | TCTTGGAATGTTTCCAGA−196 | |
| blr7895 | Rubrerythrin-like protein | 2.7 | ATTTAGAATCATTCTAAA−20 | ||
| Decreased expression under iron repletion | |||||
| bll7076 | hmuR | Hemin receptor precursor | −50.8 | ATTTACAATCGATATAAA−149 | tonB, exbD, exbB, bll7074, bll7075 |
| blr3555 | fcuA | Probable ferrichrome receptor precursor | −49.8 | CTTTGGAAGTTTTCCAAA−232 | bsr3556 |
| bll7968 | Probable TonB-dependent receptor | −35.6 | GCTTAGAACCATTCAACA−98 | bll7967, bll7966e | |
| blr4504 | TonB-dependent receptor | −27.0 | GTCTAGAAGGATTCCAAA−151 | blr4505 | |
| blr7077 | hmuT | Hemin ABC transporter, hemin-binding protein | −18.1 | GTTTATATCGATTGTAAA−284 | hmuU, hmuV |
| blr3904 | Probable iron transport protein | −16.1 | TTTTAGAACGACTCCAAT−166 | blr3905 blr3906 blr3907 blr3908 | |
| blr5540 | Hypothetical protein | −12.7 | GATCACAATCGGTCTAAA−61 | blr5541 | |
| bll0597 | Similar to nickel-dependent hydrogenase, cytochrome B subunit | −6.5 | CGTTAGAATATCTCTAAA−460 | ||
| blr0697 | Hypothetical protein | −3.7 | TTTCAGAAGGCTTGTGAA−38 | ||
| blr7418 | Hypothetical glutathione S-transferase like protein | −2.9 | TTTTCGAGTTTTTCTATT−100 | ||
| bll2216 | Transcriptional regulatory protein TetR family | −2.5 | ATTTGGAATTCGTCAAAA−55 |
Fe-regulated genes were identified by microarray analysis using the BJAPETHa520090 gene chip and cDNA derived from B. japonicum cells grown under Fe-depleted (no Fe supplement) or Fe-replete (50 μM FeSO4) conditions. Change values (n-fold) are based on four biological replicas for each growth condition. A comprehensive list of all Fe-regulated genes is shown in Table S2 in the supplemental material.
ICE-like motifs were identified as described in the text. A complete list of all ICE-like motifs found in this search is presented in Table S1 in the supplemental material. Numbers refer to the distance of the 3′ nucleotide from the translational start codon of the ICE-associated gene.
Genes considered to be part of an operon are oriented in the same direction and separated by less than 32 nucleotides or by less than 100 nucleotides in case they are functionally related.
The ICE motif is associated with blr2217, which is probably organized in an operon with blr2219, yet no significant iron regulation was found for blr2217.
Expression of bll7967 and bll7966 was iron regulated; however, change (-fold) and P values were below the standard cutoff values.
DISCUSSION
The work reported here has expanded our knowledge of the B. japonicum regulatory circuitry that responds to external iron. The new and important facets are concerned with the functional analysis of the cis-acting ICE motif, the identification of the Irr protein as the putative trans-acting factor binding to ICEs, and the identification of additional iron-regulated target genes, many of them associated with an ICE-like motif.
In a previous study, the ICE motif was shown to be involved in transcriptional activation of B. japonicum heme uptake genes when iron was limited (43) (see the introduction). It was suggested that the ICE represents the binding site for a transcription activator protein, yet the element was only poorly characterized and the identity of the postulated binding protein remained unknown. 5′-end deletion analysis indicated that both half-sites of ICEhmu are required and sufficient for maximal iron regulation of hmuR. As shown for bll6680 and blr7895, ICE motifs can be involved also in repression under low-iron conditions, and at least in the case of ICE6680, both half-sites are critical for this type of negative control.
The B. japonicum ferric uptake regulator Fur (encoded by bll0797) is not directly involved in ICE-dependent regulation of hmuR and bll6680, as concluded from the retained regulation in the fur mutant strain GEM4. Likewise, lack of Fur did not eliminate repression of bll6680 under low-iron conditions (data not shown). Somewhat unexpectedly, iron regulation of hmuR was not completely abolished in both backgrounds when ICEhmu was partially (pRJ8723) or completely (pRJ8724) deleted. This may point to the existence of an additional ICE- and Fur-independent iron control mechanism.
The potential involvement of the B. japonicum Irr protein in ICE-dependent iron control could not be tested directly by genetic means, as we failed to delete the respective gene (bll0768) in our B. japonicum wild-type strain 110spc4. While the reason for this remains unknown, it contrasts with what was previously described for a different B. japonicum strain (LO) from which a mutant derivative carrying a Tn5 insertion in the 5′ portion of the irr gene had been isolated (23). Therefore, Irr binding to ICEhmu, ICE6680, and ICE7895 was probed in a yeast one-hybrid system and with electrophoretic mobility shift assays. While Irr bound to all three targets in vivo, specific in vitro binding of Irr was observed only for ICE6680 and ICE7895. This may reflect different binding affinities between negatively and positively regulated Irr target promoters. No indication for an interaction between Fur and the ICE was found in the yeast reporter system, which is in accordance with the lacZ expression studies in the B. japonicum fur mutant. Collectively, the results from the ICE deletion analysis and the interaction studies with Irr provide compelling evidence that the Irr protein is the regulator that mediates ICE-dependent, positive or negative iron control. This conclusion is well compatible with the elevated activation of hmuR in the fur mutant background (Table 3), as the irr gene is under negative control by Fur (14, 25).
The Irr protein of B. japonicum was originally characterized as a repressor of the heme biosynthesis gene hemB in iron-limited cells (23). An increase of iron supply leads to hemB derepression. In addition to transcriptional control by Fur, the level of Irr protein is controlled at the posttranslational level via iron-dependent degradation mediated by heme (49). Specifically, it was shown that in the presence of iron, Irr interacts directly with the heme biosynthesis enzyme ferrochelatase, which leads to inactivation and subsequent heme-dependent degradation of Irr (50, 65). Since heme acts as an effector molecule in Irr degradation, B. japonicum possesses means to coordinate iron availability with heme biosynthesis. In light of the dual control (transcriptional and posttranslational) which is imposed on Irr, there is no need for additional regulation of its DNA binding properties by iron and/or heme. Indeed, heme or iron had no effect on in vitro binding of Irr to ICE6680 and ICE7895.
Given that B. japonicum is able to utilize external heme as an iron source, it makes sense that cells induce heme uptake via Irr when Fe becomes limiting. As opposed to repression of hemB, bll6680, and blr7895, the hmu heme uptake genes appear to be activated by Irr under low-iron conditions. The same type of Irr-dependent regulation was observed previously for uptake of ferric iron, yet the respective genes had not been characterized in that study (23). Based on our microarray results, one may envisage the fcuA ferrichrome receptor gene (blr3555) and/or the putative iron uptake system encoded by the blr3904 to -3908 gene cluster as candidate genes for this phenotypic property, because they are repressed by iron and associated with ICE-like motifs (Table 5). Target-dependent positive or negative control by iron regulatory proteins is not unprecedented, as it had been described previously for the N. meningitidis Fur and the Mycobacterium tuberculosis IdeR regulators (9, 18). Notably, the latter is an essential protein in M. tuberculosis (52), which may also hold true for Irr in B. japonicum.
The identification of new ICEs and ICE-associated genes is another advancement reported here. Given the lack of any ICE consensus sequences at the onset of this work, we looked for B. japonicum genome sequences that shared closest similarity with the ICE near hmuR. Two of the four initially identified ICE-like sequences were of immediate interest because they are associated with genes that encode putative proteins involved in iron metabolism (bll6680 for bacterioferritin and blr7895 for rubrerythrin). Having determined the transcription start sites of bll6680 and blr7895, it became evident that ICEs and promoters overlapped, a situation that is reminiscent of negatively regulated genes (Fig. 2). A repression mechanism at these genes is indeed likely, because we could demonstrate in vivo and in vitro binding of Irr to ICE6680 and ICE7895 (Table 4; Fig. 4).
The predicted functions of bacterioferritin and rubrerythrin are in accordance with the need to negatively regulate the corresponding genes in response to iron limitation. Bacterioferritin is a well-known iron-mineralizing and storage protein (7, 8) that should be synthesized only when the iron supply tends to exceed the cellular demand for iron incorporation into other proteins. Rubrerythrin, which ligates three iron atoms at two different sites, is also a structurally and biophysically well-characterized protein (33, 35); its function in cells, however, is much less clear. The current hypothesis is that rubrerythrin provides oxidative-stress protection via catalytic reduction of intracellular hydrogen peroxide (37, 56, 61), although this function was recently disputed (30). It is likely that such a function becomes more relevant at increasing iron concentrations with the concomitant risk that Fe2+ or Fe3+ reacts with H2O2 to form hydrogen peroxide radicals (Fenton reaction) (58). Conversely, it would make physiological sense if cells down-regulated rubrerythrin gene expression under iron limitation, which in fact holds true for blr7895 (Fig. 3; Table S2 in the supplemental material).
A more global search for ICE-like motifs in the B. japonicum genome yielded a large number (172) of putative ICE motifs whose significance is difficult to evaluate. Yet, about 15% of them are linked to genes which either encode iron-containing proteins or whose function is related to iron metabolism. To identify those ICE-associated genes which are iron regulated, microarray analysis was performed using a recently developed B. japonicum whole-genome Affymetrix gene chip (F. Hauser et al., unpublished material). Strikingly, 41% (7 of 17) of the genes showing increased expression under high-iron conditions and 40% (27 of 68) of the genes repressed under these conditions are either directly associated with a predicted ICE-like motif or may belong to a putative operon that is preceded by such a motif. This suggests that ICE-dependent regulation through the Irr protein is a widely used mechanism for cellular iron homeostasis in B. japonicum and possibly also in other α-proteobacteria encoding an Irr homolog (62). It also may indicate that direct repression by the Fur protein is less common in B. japonicum than, e.g., in E. coli or Bacillus subtilis (4, 39). The sole B. japonicum target gene for which this regulatory function of Fur has been well documented is the irr gene (13), and only indirect evidence suggests Fur-dependent repression also at the hemA (25) and the fegA genes (34). This situation is reminiscent to that in R. leguminosarum and Sinorhizobium meliloti, where RirA, and not Fur, is the general iron regulator (59, 62).
Unexpectedly, the microarray analysis failed to detect iron regulation of bll6680, although this was clearly observed in the primer extension experiment (Fig. 3A) and, albeit weakly, also with the bll6680′-′lacZ fusion (Fig. 5). It turns out that bll6680 indeed showed increased expression under high-iron conditions in all four replicates of the microarray experiments, yet the average change value was below our cutoff (≥2-fold change). This means that the number of genes listed in Table S2 is probably an underestimation which does not include all iron-regulated genes.
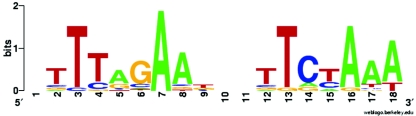
The ICE consensus sequence (Fig. 6) which was defined on the basis of all putative ICE motifs associated with iron-regulated genes (Table 5) reveals that critical nucleotides of the ICE comprise not only the TTTA-N9-TAAA motif which was used in the initial search but rather a more extended palindrome (TTTAGAA-N3-TTCTAAA) with individual positions showing different degrees of invariance. It is interesting to note that ICE6680 and ICE7895, which both bound to Irr in vitro, perfectly match the consensus sequence, whereas ICEhmu, which showed no consistent in vitro binding of Irr (data not shown), deviates from the consensus sequence at three positions (TTTACAA-N3-ATATAAA; italicized nucleotides mark deviations).
FIG. 6.
ICE sequence logo created by using “WebLogo” version 2.8. The consensus motif is based on all individual ICE motifs listed in Table 4, plus that of bll6680 (ICE6680).
It is somewhat puzzling that we found no convincing ICE-like DNA sequence in the promoter region of the irr-controlled hemB gene. To our knowledge, Irr binding to an operator near hemB has not been shown directly, which leaves open the formal possibility that an Irr-dependent unknown regulator controls hemB. As the nucleotides of the ICE that make contact with Irr have not been ascertained precisely, the ICE consensus sequence proposed in Fig. 6 must be regarded as tentative.
Supplementary Material
Acknowledgments
We thank Mark O'Brian (State University of New York at Buffalo) for providing the B. japonicum strain GEM4. The technical assistance of Olivera Volareviç and Sarah Wilhelm is gratefully acknowledged. We highly appreciate the advice of Claude Jakob on experiments with yeast and the help of Achim Stocker with the ion-exchange chromatography. We also thank Andrea Patrignani and Ralph Schlapbach (Functional Genomics Center Zürich [FGCZ]) for advice and assistance in the microarray experiments.
This work was supported by a grant from the Swiss National Foundation for Scientific Research.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alvarez-Morales, A., M. Betancourt-Alvarez, K. Kaluza, and H. Hennecke. 1986. Activation of the Bradyrhizobium japonicum nifH and nifDK operons is dependent on promoter-upstream DNA sequences. Nucleic Acids Res. 14:4207-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., H. Hennecke, and H. M. Fischer. 1996. Two different mechanisms are involved in the heat-shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol. Microbiol. 19:827-839. [DOI] [PubMed] [Google Scholar]
- 4.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 5.Benos, P. V., M. L. Bulyk, and G. D. Stormo. 2002. Additivity in protein-DNA interactions: how good an approximation is it? Nucleic Acids Res. 30:4442-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, J., M. N. Oza, and J. R. Murphy. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. USA 87:5968-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, V. 1997. Avoidance of iron toxicity through regulation of bacterial iron transport. Biol. Chem. 378:779-786. [PubMed] [Google Scholar]
- 8.Carrondo, M. A. 2003. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 22:1959-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delany, I., R. Rappuoli, and V. Scarlato. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 52:1081-1090. [DOI] [PubMed] [Google Scholar]
- 10.Diaz-Mireles, E., M. Wexler, G. Sawers, D. Bellini, J. D. Todd, and A. W. Johnston. 2004. The Fur-like protein Mur of Rhizobium leguminosarum is a Mn2+-responsive transcriptional regulator. Microbiology 150:1447-1456. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson, A. D., and J. Deisenhofer. 2004. Metal import through microbial membranes. Cell 116:15-24. [DOI] [PubMed] [Google Scholar]
- 12.Fischer, H. M., M. Babst, T. Kaspar, G. Acuña, F. Arigoni, and H. Hennecke. 1993. One member of a groESL-like chaperonin multigene family in Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 12:2901-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman, Y. E., and M. R. O'Brian. 2004. The ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum is an iron-responsive transcriptional repressor in vitro. J. Biol. Chem. 279:32100-32105. [DOI] [PubMed] [Google Scholar]
- 14.Friedman, Y. E., and M. R. O'Brian. 2003. A novel DNA-binding site for the ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum. J. Biol. Chem. 278:38395-38401. [DOI] [PubMed] [Google Scholar]
- 15.Gaballa, A., T. Wang, R. W. Ye, and J. D. Helmann. 2002. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 184:6508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genco, C. A., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 17.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gold, B., G. M. Rodriguez, S. A. Marras, M. Pentecost, and I. Smith. 2001. The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol. Microbiol. 42:851-865. [DOI] [PubMed] [Google Scholar]
- 19.Gonnet, G. H., M. T. Hallett, C. Korostensky, and L. Bernardin. 2000. Darwin v. 2.0: an interpreted computer language for the biosciences. Bioinformatics 16:101-103. [DOI] [PubMed] [Google Scholar]
- 20.Guerinot, M. L. 1991. Iron uptake and metabolism in the rhizobia legume symbioses. Plant Soil 130:199-209. [Google Scholar]
- 21.Guerinot, M. L. 1994. Microbial iron transport. Annu. Rev. Microbiol. 48:743-772. [DOI] [PubMed] [Google Scholar]
- 22.Hahn, M., and H. Hennecke. 1986. Genomstruktur von Bradyrhizobium japonicum: gemeinsames Vorkommen von repetetiven Sequenzen und Genen für die Wurzelknöllchensymbiose. Dissertation ETHZ Nr. 8104. Eidgenössische Technische Hochschule, Zürich, Switzerland.
- 23.Hamza, I., S. Chauhan, R. Hassett, and M. R. O'Brian. 1998. The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem. 273:21669-21674. [DOI] [PubMed] [Google Scholar]
- 24.Hamza, I., R. Hassett, and M. R. O'Brian. 1999. Identification of a functional fur gene in Bradyrhizobium japonicum. J. Bacteriol. 181:5843-5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamza, I., Z. Qi, N. D. King, and M. R. O'Brian. 2000. Fur-independent regulation of iron metabolism by Irr in Bradyrhizobium japonicum. Microbiology 146:669-676. [DOI] [PubMed] [Google Scholar]
- 26.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 27.Hantke, K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182:288-292. [DOI] [PubMed] [Google Scholar]
- 28.Hauser, F., A. Lindemann, S. Vuilleumier, A. Patrignani, R. Schlapbach, H. M. Fischer, and H. Hennecke. 17 November 2005. Design and validation of a partial-genome microarray for transcriptional profiling of the Bradyrhizobium japonicum symbiotic gene region. Mol. Genet. Genomics doi: 10.1007/s00438-005-0059-7. [DOI] [PubMed]
- 29.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jean, D., V. Briolat, and G. Reysset. 2004. Oxidative stress response in Clostridium perfringens. Microbiology 150:1649-1659. [DOI] [PubMed] [Google Scholar]
- 31.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 32.Lawrence, C. E., S. F. Altschul, M. S. Boguski, J. S. Liu, A. F. Neuwald, and J. C. Wootton. 1993. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science 262:208-214. [DOI] [PubMed] [Google Scholar]
- 33.LeGall, J., B. C. Prickril, I. Moura, A. V. Xavier, J. J. Moura, and B. H. Huynh. 1988. Isolation and characterization of rubrerythrin, a non-heme iron protein from Desulfovibrio vulgaris that contains rubredoxin centers and a hemerythrin-like binuclear iron cluster. Biochemistry 27:1636-1642. [DOI] [PubMed] [Google Scholar]
- 34.LeVier, K., and M. L. Guerinot. 1996. The Bradyrhizobium japonicum fegA gene encodes an iron-regulated outer membrane protein with similarity to hydroxamate-type siderophore receptors. J. Bacteriol. 178:7265-7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, M., M. Y. Liu, J. LeGall, L. L. Gui, J. Liao, T. Jiang, J. P. Zhang, D. C. Liang, and W. R. Chang. 2003. Crystal structure studies on rubrerythrin: enzymatic activity in relation to the zinc movement. J. Biol. Inorg. Chem. 8:149-155. [DOI] [PubMed] [Google Scholar]
- 36.Liu, J., T. E. Wilson, J. Milbrandt, and M. Johnston. 1993. Identifying DNA-binding sites and analyzing DNA-binding domains using a yeast selection system. Methods 5:125-137. [Google Scholar]
- 37.Lumppio, H. L., N. V. Shenvi, A. O. Summers, G. Voordouw, and D. M. Kurtz, Jr. 2001. Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J. Bacteriol. 183:101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 40.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Minton, N. P. 1984. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene 31:269-273. [DOI] [PubMed] [Google Scholar]
- 42.Nadler, K. D., A. W. B. Johnston, J. W. Chen, and T. R. John. 1990. A Rhizobium leguminosarum mutant defective in symbiotic iron acquisition. J. Bacteriol. 172:670-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nienaber, A., H. Hennecke, and H. M. Fischer. 2001. Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol. Microbiol. 41:787-800. [DOI] [PubMed] [Google Scholar]
- 44.Nienaber, A., A. Huber, M. Göttfert, H. Hennecke, and H. M. Fischer. 2000. Three new NifA-regulated genes in the Bradyrhizobium japonicum symbiotic gene region discovered by competitive DNA-RNA hybridization. J. Bacteriol. 182:1472-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 46.Noya, F., A. Arias, and E. Fabiano. 1997. Heme compounds as iron sources for nonpathogenic Rhizobium bacteria. J. Bacteriol. 179:3076-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ochsner, U. A., and M. L. Vasil. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. USA 93:4409-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2001. Comparative analysis of FUR regulons in gamma-proteobacteria. Nucleic Acids Res. 29:5195-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qi, Z., I. Hamza, and M. R. O'Brian. 1999. Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc. Natl. Acad. Sci. USA 96:13056-13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qi, Z., and M. R. O'Brian. 2002. Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol. Cell 9:155-162. [DOI] [PubMed] [Google Scholar]
- 51.Regensburger, B., and H. Hennecke. 1983. RNA polymerase from Rhizobium japonicum. Arch. Microbiol. 135:103-109. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Simon, R., U. Priefer, and A. Pühler. 1983. Vector plasmids for in vivo and in vitro manipulation of Gram-negative bacteria, p. 98-106. In A. Pühler (ed.), Molecular genetics of the bacteria-plant interaction. Springer-Verlag, Heidelberg, Germany.
- 55.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 56.Sztukowska, M., M. Bugno, J. Potempa, J. Travis, and D. M. Kurtz, Jr. 2002. Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis. Mol. Microbiol. 44:479-488. [DOI] [PubMed] [Google Scholar]
- 57.Todd, J. D., M. Wexler, G. Sawers, K. H. Yeoman, P. S. Poole, and A. W. B. Johnston. 2002. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology 148:4059-4071. [DOI] [PubMed] [Google Scholar]
- 58.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 59.Viguier, C., P. Ó. Cuív, P. Clarke, and M. O'Connell. 2005. RirA is the iron response regulator of the rhizobactin 1021 biosynthesis and transport genes in Sinorhizobium meliloti 2011. FEMS Microbiol. Lett. 246:235-242. [DOI] [PubMed] [Google Scholar]
- 60.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 61.Weinberg, M. V., F. E. Jenney, Jr., X. Cui, and M. W. Adams. 2004. Rubrerythrin from the hyperthermophilic archaeon Pyrococcus furiosus is a rubredoxin-dependent, iron-containing peroxidase. J. Bacteriol. 186:7888-7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wexler, M., J. D. Todd, O. Kolade, D. Bellini, A. M. Hemmings, G. Sawers, and A. W. B. Johnston. 2003. Fur is not the global regulator of iron uptake genes in Rhizobium leguminosarum. Microbiology 149:1357-1365. [DOI] [PubMed] [Google Scholar]
- 63.Wexler, M., K. H. Yeoman, J. B. Stevens, N. G. de Luca, G. Sawers, and A. W. B. Johnston. 2001. The Rhizobium leguminosarum tonB gene is required for the uptake of siderophore and haem as sources of iron. Mol. Microbiol. 41:801-816. [DOI] [PubMed] [Google Scholar]
- 64.Winteler, H. V., and D. Haas. 1996. The homologous regulators ANR of Pseudomonas aeruginosa and FNR of Escherichia coli have overlapping but distinct specificities for anaerobically inducible promoters. Microbiology 142:685-693. [DOI] [PubMed] [Google Scholar]
- 65.Yang, J., K. Ishimori, and M. R. O'Brian. 2005. Two heme binding sites are involved in the regulated degradation of the bacterial iron response regulator (Irr) protein. J. Biol. Chem. 280:7671-7676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.