Abstract
High-affinity vanadate transport systems have not heretofore been identified in any organism. Anabaena variabilis, which can fix nitrogen by using an alternative V-dependent nitrogenase, transported vanadate well. The concentration of vanadate giving half-maximum V-nitrogenase activity when added to V-starved cells was about 3 × 10−9 M. The genes for an ABC-type vanadate transport system, vupABC, were found in A. variabilis about 5 kb from the major cluster of genes encoding the V-nitrogenase, and like those genes, the vupABC genes were repressed by molybdate; however, unlike the V-nitrogenase genes the vanadate transport genes were expressed in vegetative cells. A vupB mutant failed to grow by using V-nitrogenase unless high levels of vanadate were provided, suggesting that there was also a low-affinity vanadate transport system that functioned in the vupB mutant. The vupABC genes belong to a family of putative metal transport genes that include only one other characterized transport system, the tungstate transport genes of Eubacterium acidaminophilum. Similar genes are not present in the complete genomes of other bacterial strains that have a V-nitrogenase, including Azotobacter vinelandii, Rhodopseudomonas palustris, and Methanosarcina barkeri.
Vanadium, molybdenum, and tungsten cofactors are essential for a variety of enzymes. The three oxyanions are very similar in size and structure, with some ability to substitute for each other in certain enzymes (15, 38, 44). Mo serves as a cofactor for many enzymes involved in metabolism in the nitrogen, sulfur, and carbon cycles (18). For all molybdoenzymes except nitrogenase, which converts N2 to ammonium, Mo is incorporated into the apoenzyme as a Mo cofactor that comprises a mononuclear Mo atom coordinated to the sulfur atoms of a pterin called molybdopterin (34). Tungstoenzymes, including dehydrogenases, hydratases, and oxidoreductases that catalyze the oxidation of aldehydes, contain a tungstopterin cofactor similar to molybdopterin and are more common in hyperthermophilic bacteria and archaea with anaerobic metabolism than are molybdoenzymes (16, 21, 24, 28, 39-41, 53). Eubacterium acidaminophilum has two tungstoenzymes, viologen-dependent formate dehydrogenase and aldehyde dehydrogenase, and tungstate uptake is mediated by a specific ABC transporter encoded by the tupABC genes (31, 32). Similar putative tungstate transport genes have been identified in the genomes of many other bacteria, although tungstoenzymes have not been characterized in most of these strains. Vanadium cofactors have been identified for three types of enzymes, V-dependent haloperoxidases, found in a variety of algae and fungi (25, 26); V-dependent nitrate reductases, found in two strains of bacteria (2-4); and V-nitrogenases, found in a several strains of nitrogen-fixing bacteria (5, 11, 45, 47). In a completely different role, vanadium functions in anaerobic respiration as an electron acceptor in several strains of bacteria (8, 29, 36, 37). However, to date no system for the transport of vanadium has been identified in any organism.
In Anabaena variabilis, vanadium functions in nitrogen fixation. Nitrogen fixation is mediated by a family of highly conserved nitrogenases that have a unique metal cofactor containing homocitrate (12, 22, 34). Most of these enzymes have a molybdenum cofactor; however, a few bacteria have an alternative V-nitrogenase that has vanadium in place of molybdenum in the cofactor (5, 11, 45, 47). Most cyanobacteria have the molybdoenzyme nitrate reductase, and many cyanobacteria fix nitrogen with a Mo-nitrogenase; hence, molybdate is important for their growth. A. variabilis ATCC 29413, unlike other commonly studied laboratory strains of cyanobacteria, has three nitrogenases: two Mo-nitrogenases and one V-nitrogenase. One Mo-nitrogenase functions only in the microaerobic environment of specialized cells called heterocysts in filaments grown in the absence of fixed nitrogen, while the other functions only under strictly anaerobic conditions in vegetative cells and in heterocysts (14, 48, 49, 54). Their expression is not dependent on the availability of molybdate; however, no functional nitrogenase is made unless molybdate is present. The vnf genes, which encode V-nitrogenase, are completely repressed by molybdate, and their expression does not require vanadate (45, 47). Under normal aerobic growth conditions in a medium containing molybdate, only the heterocyst-specific Mo-nitrogenase genes are expressed. If such cells are starved for molybdate, the vnf genes are expressed.
Bacteria that use molybdoenzymes typically have a high-affinity molybdate transport system (17, 43). In A. variabilis, molybdate transport is mediated by a high-affinity ABC transport system including ModA, the periplasmic molybdate-binding protein, and ModBC, a fused protein that combines the membrane-spanning permease component and the cytoplasmic ATPase in one protein (50, 55). Despite the similarity of molybdate and vanadate, the known molybdate transport systems do not transport vanadate (43, 50). We describe here the identification of genes that encode a high-affinity vanadate transport system in A. variabilis.
MATERIALS AND METHODS
Strains and growth conditions.
A. variabilis FD and strains derived from that strain were grown photoautotrophically in liquid cultures in an eightfold dilution of the medium of Allen and Arnon (1), AA/8, in some instances supplemented with 2.5 mM NaNO3 and 2.5 mM KNO3 (AA/8-nitrate) or with 5.0 mM NH4Cl and 10 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES), pH 7.2, at 30°C with illumination at 50 to 80 microeinsteins m−2 s−1. Mo-free medium was prepared from stocks scrubbed free of contaminating Mo with activated charcoal (42). The microelement stock was prepared without added Mo but was not treated with activated charcoal. To remove traces of Mo, glassware was treated with 1% Count-Off (New England Nuclear) and 10 mM EDTA for 24 h and then thoroughly rinsed with deionized water purified through a Millipore water purification system. Cyanobacteria were subcultured in Mo-free AA/8 for at least 15 generations to deplete internal Mo reserves. Growth experiments were performed three times, and a representative graph is provided.
Vanadate transport assay.
Mo- and V-starved cultures of strain FD produced heterocysts but grew poorly, with very low levels of nitrogenase as measured by acetylene reduction. In preliminary experiments, we determined that upon addition of vanadate these starved cultures showed a sharp increase in V-nitrogenase activity (as measured by acetylene reduction to ethylene and ethane) within 30 min and that the increase in enzyme activity continued linearly for at least 2 h. Na3VO4 at concentrations of 5 × 10−10 M to 5 × 10−8 M was added to Mo- and V-starved cells (optical density at 720 nm = 0.3) 60 min before the measurement of acetylene reduction to ethylene and ethane as described previously (45). A plot of V-nitrogenase activity (as measured by acetylene reduction) versus the concentration of vanadate produced a hyperbola that was analyzed by nonlinear regression to determine the approximate Km for transport of vanadate.
Cloning of genes and construction of mutants.
The vupABC region was amplified by PCR from A. variabilis DNA with primers vupBmut4-L (5′-GCAGTCCGCCATCAATCAGTCAA-3′) and vupBmut4-R (5′-ACACCCCGCACAATCCGTTCTAC-3′) and inserted into TA vector pBP238 (Invitrogen) to create plasmid pBP238. The vupABC region was excised from pCR2.1 and cloned into pUC18 with EcoRI to create pBP243. The Nmr cassette from pRL648 (7) was excised with SmaI and inserted into the unique HpaI site of vupB in pBP243 to create pBP249. The 5-kb XhoI fragment of pRL1075 (6), which is required for conjugation of plasmids to A. variabilis, was inserted into the SalI site of pBP249 to create pBP250. Replacement of the wild-type vupB gene in the chromosome of strain FD by the mutant vupB allele in pBP250 was accomplished with conjugative nonreplicative plasmids as described previously (30). The mutant was segregated as described previously (45, 46, 51) and tested by PCR to verify that no wild-type copies of the gene remained.
Transcript analysis.
Northern blot analysis was performed with 20 μg of RNA as described previously (55) with PCR-generated probes for vupA or vupB (primers WabcA-L [5′-AGCCAACGCTCAATCTCCTA-3′], WabcA-R [5′-CTCTTACCCCCATCAGCAAA-3′], WabcB-L [5′-GTTGGCTTGGTGGTGAGTCT-3′], and WabcB-R [5′-CGACACCACAATGCTGTAGG-3′]).
For reverse transcription (RT)-PCR, RNA was extracted from a 50-ml culture (optical density at 720 nm = 0.15) in 400 μl of TriReagent (Sigma) containing 200 mg of acid-washed 150- to 212-μm glass beads. Cells were broken by 2 min of rapid shaking with an amalgamator, followed by a 10-min incubation at 55°C. The extract was removed from the glass beads after centrifugation, and the beads were washed with an additional 300 μl of TriReagent, which was combined with the original 400 μl. The sample was extracted with 300 μl of chloroform for 10 min at room temperature and centrifuged, and the RNA in the aqueous phase was precipitated by addition of 175 μl High Salt Precipitation Solution (0.8 M sodium citrate, 1.2 M NaCl) and 175 μl isopropanol. The RNA pellet was washed once in 0.5 ml ice-cold 70% ethanol and air dried, and the RNA was dissolved in 34 μl of RNase-free water plus 1 μl RNasin (Promega). The RNA was further treated to remove DNA by the Ambion DNA-free procedure.
RT-PCRs were done with 25-μl single-tube reaction mixtures that contained RT-PCR buffer (20 mM Tris-HCl, pH 8.33, 50 mM KCl, 2.5 mM MgCl), 5 pmol of WabcA primers, 250 ng RNA, 50 U SuperScript II (Invitrogen), and 1.0 U Taq polymerase (Invitrogen). The RT-PCR thermocycler program used for the WabcA primers was 45°C for 30 min, 94°C for 2 min, 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; 40 cycles of steps 3 to 5; and then 72°C for 10 min.
Nucleotide sequence accession numbers.
Proteins were aligned with ClustalX (10). The accession numbers are as follows: A. variabilis VupA, ZP_00160423; VupB, ZP_00160424; VupC, ZP_00351494; E. acidaminophilum TupA, CAC40782; TupB, CAC40783; TupC, CAC40784; G. metallireducens TupA, ZP_00534807; TupB, ZP_00534806; TupC, ZP_00534805; S. oneidensis TupA, NP_720235; TupB, NP_720236; TupC, NP_720237.
RESULTS AND DISCUSSION
Kinetics of vanadate transport.
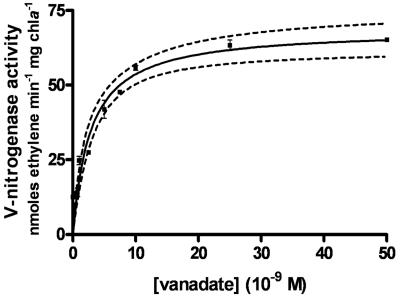
In the absence of Mo, A. variabilis can fix nitrogen under aerobic conditions with V-nitrogenase in media with concentrations of vanadate as low as 10−8 M, suggesting that there is a high-affinity vanadate transport system. Because high-specific-activity [49V]vanadate is not easily available, we could not directly measure transport of vanadate. Therefore, we used an indirect method, i.e., stimulation of V-nitrogenase activity by addition of vanadate to cells previously starved for molybdate and vanadate, to estimate the kinetics of vanadate transport in A. variabilis. Plotting V-nitrogenase activity (as measured by acetylene reduction) after the addition of various concentrations of vanadate versus the concentration of vanadate produced a hyperbola that was analyzed by nonlinear regression, giving an estimate of the concentration of vanadate giving a half-maximum cellular response for transport of 2.8 × 10−9 M (95% confidence interval, 1.8 × 10−9 to 3.8 × 10−9 M) (Fig. 1). In comparison, the molybdate transport system of this strain has a Km of about 3 × 10−10 M (50), perhaps reflecting the greater availability of vanadium compared to molybdenum in some environments where both metals are scarce (35).
FIG. 1.
Vanadate transport in wild-type strain FD. Strains were grown as previously described, in molybdate-free AA/8 medium (1, 50). The kinetics of vanadate transport was measured indirectly by the ability of vanadate to stimulate V-nitrogenase activity in Mo- and V-starved cells. Na3VO4 at various concentrations was added to starved cells 60 min before the measurement of acetylene reduction to ethylene and ethane. Ethane values were about 4% of the ethylene values at all concentrations of vanadate. The graph represents a nonlinear regression analysis with the 95% confidence interval shown by the dashed lines. chla, chlorophyll a.
Vanadate transport genes.
The genome of A. variabilis has been sequenced (http://genome.jgi-psf.org/finished_microbes/anava/anava.home.html); however, because no genes for vanadate transport have been identified in any organism, we could not search for genes homologous to known vanadate transport genes. However, we analyzed the region of the genome near the genes encoding V-nitrogenase for putative transport genes. The region of the genome containing the genes for V-nitrogenase, vnfDG, vnfK, vnfE, and vnfN, is shown in Fig. 2. The vnfH gene (not shown in Fig. 2) is also in this region, 22 kb downstream of vnfN (B. S. Pratte and T. Thiel, submitted for publication). Genes encoding a putative ABC transport system were identified about 5 kb downstream from vnfDGKEN. The gene most similar to a periplasmic metal-binding protein is vupA, the permease component gene is vupB, and the gene encoding the ATP-binding protein is vupC. The vupB and vupC genes overlap by 2 nucleotides, indicating that they are cotranscribed. The vupA and vupB genes have good similarity to the tupAB genes that mediate tungstate transport in E. acidaminophilum (31).
FIG. 2.
Map of the region of the genome containing the vnf genes encoding V-nitrogenase in A. variabilis. Genes designated vnfDG, vnfK, vnfE, and vnfN were identified and characterized previously (45, 47). The vupABC genes are ABC transport genes shown here to function in vanadate transport. Genes not identified had very weak similarity to known genes with no apparent relevance to nitrogen fixation. NifV-type products synthesize homocitrate, a part of nitrogenase metal cofactors.
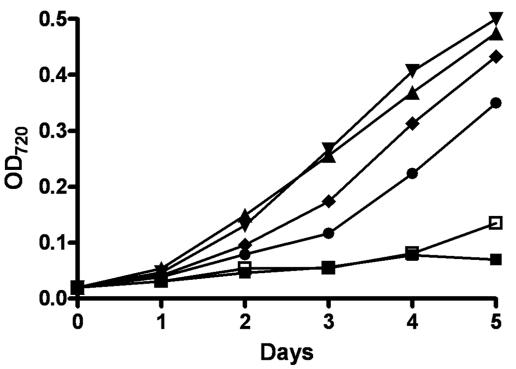
The vupB mutant strain, BP250, was unable to grow in the absence of molybdate using V-nitrogenase with the normal concentration of vanadate (10−6 M). We compared the ability of the parent and the mutant to grow with V-nitrogenase at various concentrations of vanadate. Parent strain FD grew equally well with 10−3 to 10−8 M vanadate in molybdate-free medium (Fig. 3; for legibility, only the highest and lowest concentrations are shown) but did not grow well in the absence of molybdate and vanadate. In contrast, BP250 did not grow at concentrations of vanadate at or below 10−5 M (Fig. 3). Growth with vanadate at 10−6, 10−7, and 10−8 M was also tested, but the mutant did not grow at these concentrations (data not shown). The vupB mutant grew well with 10−3 M vanadate and also grew with 10−4 M vanadate (Fig. 3). We also determined that all cultures that grew with vanadate in the absence of molybdate reduced acetylene to both ethylene and ethane, which is characteristic of V-nitrogenases (data not shown). Thus, the vupB mutant was unable to use V-nitrogenase unless it was provided with a very high concentration of vanadate, consistent with the role of VupABC in vanadate transport. The ability of the vupB mutant strain to transport vanadate at 10−3 to 10−4 M is similar to the ability of molybdate transport mutants of A. variabilis to transport molybdate at those high concentrations of molybdate (55). It is possible that both vanadate and molybdate are transported by the same low-affinity system.
FIG. 3.
Growth of wild-type strain FD and the vupB mutant using V-nitrogenase. The vupB mutant, BP250, and FD (wild type) were first starved in Mo- and V-free AA/8 and then grown in AA/8 lacking molybdate with vanadate at various concentrations: FD with 10−3 M vanadate, ▾; FD with 10−8 M vanadate, ▴; FD with no vanadate, ▪; BP250 with 10−3 M vanadate, ♦; BP250 with 10−4 M vanadate, •; BP250 with 10−5 M vanadate, □. OD720, optical density at 720 nm.
Transport of tungstate.
Because the vupAB genes showed some similarity to tungstate transport genes in E. acidaminophilum (31), it was possible that the VupABC system also transported tungstate. Tungstate is transported by the high-affinity molybdate transport system (50); therefore, we measured the effect of tungstate on the transport of vanadate in strain KA12, a modA mutant impaired in high-affinity molybdate (and tungstate) transport (55). The transport of vanadate at 10−8 M, as measured by its stimulation of V-nitrogenase in Mo- and V-starved cells, was not inhibited by the addition of a 5-fold, 10-fold, or 100-fold excess of tungstate. Acetylene reduction values 1 h after the addition of 10−8 M vanadate to Mo- and V-starved cells, with or without excess tungstate, were in the range of 45 to 55 nmol ethylene mg chlorophyll a−1 min−1. Thus, tungstate is transported by the high-affinity molybdate transport system but not by the high-affinity vanadate transport system.
Transcript analysis.
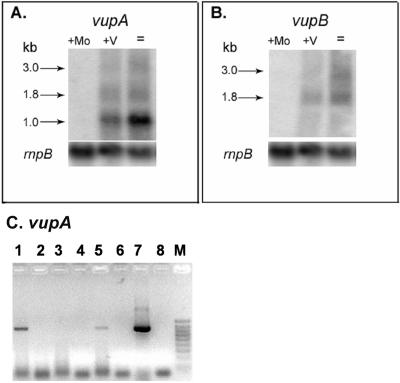
By Northern blot analysis, we determined that the vupA and vupB genes were expressed only in the absence of molybdate, in the presence or absence of vanadate (Fig. 4). This same pattern of expression was also observed for the vnfDGK and vnfEN genes (45, 47). The repression of the vupAB genes by molybdate was consistent with their role in providing vanadate for the alternative V-nitrogenase system. There appeared to be a slight repression of transcription by vanadate in Mo- and V-starved cells; however, vanadate did not repress strongly. The predicted sizes for the vupA, vupAB, and vupABC transcripts were about 1.0, 1.8, and 3.0 kb, respectively. The genes appeared to constitute an operon, although the smallest transcript, encoding only vupA, was much stronger than the two larger transcripts. Similarly, the modA gene in A. variabilis, which encodes the molybdate-binding protein, is much more highly expressed than modBC (55). This may reflect a need by the cell for more metal-binding protein in the periplasm than for the permease component and ATPase components. Although vanadate is required only for V-nitrogenase, which is expressed in heterocysts, the presence of an RT-PCR product for vupA in cells grown with ammonia in the absence of molybdate indicated that the vup genes are expressed in vegetative cells (Fig. 4C). There was not much RT-PCR product for vupA in ammonia-grown cells, which could reflect weak transcription in vegetative cells or might be the result of trace amounts of molybdate in the medium that could not be depleted by the cells by nitrogen fixation, and thus there might be some molybdate inhibition of transcription of vupABC. It appeared, however, that while the vnf and vup genes were both regulated by molybdate, only expression of the vnf genes was completely repressed by ammonia.
FIG. 4.
Transcript analysis of vupAB. (A and B) For Northern blot assays of vupA and vupB transcripts, RNA was isolated from cultures of A. variabilis grown in AA/8 with or without Mo or V and was hybridized with 32P-labeled probes to vupA (A) or vupB (B) as described previously (55). +Mo, with molybdate; +V, Mo-free medium with V; =, Mo- and V-free medium. Arrows indicate sizes of the hybridizing bands. The lower parts of panels A and B show the hybridization signal with a probe for the constitutively expressed rnpB gene (52). (C) RT-PCR performed with RNA isolated from cells grown in Mo-free AA/8 with vanadate (lanes 1 and 2), with molybdate (lanes 3 and 4), or in Mo-free AA/8 with NH4Cl to repress heterocysts (lanes 5 and 6). Lanes 1, 3, and 5 contained complete RT-PCR reagents. Control reactions shown in lanes 2, 4, and 6 lacked only reverse transcriptase. The PCR product made from DNA with the WabcA primers is shown in lane 7. The control reaction mixture shown in lane 8 had no template. The markers shown in lane M were Bioline Hyperladder IV (100 to 1,000 bp).
Origins of vupABC genes.
Consistent with their role in providing vanadate for V-nitrogenase, the vupABC cluster is not present in the genomes of other nitrogen-fixing cyanobacteria, such as Anabaena sp. strain PCC 7120 (20) and Nostoc punctiforme ATCC 29133 (33), which have only a Mo-nitrogenase. These genes have no similarity to other cyanobacterial transporters, suggesting that they were acquired by lateral transfer from other bacteria. The vupABC genes are most similar to a family of putative tungstate transport genes, exemplified by the tupABC genes of E. acidaminophilum, which have been shown to be required for the transport of tungstate in that strain (31). The similarity of vupA/tupA and vupB/tupB is much higher (58% and 61%, respectively) than vupC/tupC (31%). Many other transport genes found in microbial genomes have been tentatively identified as putative tungstate transporters based on their similarity to the tupABC genes of E. acidaminophilum; however, their function is not known.
Geobacter metallireducens and Shewanella oneidensis can grow with a number of metals, including vanadium, as terminal electron acceptors for anaerobic respiration (8, 9, 36, 37). The complete genomes of these two strains have genes similar to tupABC of E. acidaminophilum and the vupABC genes of A. variabilis. S. oneidensis takes up vanadate; however, the concentrations needed for growth using vanadate for anaerobic respiration are 5 to 10 mM (9), much higher than is required for strains that use vanadate to synthesize enzyme cofactors. It appears likely that the tupABC genes and vupABC genes share a common ancestor and that the putative tungstate transport genes found in many bacterial genomes, including possibly the genes in G. metallireducens and S. oneidensis, function for transport of tungsten or vanadium, or possibly both.
In contrast, genes similar to tupABC/vupABC are not present in the complete genomes of three other bacterial strains that have a V-nitrogenase, including Azotobacter vinelandii, Rhodopseudomonas palustris, and Methanosarcina barkeri (11, 13, 19, 23, 27). The complete genome of R. palustris is finished, and that of A. vinelandii is in the draft stage. In the genome annotation of R. palustris (http://genome.jgi-psf.org/finished_microbes/rhopa/rhopa.home.html), genes for an ABC transporter, similar to phosphonate transporters and located near the vnf genes, have been designated as a putative vanadate transport system. Although to date there are no experimental data to support this designation, there are similar phosphonate transporter genes in A. vinelandii. The genomes of A. variabilis, N. punctiforme, and Anabaena sp. strain PCC 7120 also have putative phosphonate transport genes (all2356 to all2358 in Anabaena sp. strain PCC 7120, which are 98% identical to the genes in A. variabilis); however, these genes show very weak similarity to those of R. palustris (about 16% identity, 25% similarity). If the putative vanadate transport genes of R. palustris are shown to function in that capacity, it would suggest that vanadate transport systems have evolved at least twice from different ancestral genes.
Acknowledgments
This work was supported by National Science Foundation grant MCB-0416663 and USDA grant 99-35100-7582.
REFERENCES
- 1.Allen, M. B., and D. I. Arnon. 1955. Studies on nitrogen-fixing blue-green algae. I. Growth and nitrogen fixation by Anabaena cylindrica Lemm. Plant Physiol. 30:366-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antipov, A. N., N. N. Lyalikova, T. V. Khijniak, and N. P. L'Vov. 1998. Molybdenum-free nitrate reductases from vanadate-reducing bacteria. FEBS Lett. 441:257-260. [DOI] [PubMed] [Google Scholar]
- 3.Antipov, A. N., N. N. Lyalikova, T. V. Khiznjak, and N. P. L'Vov. 1999. Some properties of dissimilatory nitrate reductases lacking molybdenum and molybdenum cofactor. Biochemistry (Moscow) 64:483-487. [PubMed] [Google Scholar]
- 4.Antipov, A. N., D. Y. Sorokin, N. P. L'Vov, and J. G. Kuenen. 2003. New enzyme belonging to the family of molybdenum-free nitrate reductases. Biochem. J. 369:185-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop, P. E., and R. Premakumar. 1992. Alternative nitrogen fixation systems, p. 736-762. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman & Hall, Inc., New York, N.Y.
- 6.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 7.Black, T. A., and C. P. Wolk. 1994. Analysis of a Het mutation in Anabaena sp. strain PCC 7120 implicates a secondary metabolite in the regulation of heterocyst spacing. J. Bacteriol. 176:2282-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpentier, W., L. De Smet, J. Van Beeumen, and A. Brige. 2005. Respiration and growth of Shewanella oneidensis MR-1 using vanadate as the sole electron acceptor. J. Bacteriol. 187:3293-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpentier, W., K. Sandra, I. De Smet, A. Brige, L. De Smet, and J. Van Beeumen. 2003. Microbial reduction and precipitation of vanadium by Shewanella oneidensis. Appl. Environ. Microbiol. 69:3636-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien, Y. T., V. Auerbuch, A. D. Brabban, and S. H. Zinder. 2000. Analysis of genes encoding an alternative nitrogenase in the archaeon Methanosarcina barkeri 227. J. Bacteriol. 182:3247-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dos Santos, P. C., D. R. Dean, Y. Hu, and M. W. Ribbe. 2004. Formation and insertion of the nitrogenase iron-molybdenum cofactor. Chem. Rev. 104:1159-1173. [DOI] [PubMed] [Google Scholar]
- 13.Eady, R. R. 1995. Vanadium nitrogenases of Azotobacter. Met. Ions Biol. Syst. 31:363-405. [PubMed] [Google Scholar]
- 14.Elhai, J., and C. P. Wolk. 1990. Developmental regulation and spatial pattern of expression of the structural genes for nitrogenase in the cyanobacterium Anabaena. EMBO J. 9:3379-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garner, C. D., and L. J. Stewart. 2002. Tungsten-substituted molybdenum enzymes. Met. Ions Biol. Syst. 39:699-726. [PubMed] [Google Scholar]
- 16.Graentzdoerffer, A., D. Rauh, A. Pich, and J. R. Andreesen. 2003. Molecular and biochemical characterization of two tungsten- and selenium-containing formate dehydrogenases from Eubacterium acidaminophilum that are associated with components of an iron-only hydrogenase. Arch. Microbiol. 179:116-130. [DOI] [PubMed] [Google Scholar]
- 17.Grunden, A. M., and K. T. Shanmugam. 1997. Molybdate transport and regulation in bacteria. Arch. Microbiol. 168:345-354. [DOI] [PubMed] [Google Scholar]
- 18.Hille, R. 2002. Molybdenum and tungsten in biology. Trends Biochem. Sci. 27:360-367. [DOI] [PubMed] [Google Scholar]
- 19.Jacobitz, S., and P. E. Bishop. 1992. Regulation of nitrogenase-2 in Azotobacter vinelandii by ammonium, molybdenum, and vanadium. J. Bacteriol. 174:3884-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213. [DOI] [PubMed] [Google Scholar]
- 21.Kletzin, A., and M. W. Adams. 1996. Tungsten in biological systems. FEMS Microbiol. Rev. 18:5-63. [DOI] [PubMed] [Google Scholar]
- 22.Kuchar, J., and R. P. Hausinger. 2004. Biosynthesis of metal sites. Chem. Rev. 104:509-525. [DOI] [PubMed] [Google Scholar]
- 23.Larimer, F. W., P. Chain, L. Hauser, J. Lamerdin, S. Malfatti, L. Do, M. L. Land, D. A. Pelletier, J. T. Beatty, A. S. Lang, F. R. Tabita, J. L. Gibson, T. E. Hanson, C. Bobst, J. L. Torres, C. Peres, F. H. Harrison, J. Gibson, and C. S. Harwood. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 22:55-61. [DOI] [PubMed] [Google Scholar]
- 24.Laukel, M., L. Chistoserdova, M. E. Lidstrom, and J. A. Vorholt. 2003. The tungsten-containing formate dehydrogenase from Methylobacterium extorquens AM1: purification and properties. Eur. J. Biochem. 270:325-333. [DOI] [PubMed] [Google Scholar]
- 25.Littlechild, J. 1999. Haloperoxidases and their role in biotransformation reactions. Curr. Opin. Chem. Biol. 3:28-34. [DOI] [PubMed] [Google Scholar]
- 26.Littlechild, J., E. Garcia-Rodriguez, A. Dalby, and M. Isupov. 2002. Structural and functional comparisons between vanadium haloperoxidase and acid phosphatase enzymes. J. Mol. Recognit. 15:291-296. [DOI] [PubMed] [Google Scholar]
- 27.Luque, F., L. A. Mitchenall, M. Chapman, R. Christine, and R. N. Pau. 1993. Characterization of genes involved in molybdenum transport in Azotobacter vinelandii. Mol. Microbiol. 7:447-459. [DOI] [PubMed] [Google Scholar]
- 28.L'Vov, N. P., A. N. Nosikov, and A. N. Antipov. 2002. Tungsten-containing enzymes. Biochemistry (Moscow) 67:196-200. [DOI] [PubMed] [Google Scholar]
- 29.Lyalikova, N. N., and N. A. Yurkova. 1992. Role of microorganisms in vanadium concentration and dispersion. Geomicrobiology 10:15-26. [Google Scholar]
- 30.Lyons, E. M., and T. Thiel. 1995. Characterization of nifB, nifS, and nifU genes in the cyanobacterium Anabaena variabilis: NifB is required for the vanadium-dependent nitrogenase. J. Bacteriol. 177:1570-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makdessi, K., J. R. Andreesen, and A. Pich. 2001. Tungstate uptake by a highly specific ABC transporter in Eubacterium acidaminophilum. J. Biol. Chem. 276:24557-24564. [DOI] [PubMed] [Google Scholar]
- 32.Makdessi, K., K. Fritsche, A. Pich, and J. R. Andreesen. 2004. Identification and characterization of the cytoplasmic tungstate/molybdate-binding protein (Mop) from Eubacterium acidaminophilum. Arch. Microbiol. 181:45-51. [DOI] [PubMed] [Google Scholar]
- 33.Meeks, J. C., J. Elhai, T. Thiel, M. Potts, F. Larimer, J. Lamerdin, P. Predki, and R. Atlas. 2002. An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Photosynth. Res. 70:85-106. [DOI] [PubMed] [Google Scholar]
- 34.Mendel, R. R., and G. Schwarz. 2002. Biosynthesis and molecular biology of the molybdenum cofactor (Moco). Met. Ions Biol. Syst. 39:317-368. [PubMed] [Google Scholar]
- 35.Morris, A. W. 1975. Dissolved molybdenum and vanadium in the northeast Atlantic Ocean. Deep-Sea Res. 22:49-54. [Google Scholar]
- 36.Myers, J. M., W. E. Antholine, and C. R. Myers. 2004. Vanadium(V) reduction by Shewanella oneidensis MR-1 requires menaquinone and cytochromes from the cytoplasmic and outer membranes. Appl. Environ. Microbiol. 70:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortiz-Bernad, I., R. T. Anderson, H. A. Vrionis, and D. R. Lovley. 2004. Vanadium respiration by Geobacter metallireducens: novel strategy for in situ removal of vanadium from groundwater. Appl. Environ. Microbiol. 70:3091-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pau, R. N., and D. M. Lawson. 2002. Transport, homeostasis, regulation, and binding of molybdate and tungstate to proteins. Met. Ions Biol. Syst. 39:31-74. [PubMed] [Google Scholar]
- 39.Rauh, D., A. Graentzdoerffer, K. Granderath, J. R. Andreesen, and A. Pich. 2004. Tungsten-containing aldehyde oxidoreductase of Eubacterium acidaminophilum. Eur. J. Biochem. 271:212-219. [DOI] [PubMed] [Google Scholar]
- 40.Roy, R., and M. W. Adams. 2002. Characterization of a fourth tungsten-containing enzyme from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 184:6952-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy, R., and M. W. Adams. 2002. Tungsten-dependent aldehyde oxidoreductase: a new family of enzymes containing the pterin cofactor. Met. Ions Biol. Syst. 39:673-697. [PubMed] [Google Scholar]
- 42.Schneider, K., A. Muller, U. Schramm, and W. Klipp. 1991. Demonstration of a molybdenum- and vanadium-independent nitrogenase in a nifHDK-deletion mutant of Rhodobacter capsulatus. Eur. J. Biochem. 195:653-661. [DOI] [PubMed] [Google Scholar]
- 43.Self, W. T., A. M. Grunden, A. Hasona, and K. T. Shanmugam. 2001. Molybdate transport. Res. Microbiol. 152:311-321. [DOI] [PubMed] [Google Scholar]
- 44.Siemann, S., K. Schneider, M. Oley, and A. Muller. 2003. Characterization of a tungsten-substituted nitrogenase isolated from Rhodobacter capsulatus. Biochemistry 42:3846-3857. [DOI] [PubMed] [Google Scholar]
- 45.Thiel, T. 1993. Characterization of genes for an alternative nitrogenase in the cyanobacterium Anabaena variabilis. J. Bacteriol. 175:6276-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiel, T. 1994. Genetic analysis of cyanobacteria, p. 581-611. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 47.Thiel, T. 1996. Isolation and characterization of the vnfEN genes of the cyanobacterium Anabaena variabilis. J. Bacteriol. 178:4493-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiel, T., E. M. Lyons, and J. C. Erker. 1997. Characterization of genes for a second Mo-dependent nitrogenase in the cyanobacterium Anabaena variabilis. J. Bacteriol. 179:5222-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thiel, T., E. M. Lyons, J. C. Erker, and A. Ernst. 1995. A second nitrogenase in vegetative cells of a heterocyst-forming cyanobacterium. Proc. Natl. Acad. Sci. USA 92:9358-9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thiel, T., B. Pratte, and M. Zahalak. 2002. Transport of molybdate in the cyanobacterium Anabaena variabilis ATCC 29413. Arch. Microbiol. 179:50-56. [DOI] [PubMed] [Google Scholar]
- 51.Thiel, T., and C. P. Wolk. 1987. Conjugal transfer of plasmids to cyanobacteria. Methods Enzymol. 153:232-243. [DOI] [PubMed] [Google Scholar]
- 52.Vioque, A. 1997. The RNase P RNA from cyanobacteria: short tandemly repeated repetitive (STRR) sequences are present within the RNase P RNA gene in heterocyst-forming cyanobacteria. Nucleic Acids Res. 25:3471-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vorholt, J. A., and R. K. Thauer. 2002. Molybdenum and tungsten enzymes in C1 metabolism. Met. Ions Biol. Syst. 39:571 619. [PubMed] [Google Scholar]
- 54.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), Molecular biology of the cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 55.Zahalak, M., B. Pratte, K. J. Werth, and T. Thiel. 2004. Molybdate transport and its effect on nitrogen utilization in the cyanobacterium Anabaena variabilis ATCC 29413. Mol. Microbiol. 51:539-549. [DOI] [PubMed] [Google Scholar]