Abstract
Methanosphaera stadtmanae has the most restricted energy metabolism of all methanogenic archaea. This human intestinal inhabitant can generate methane only by reduction of methanol with H2 and is dependent on acetate as a carbon source. We report here the genome sequence of M. stadtmanae, which was found to be composed of 1,767,403 bp with an average G+C content of 28% and to harbor only 1,534 protein-encoding sequences (CDS). The genome lacks 37 CDS present in the genomes of all other methanogens. Among these are the CDS for synthesis of molybdopterin and for synthesis of the carbon monoxide dehydrogenase/acetyl-coenzyme A synthase complex, which explains why M. stadtmanae cannot reduce CO2 to methane or oxidize methanol to CO2 and why this archaeon is dependent on acetate for biosynthesis of cell components. Four sets of mtaABC genes coding for methanol:coenzyme M methyltransferases were found in the genome of M. stadtmanae. These genes exhibit homology to mta genes previously identified in Methanosarcina species. The M. stadtmanae genome also contains at least 323 CDS not present in the genomes of all other archaea. Seventy-three of these CDS exhibit high levels of homology to CDS in genomes of bacteria and eukaryotes. These 73 CDS include 12 CDS which are unusually long (>2,400 bp) with conspicuous repetitive sequence elements, 13 CDS which exhibit sequence similarity on the protein level to CDS encoding enzymes involved in the biosynthesis of cell surface antigens in bacteria, and 5 CDS which exhibit sequence similarity to the subunits of bacterial type I and III restriction-modification systems.
There are two types of methanogenic archaea, those belonging to the order Methanosarcinales, which contain cytochromes and which can use methanol, methyl amines, acetate, and/or CO2 plus H2 as methanogenic substrates, and those belonging to the orders Methanobacteriales, Methanomicrobiales, Methanococcales, and Methanopyrales, which are devoid of cytochromes and which can use CO2 plus H2 and/or formate only to fuel anaerobic growth (95, 102). The energy metabolism of both types of methanogens has been investigated in detail (17). However, there are still a few pertinent questions. For example, why is the growth yield on H2 and CO2 of methanogens lacking cytochromes considerably lower (<50%) than that of cytochrome-containing methanogens? The growth yield on H2 and CO2 of Methanobrevibacter arboriphilus is 1.3 g/mol methane, whereas that ofMethanosarcina barkeri is 7.3 g/mol (101). Could the reason for this be that in cytochrome-containing methanogens two steps in the reduction of CO2 to methane, methyl transfer from methyl-tetrahydromethanopterin (methyl-H4MPT) to coenzyme M and reduction of the heterodisulfide coenzyme M-S-S-coenzyme B (CoM-S-S-CoB) with H2, are coupled with energy conservation, whereas in methanogens without cytochromes only one step, the methyltransfer reaction, is coupled? Indeed, methanogens with cytochromes contain a heterodisulfide reductase (HdrDE) that is anchored via a cytochrome b (HdrE) in the cytoplasmic membrane (35) and is electronically linked via methanophenazine to a cytochrome b (VhoC)-containing hydrogenase complex (VhoACG), whose active site faces toward the periplasm (14, 16), whereas in methanogens without cytochromes, the corresponding hydrogenase (MvhADG) is a cytoplasmic enzyme (74, 82) and the HdrABC complex is recovered mainly in the cytoplasmic fraction (35). There is, however, evidence that HdrABC could be anchored via its HdrB subunit in the cytoplasmic membrane (36, 50). Since HdrB lacks recognizable transmembrane helices, this question is still a matter of dispute.
There is only one indication that the Hdr-catalyzed reaction in methanogens without cytochromes is coupled to energy conservation. It comes from the finding that there is one methanogen belonging to the order Methanobacteriales that can grow on methanol and H2. This organism, Methanosphaera stadtmanae (64, 65), thrives in the human intestine, where methanol is a product of pectin degradation by Bacteroides species and other anaerobic bacteria (18, 40). With cell suspensions of M. stadtmanae it has been shown that the reduction of methanol with H2 to methane is coupled to ADP phosphorylation via the electrochemical proton potential (87). However, correct interpretation of this finding will not be possible as long as the enzymes involved are not known.
M. stadtmanae, a member of the Methanobacteriales, is unusual because it is able to grow on methanol and H2 as energy substrates, a property otherwise restricted to members of the Methanosarcinales. It is also remarkable that M. stadtmanae can neither oxidize methanol to CO2 nor reduce CO2 to methane. This organism is also not capable of autotrophic growth on CO2, requiring acetate and CO2 as main carbon sources. Labeling studies have revealed that acetate and CO2 are assimilated via reductive carboxylation of acetyl-CoA to pyruvate, from which most cell compounds are formed (4, 10, 11, 51, 62). Interestingly, Methanobrevibacter smithii, the only other human methanogenic commensal, is also dependent on acetate for biosyntheses. However, this organism grows on H2 and CO2 rather than on methanol and H2 as energy sources (5, 66). These two archaea, both belonging to the order Methanobacteriales, also differ significantly in lipid composition (89).
M. stadtmanae has attracted additional interest because it is the first human archaeal commensal whose genome has been sequenced. Such commensals not only are able to survive in the human gastrointestinal tract, which is protected by a highly active immune system, but also stimulate the development of a healthy intestinal epithelium and immune system (58). In Western countries the increasing number of chronic inflammatory diseases of the intestine (referred to as inflammatory bowel disease) indicates that there is perturbation of the precarious commensal relationship in the human intestine. However, the molecular mechanisms of commensalism are still not fully understood. Studying the genetic information encoded in the small genome of M. stadtmanae may provide clues that will increase our understanding of adaptations of this organism that allow it to live as a commensal (2, 25, 55).
We describe here the complete genome sequence of M. stadtmanae and compare it with the genome sequences of all other methanogenic archaea sequenced so far. The comparison revealed not only why this archaeon is dependent on H2, methanol, acetate, and CO2 for growth but also that the archaeon has several unique features that probably reflect its adaptation to the human intestinal environment.
MATERIALS AND METHODS
M. stadtmanae strain DSZM 3091 was obtained from the Deutsche Sammlung von Zellkulturen und Mikroorganismen, Braunschweig, Germany. This methanogenic archaeon was grown on the medium described by Sparling et al. (87, 88), which also contained 0.5 g/liter sodium formate and 10% rumen fluid. The medium was prepared in an anaerobic chamber (95% N2-5% H2) and was reduced prior to autoclaving with 0.28 g Na2S · 9H2O per liter and 0.14 g l-cysteine-HCl per liter. Methanol was added after autoclaving to a concentration of 0.4% (vol/vol). Cells were routinely grown in 2-liter bottles containing 1 liter of medium and H2-CO2 (80:20) at a pressure of 2 × 105 Pa as the gas phase with continuous shaking at 37°C. When an optical density at 578 nm of approximately 2.5 was reached, the cells were harvested by centrifugation.
Sequencing strategy.
Total genomic DNA of M. stadtmanae was extracted and sheared. Several shotgun libraries were constructed using 3- to 5-kb size fractions. The fragments were cloned into vector pCR4-TOPO (Invitrogen). Insert ends of the recombinant plasmids were sequenced using dye terminator chemistry with MegaBACE 1000 and 4000 (GE Healthcare) and ABI Prism 377 (Applied Biosystems) automated DNA sequencers. Sequences were processed with Phred and assembled into contigs using the Phrap assembly tool (http://www.phrap.org). Sequence editing was done using GAP4, which is part of the Staden software package (90); 8.7-fold coverage was obtained after assembly of 21,555 sequences. In order to solve problems with misassembled regions caused by repetitive sequences and to close remaining sequence gaps, PCR-based techniques and primer walking with recombinant plasmids were used.
Gene prediction and annotation.
The initial gene prediction was accomplished using YACOP (93). The output was verified and edited manually using criteria such as the presence of a ribosome binding site, GC frame plot analysis, and similarity to known protein-encoding sequences (CDS). Annotation was done using the ERGO tool (Integrated Genomics) (www.integratedgenomics.com) with a two-step approach. Initially, all proteins were searched against a nonredundant database consisting of all publicly available sequence data by using FASTA3, which resulted in automatic annotation. All predictions were verified and modified manually by comparing the protein sequences with the Swiss-Prot, GenBank, ProDom, COG, and Prosite public databases. All coding sequences were searched for similarities to protein families and domains using CD-search (57). TMpred was used to predict transmembrane helices within the CDS (39). The SIGI tool (59) was used for score-based identification of genomic islands, referred to as alien CDS, and for identification of potentially highly expressed CDS.
Comparative genomics.
For comparative analysis, each CDS of one genome was searched against all CDS of another genome by using the BLAST program (1). For these unidirectional BLAST comparisons a strict cutoff value between e−15 and e−20 was used. BLAST hits were considered an indication of homology. For identification of CDS specific for all methanogens but missing in M. stadtmanae, BLAST comparisons (e−15) with the genomes of Methanocaldococcus jannaschii (9), Methanococcus maripaludis (38), Methanopyrus kandleri (85), Methanosarcina acetivorans (27), and Methanosarcina mazei (15) were carried out using all CDS of Methanobacterium thermoautotrophicus (86) as query sequences. CDS of M. stadtmanae not present in the genomes of other archaea were identified by searching each CDS encoding more than 100 amino acids against the genomes mentioned above plus those of Aeropyrum pernix (44), Archaeoglobus fulgidus (48), Haloarcula marismortui (3), Halobacterium sp. strain NRC-1 (70), Picrophilus torridus (26), Pyrobaculum aerophilum (24), Pyrococcus abyssi (12), Pyrococcus furiosus (76), Pyrococcus horikoshii (45), Sulfolobus solfataricus (83), Sulfolobus tokodaii (43), Thermoplasma acidophilum (77), and Thermoplasma volcanium (46). The CDS without significant BLAST hits (e−20) were searched with the same cutoff against the NCBI protein database (ftp.ncbi.nih.gov/BLAST/db/nr.tar.gz).
Purification of enzymes.
Frozen cells (10 g [wet weight]) of M. stadtmanae were routinely suspended in 10 ml potassium phosphate (pH 7.0), and the suspension was then passed through a French pressure cell at 108 Pa. After removal of cell debris and the membrane fraction by ultracentrifugation, a supernatant was obtained, from which MtaA and the MtaBC complex (methanol:coenzyme M methyltransferase), the HdrABC complex (heterodisulfide reductase), and the MvhADG complex (non-F420-reducing hydrogenase) were purified. Protein concentrations were determined by the Bradford method using bovine serum albumin as the standard.
(i) Methanol:coenzyme M methyltransferase.
The MtaBC complex was purified by chromatography on DEAE-Sepharose, Q-Sepharose, and hydroxyapatite (49). Purification was monitored by determining the characteristic absorption spectrum of the corrinoid protein using dicyanocobalamin as the standard (ɛ580-640 = 7.3 mM−1 cm−1). Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) revealed the presence of two polypeptides with apparent molecular masses of 50 kDa and 30 kDa. The N-terminal amino acid sequences of the 50- and 30-kDa subunits were determined by Edman degradation to be SCKYFTKMENASADEMVFG and MD(or S)S(or P)P(or L)L(or E)E(or K)K(or Y)YGKLTL(or A or H)Y, respectively.
MtaA was purified by ammonium sulfate (70%) precipitation, hydrophobic chromatography on Phenyl-Sepharose, gel filtration on Superdex 200, and anion-exchange chromatography on MonoQ (49). The N-terminal amino acid sequence was MDLIENLKAALNGEXVXKVPAISATAAAVEEAFPAANVS.
(ii) Heterodisulfide reductase.
The HdrABC complex was purified 40-fold by chromatography on DEAE-Sepharose, Q-Sepharose, Phenyl-Sepharose, and Superdex 200 (75). Purification was monitored by measuring the specific rate of heterodisulfide reduction with reduced methyl viologen. SDS-PAGE revealed the presence of three polypeptides with apparent molecular masses of 71 kDa, 33 kDa, and 21 kDa; the N-terminal amino acid sequences of these subunits were determined by Edman degradation to be XNNEXVVIGVYTHXXXXNV, XAYAYFLGCIMNNXYPGIEKS, and XTLLNESEYITDKDVDPTFK, respectively (75).
(iii) Non-F420-reducing hydrogenase.
The MvhADG complex was purified by chromatography on DEAE-Sepharose, hydroxyapatite, and Superdex 200 (75). Purification was monitored by measuring the specific activity of methyl viologen reduction with H2 at pH 7.6. SDS-PAGE revealed the presence of three polypeptides with apparent molecular masses of 55 kDa, 38 kDa, and 30 kDa. The N-terminal amino acid sequence of the 55 kDa subunit was determined by Edman degradation to be VELTLEPXTXIE (75).
Quantification of corrinoids.
Corrinoids were extracted from enzyme and cell samples with KCN as described by Fischer et al. (23). After centrifugation of the extract, 20 to 80 μl of the supernatant was analyzed by high-performance liquid chromatography using a Phenomenex phenyl-hexyl reverse-phase column. Dicyanocobalamin was used as the standard (ɛ580-640 = 7.3 mM−1 cm−1) The corrinoid was identified as hydroxybenzimidazolyl cobamide (factor III) by matrix-assisted laser desorption ionization—time of flight mass spectrometry (49).
Nucleotide sequence accession number.
The M. stadtmanae genome sequence has been deposited in the EMBL/GenBank/DDBJ database under accession number CP000102.
RESULTS AND DISCUSSION
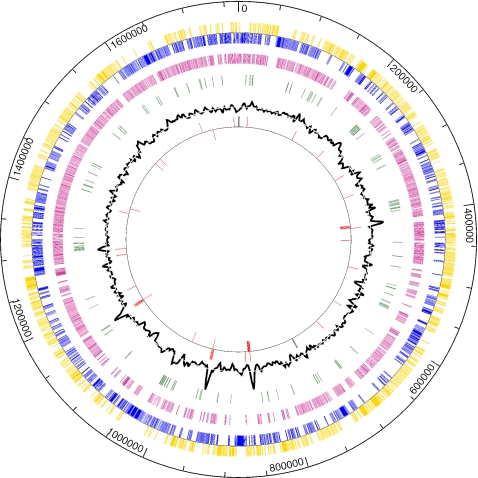
M. stadtmanae contains one circular chromosome (Fig. 1), and plasmids were not found. After a short description of the general features, the genome of M. stadtmanae is analyzed below mainly with respect to energy metabolism, carbon assimilation into C1 units, and the genes unique to this human intestinal archaeon. The results of the analysis of the carbon and energy metabolism of the C1 units are summarized in Fig. 2.
FIG. 1.
Circular map of the chromosome of M. stadtmanae. Rings from the outside to the inside: scale (in base pairs), protein-encoding sequences (blue or yellow), CDS with gene homologues in M. thermautotrophicus (magenta), putative highly expressed CDS (green), G+C content variation (higher values on the outside), rRNA and tRNA coding sequences (red), and cdc6 genes (black).
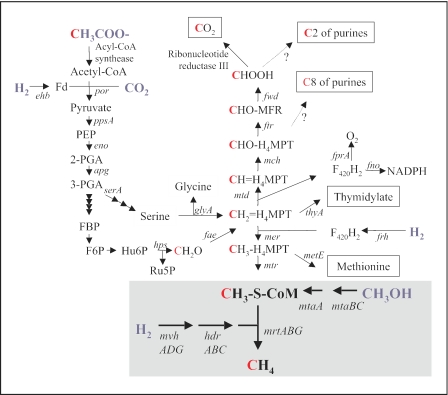
FIG. 2.
Metabolic pathways involved in methanol reduction to methane with H2 and in C1 unit biosynthesis from C-2 of acetate in M. stadtmanae. The genes encode the following enzymes (see text): ehb, energy-converting hydrogenase; por, pyruvate:ferredoxin oxidoreductase; ppsA, phosphoenolpyruvate synthase; eno, enolase; apg, phosphoglycerate mutase; serA, phosphoglycerate dehydrogenase; glyA, serine:H4MPT hydroxymethyl transferase; hps, hexulose phosphate synthase; fae, formaldehyde-activating enzyme; fwd, formylmethanofuran dehydrogenase; ftr, formylmethanofuran:H4MPT formyltransferase; mch, methenyl-H4MPT cyclohydrolase; fprA, F420H2 oxidase; fno, F420H2:NADP oxidoreductase; mtd, methylene-H4MPT dehydrogenase; thyA, thymidylate synthase; frh, F420-reducing hydrogenase; mer, methylene-H4MPT reductase; metE, methionine synthase; mtr, methyl-H4MPT:coenzyme M methyltransferase; mta, methanol:coenzyme M methyltransferase; mvh, non-F420-reducing hydrogenase; hdr, heterodisulfide reductase; and mrt, methyl-coenzyme M reductase. PEP, phosphoenolpyruvate; 2-PGA, 2-phosphoglycerate; 3-PGA, 3-phosphoglycerate; FBP, fructose bisphosphate; F6P, fructose-6-phosphate; Hu6P, 3-hexulose-6-phosphate; CHO-MFR, formylmethanofuran; Ru5P, ribulose-5-phosphate.
General features.
The circular chromosome consists of 1,767,403 bp and has the lowest G+C content of all archaeal genomes sequenced so far. Other features of the genome are shown in Table 1. The number of protein-encoding sequences (1,534 CDS) is the lowest number obtained for all methanogens (M. kandleri has 1,687 CDS). The genome contains open reading frames for 40 tRNAs. One of these, coding for a Met-tRNA, contains a putative intron that is 34 bp long (Msp0230). Selenocysteinyl-tRNAs were not found. Four rRNA operons are present in the genome of M. stadtmanae, the highest number found in archaeal genomes (rrnA to rrnD). Three rRNA operons contain a gene for an alanyl-tRNA between the 16S rRNA and 23S rRNA genes (rrnB to rrnD). A cumulative GC skew analysis revealed the presence of only one origin of replication in the neighborhood of one of two CDS for Cdc6 (involved in replication initiation) (Msp0001),for the small subunit of DNA polymerase II (Msp1584), for two helicases (Msp0005 and Msp0007), and for two genes for B12 biosynthesis (Msp1587 and Msp1588). The genome of M. stadtmanae contains four 1,528-bp insertion elements, all of which include either one of three highly homologous CDS (Msp0017, Msp0233, and Msp0471) or a pseudogene (Msp1439), The CDS show up to 29% identity to a CDS identified in an insertion sequence of Methanobrevibacter smithii (31), a close relative of M. stadtmanae. Additionally, a complete CDS (Msp1478) and a pseudogene (Msp1400) for two putative transposases and a putative integrase gene (Msp1355) were detected, but there is no evidence for the presence of a prophage. A 4.8-kbp genome segment in which a 30-bp sequence is repeated 59 times is noteworthy. This segment includes 10 short CDS (Msp0948 to Msp0957), which may not code for functional proteins.
TABLE 1.
General features of the M. stadtmanae genome
| Characteristic | Value |
|---|---|
| Genome length (bp) | 1,767,403 |
| % Coding regions | 84 |
| G+C content (%) | 28 |
| No. of rRNA operons | 4 |
| No. of tRNAs | 40 |
| No. of tRNAs with introns | 1 |
| No. of protein-encoding sequences | 1,534 |
| No. of CDS encoding hypothetical proteins | 287 |
| No. of CDS encoding conserved hypothetical proteins | 281 |
| No. of CDS encoding proteins with functional assignments | 966 |
| No. of CDS encoding predicted membrane-spanning proteins | 448 |
| No. of highly expressed CDSa | 116 |
| No. of alien CDS | 18 |
| No. of CDS encoding proteins with a twin-arginine motif | 0 |
| No. of CDS encoding aminoacyl-tRNA synthetases | 19 |
| No. of CDS encoding histones | 6 |
| No. of CDS encoding proteins with inteins | 0 |
| No. of insertion sequences | 4 |
| No. of putative transposases (fragments) | 1 (2) |
| No. of putative recombinases | 1 |
| No. of CDS with gene homologues in the genome of M. thermautotrophicus (e−15) | 1,002 |
| No. of CDS in the genome of M. thermoautotrophicus with gene homologues in the genomes of all other methanogens except M. stadtmanae (e−15)b | 37 |
| No. of CDS (>100 amino acids) without gene homologues in the genomes of other archaea (e−20) | 323 |
| No. of CDS (>100 amino acids) with gene homologues only in nonarchaeal genomes (<e−20)c | 73 |
(i) CDS shared with Methanothermobacter.
M. thermoautotrophicus is the closest relative of M. stadtmanae whose genome sequence has been published. A direct BLAST comparison identified gene homologues in M. thermautotrophicus for 1,002 CDS of the M. stadtmanae genome. Detailed analysis of the shared CDS indicated that the two organisms have very similar machinery for DNA repair, replication, transcription, translation, cell division, and protein folding. The two archaea also have related pathways for biosynthesis of isoprenoid lipids, amino acids, carbohydrates, nucleotides, and cofactors starting from acetyl-CoA. The two archaea do not contain CDS for selenoproteins, for cytochromes, for flagellar proteins, for tetrahydrofolate-specific enzymes, and for citrate synthase, aconitase, and isocitrate dehydrogenase. Consistently, archaea belonging to the Methanobacteriales are not dependent on selenium for growth, do not contain cytochromes, are nonmotile, do not contain folate, and synthesize 2-oxoglutarate from oxaloacetate via malate, fumarate, and succinate and not via citrate, aconitate, and isocitrate.
(ii) Absence of CDS found in all other methanogens.
BLAST comparisons revealed that 37 CDS present in the genomes of all methanogens sequenced so far are absent in M. stadtmanae. These CDS include genes for molybdenum cofactor synthesis and CO dehydrogenase/acetyl-CoA synthase (Table 2). Also absent are genes for superoxide dismutase and catalase, which are present in some but not all methanogenic archaea. The two enzymes involved in oxygen detoxification are mentioned since they were shown to be present in Methanobrevibacter species (7, 84), which are phylogenetically closely related to M. stadtmanae and which thrive in the intestinal tracts of animals and insects (63).
TABLE 2.
Thirty-seven protein-encoding sequences found in the genome of M. thermautotrophicus and all other methanogens except M. stadtmanae
| CDS or locus | Function according to Smith et al. (function according to our annotations)a |
|---|---|
| NP_276663 | Molybdenum cofactor biosynthesis protein MoaA |
| NP_276967 | Molybdenum cofactor biosynthesis protein MoaB |
| NP_275948 | Molybdenum cofactor biosynthesis protein MoaC |
| NP_276138 | Molybdenum cofactor biosynthesis protein MoeA |
| NP_276820 | Carbon monoxide dehydrogenase, alpha subunit (CO dehydrogenase/acetyl-CoA synthase, subunit alpha) |
| NP_276822 | Carbon monoxide dehydrogenase, alpha subunit (CO dehydrogenase/acetyl-CoA synthase, subunit beta) |
| NP_276825 | Corrinoid/iron-sulfur protein, large subunit (CO dehydrogenase/acetyl-CoA synthase, subunit gamma) |
| NP_276824 | Corrinoid/iron-sulfur protein, small subunit (CO dehydrogenase/acetyl-CoA synthase, subunit delta) |
| NP_276823 | Nitrogenase reductase-related protein (CO dehydrogenase/acetyl-CoA synthase, accessory protein) |
| NP_276732 | Thiamine biosynthetic enzyme |
| NP_276299 | Phosphonoacetaldehyde methylase |
| NP_277018 | Possible protein methyltransferase (predicted RNA methylase) |
| NP_276283 | Na+/Ca+ exchanging protein related |
| NP_276665 | Formate dehydrogenase, alpha subunit homologue |
| NP_276615 | Phenylalanyl-tRNA synthetase alpha subunit |
| NP_276113 | NADP-dependent glyceraldehyde-3-phosphate dehydrogenase (predicted NAD-dependent aldehyde dehydrogenase) |
| NP_275964 | l-Isoaspartyl protein carboxyl methyltransferaseb |
| NP_277015 | Hypothetical protein (putative RNA 3′-terminal phosphate cyclase) |
| NP_275838 | Hypothetical protein (predicted ABC-type transport system, permease protein) |
| NP_276300 | Possible cation transporter |
| NP_276301 | Cell division inhibitor-related protein |
| NP_276195 | Acetyltransferase |
| NP_276886 | Stomatin-like protein |
| NP_275744 | Conserved protein (contains ferredoxin domain) |
| NP_276442 | Hypothetical proteinb |
All CDS of M. thermautotrophicus (86) were searched with the BLAST program against the genomes of all other methanogenic archaea sequenced so far. CDS of M. thermautotrophicus with BLAST hits better than e−15 in all other genomes except that of M. stadtmanae are listed. Only the first CDS is listed when there was more than one hit resulting from different CDS with high levels of sequence similarity in the M. thermautotrophicus genome.
(iii) CDS for membrane-spanning proteins.
Approximately 29% (448 CDS) of the 1,534 CDS are predicted to code for transmembrane proteins based on the presence of at least one transmembrane helix. Among these CDS are genes for more than 10 different ABC transporters. They are annotated to be involved in the transport of cobalt, phosphate, other inorganic ions, amino acids, peptides, and polyols. The genome appears to lack a CDS for a molybdate ABC transporter gene, which is consistent with the absence of homologues for molybdopterin biosynthesis genes (Table 2). Examples of genes encoding non-ABC transporters found in the genome of M. stadtmanae are CDS for putative subunits of a potassium transport system (Msp0250, Msp0723, Msp0830, Msp1223, Msp1489, and Msp1504), as well as a Co/Zn/Cd transporter (Msp0519), an Mn/Fe transporter (Msp1554), an ammonium transporter (Msp0663), and a ferrous iron transporter (Msp1441 and Msp1440). A sodium-dependent transport system for leucine that has been characterized in M. stadtmanae (88) is probably among the six predicted sodium-dependent transporters (Msp0148, Msp0513, Msp0691, Msp0738, Msp0824, and Msp1417). Genes for sodium-proton antiporters found in other methanogens (37) appear to be absent. M. barkeri has been shown to grow like M. stadtmanae on methanol plus H2 with acetate as the carbon source and requires sodium ions under these conditions (68). It would be surprising if M. stadtmanae could not regulate the intracellular sodium ion concentration by employing a sodium-proton antiporter or a related system.
(iv) CDS predicted to be highly expressed.
Codon usage analysis with the SIGI tool (59) revealed a group of 116 CDS which are predicted to be highly expressed (Table 3). Among the putative highly expressed CDS are most of the genes for the enzymes involved in energy metabolism in M. stadtmanae.
TABLE 3.
CDS in the genome of M. stadtmanae predicted to be highly expresseda
| CDS | Function | Gene | CDS | Function | Gene | |
|---|---|---|---|---|---|---|
| Msp0010 | 30S ribosomal protein S15P | rps15p | Msp0769 | Archaeal histone | ||
| Msp0017 | Conserved hypothetical protein, encoded in insertion element | Msp0789 | Rubrerythrin | |||
| Msp0818 | Methanol corrinoid protein 4 | mtaC4 | ||||
| Msp0022 | Ketol-acid reductoisomerase | Msp0819 | Methanol:cobalamin methyltransferase 4 | mtaB4 | ||
| Msp0072 | Pyridoxine biosynthesis protein Pdx1 | pdx1 | Msp0865 | 30S ribosomal protein S9P | rps9p | |
| Msp0079 | Hypothetical proteinb | Msp0866 | 50S ribosomal protein L13P | rpl13p | ||
| Msp0106 | 30S ribosomal protein S17e | rps17e | Msp0869 | 30S ribosomal protein S11P | rps11p | |
| Msp0114 | Thermosome, subunit beta | thsB | Msp0870 | 30S ribosomal protein S4P | rps4p | |
| Msp0117 | Predicted 3-hydroxy-3-methylglutaryl-CoA synthase | Msp0871 | 30S ribosomal protein S13P | rps13p | ||
| Msp0880 | 50S ribosomal protein L14e | rpl14e | ||||
| Msp0122 | Archaeal histone | Msp0882 | 50S ribosomal protein L34e | rpl34e | ||
| Msp0130 | Thermosome, subunit alpha | thsA | Msp0886 | 50S ribosomal protein L15P | rpl15p | |
| Msp0147 | Ferredoxin | Msp0887 | 50S ribosomal protein L30P | rpl30p | ||
| Msp0182 | Methylcobalamin:coenzyme M methyltransferase 4 | mtaA4 | Msp0888 | 30S ribosomal protein S5P | rps5p | |
| Msp0894 | 30S ribosomal protein S14P | rps14p | ||||
| Msp0184 | Methanol:cobalamin methyltransferase 3 | mtaB3 | Msp0895 | 50S ribosomal protein L5P | rpl5p | |
| Msp0185 | Methanol corrinoid protein 3 | mtaC3 | Msp0896 | 30S ribosomal protein S4e | rps4e | |
| Msp0186 | Methanol:cobalamin methyltransferase 2 | mtaB2 | Msp0898 | 50S ribosomal protein L14P | rpl14p | |
| Msp0187 | Methanol corrinoid protein 2 | mtaC2 | Msp0903 | 30S ribosomal protein S3P | rps3p | |
| Msp0188 | Methanol:cobalamin methyltransferase 1 | mtaB1 | Msp0906 | 50S ribosomal protein L2P | rpl2p | |
| Msp0189 | Methanol corrinoid protein 1 | mtaC1 | Msp0908 | 50S ribosomal protein L1e | rpl1e | |
| Msp0198 | DNA/RNA-binding protein AlbA | albA | Msp0960 | Predicted ABC-type polar amino acid transport system, periplasmic substrate-binding protein | ||
| Msp0229 | Putative preprotein translocase, subunit SecG | secG | ||||
| Msp0234 | Glutamine synthetase | |||||
| Msp0251 | Putative thiamine biosynthesis protein 2 | thiC2 | Msp0233 | Conserved hypothetical protein, encoded in insertion element | ||
| Msp0314 | Non-F420-reducing hydrogenase, subunit delta | mvhD | ||||
| Msp0315 | Non-F420-reducing hydrogenase, subunit gamma | mvhG | Msp1002 | Cysteine desulfurase | iscS | |
| Msp0316 | Non-F420-reducing hydrogenase, subunit alpha | mvhA | Msp1013 | Heterodisulfide reductase, subunit B1 | hdrB1 | |
| Msp0318 | Methyl-coenzyme M reductase II, subunit B | mrtB | Msp1014 | Heterodisulfide reductase, subunit C1 | hdrC1 | |
| Msp0320 | Methyl-coenzyme M reductase II, subunit G | mrtG | Msp1061 | Hypothetical protein, related to ferredoxin | ||
| Msp0321 | Methyl-coenzyme M reductase II, subunit A | mrtA | Msp1134 | A1A0 ATPase, subunit B | ahaB | |
| Msp0325 | Predicted peptidyl-prolyl cis-trans isomerase 2 | Msp1135 | A1A0 ATPase, subunit A | ahaA | ||
| Msp0327 | 50S ribosomal protein L10e | rpl10e | Msp1136 | A1A0 ATPase, subunit F | ahaF | |
| Msp0383 | Archaeal histone | Msp1139 | A1A0 ATPase, subunit K | ahaK | ||
| Msp0409 | Conserved hypothetical proteinc | Msp1141 | A1A0 ATPase, subunit H | ahaH | ||
| Msp0444 | Rubredoxin | Msp1145 | d-3-Phosphoglycerate dehydrogenase | serA | ||
| Msp0471 | Conserved hypothetical protein, encoded in insertion element | Msp1158 | Predicted Zn ribbon RNA-binding protein | |||
| Msp1174 | 50S ribosomal protein L37e | rpl37e | ||||
| Msp0477 | Predicted type I restriction-modification system, restriction subunit | Msp1243 | 50S ribosomal protein L15e | rpl15e | ||
| Msp1253 | 50S ribosomal protein L37Ae | rpl37ae | ||||
| Msp0478 | Hypothetical protein, related to type I restriction-modification system subunit | Msp1256 | Partially conserved hypothetical protein, related to prefoldin, subunit beta | |||
| Msp0479 | Putative type I restriction-modification system, methyltransferase subunit | Msp1263 | 50S ribosomal protein L12P | rpl12p | ||
| Msp1280 | 30S ribosomal protein S8 | rps8e | ||||
| Msp0518 | Archaeal histone | Msp1292 | 50S ribosomal protein L40e | rpl41e | ||
| Msp0565 | Predicted M42 glutamyl aminopeptidase | Msp1297 | 30S ribosomal protein S3Ae | rps30ae | ||
| Msp0594 | 30S ribosomal protein S19e | rps19e | Msp1352 | Translation initiation factor 5A (aIF-5A) | eif5a | |
| Msp0597 | 50S ribosomal protein L31e | rpl31e | Msp1366 | Translation elongation factor 1-alpha (EF-Tu) | tuf | |
| Msp0599 | 50S ribosomal protein LX | rplX | Msp1368 | 30S ribosomal protein S7P | rps7p | |
| Msp0600 | Conserved hypothetical protein, related to prefoldin, subunit alpha | Msp1369 | 30S ribosomal protein S12P | rps12p | ||
| Msp1416 | Glutamate dehydrogenase | gdhA | ||||
| Msp0620 | 30S ribosomal protein S27Ae | rps27ae | Msp1449 | Energy-converting hydrogenase B, subunit I | ehbI | |
| Msp0628 | 30S ribosomal protein S6e | rps6e | Msp1450 | Energy-converting hydrogenase B, subunit H | ehbH | |
| Msp0631 | 50S ribosomal protein L24e | rpl24e | Msp1457 | Energy-converting hydrogenase B, subunit A | ehbA | |
| Msp0632 | 30S ribosomal protein S28e | rps28e | Msp1476 | Heterodisulfide reductase, subunit A1 | hdrA1 | |
| Msp0633 | 50S ribosomal protein L7Ae | rpl7ae | Msp1517 | Chaperone protein DnaK | dnaK | |
| Msp0719 | Partially conserved hypothetical protein, related to AAA ATPase | Msp1528 | 30S ribosomal protein S27e | rps27e | ||
| Msp1529 | 50S ribosomal protein L44e | rpl44e | ||||
| Msp0729 | Hypothetical protein, related to nucleic acid-binding protein | Msp1542 | 50S ribosomal protein L21e | rpl21e |
Putative highly expressed CDS were obtained by codon usage analysis with the SIGI tool (59).
Additional hypothetical proteins are encoded by Msp0238, Msp0330, Msp0464, Msp0707, Msp0827, Msp1081, Msp1422, and Msp1498.
Additional conserved hypothetical proteins are encoded by Msp0425, Msp0677, and Msp1566.
Energy metabolism.
The genome of M. stadtmanae contains CDS for the following enzymes that are predicted based on their functions in other methanogens to be involved in methanol reduction with H2 to methane and ATP synthesis: methanol:coenzyme M methyltransferase (MtaABC)(reaction 1), methyl-coenzyme M reductase (MrtABG) (reaction 2), heterodisulfide reductase (HdrABC) (reaction 3), non-F420-reducing hydrogenase (MvhADG) (reaction 4), and proton-translocating ATPase (AhaABCDEFHIK) (reaction 5). Consistent with the proposed catabolic function, the codon usage of most of the genes was that of highly expressed genes (Table 3).
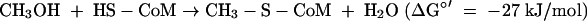 |
1 |
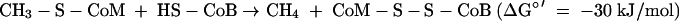 |
2 |
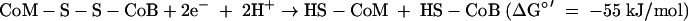 |
3 |
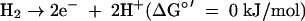 |
4 |
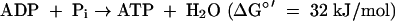 |
5 |
where HS-CoM is coenzyme M, CH3-S-CoM is methyl-coenzyme M, HS-CoB is coenzyme B, and Pi is inorganic phosphate. The Eo′ of the CoM-S-S-CoB/HS-CoM + HS-CoB couple has recently been determined to be −140 mV rather than −200 mV (97). The ΔGo′ of methanol reduction with H2 to methane and H2O is −112 kJ/mol (96).
(i) Formation of methyl-coenzyme M from methanol and coenzyme M.
In Methanosarcina species, the only other methanogens known to grow on methanol and H2, the initial step in methanol reduction to methane has been shown to be the formation of methyl-coenzyme M from methanol and coenzyme M (reaction 1). This reaction is catalyzed by three cytoplasmatic proteins, a corrinoid protein (MtaC), a methyltransferase (MtaB) that catalyzes the methylation of MtaC with methanol, and a methyltransferase (MtaA) that catalyzes methyl transfer from MtaC to coenzyme M. The corrinoid protein has to be reduced to the cob(I)alamin oxidation state to become active. This reduction is catalyzed by a methyltransferase that activates the MapAB protein (13). MtaC and MtaB form a tight complex, and the genes that encode them form a transcription unit, mtaBC. MtaA is isolated separately and is translated from a monocistronic mRNA (78). The genomes of M. barkeri, M. acetivorans, and M. mazei each contain three sets of mtaBC genes and two sets of mtaA genes, which are transcribed under different growth conditions (73).
Four mtaA, mtaB, and mtaC homologues were found in the genome of M. stadtmanae: mtaB1C1 (Msp0188 and Msp0189), mtaB2C2 (Msp0186 and Msp0187), mtaB3C3 (Msp0184 and Msp0185), mtaB4C4 (Msp0819 and Msp0818), mtaA1 (Msp0761), mtaA2 (Msp0112), mtaA3 (Msp0774), and mtaA4 (Msp0182). At the 3′ end of the mtaB4C4 cluster, the genome harbors a gene, Msp0190, that codes for a protein with sequence similarity with RamA. The iron-sulfur protein RamA has been shown to be involved in ATP-dependent reductive activation of the methylamine methyltransferase corrinoid protein in M. barkeri (T. Ferguson, T. Lienard, G. Gottschalk, and J. Krzycki, unpublished results). There is no indication that any of these polypeptides are membrane associated and could therefore be involved in proton or sodium ion translocation. Cells of M. stadtmanae grown on methanol and H2 in the laboratory were found to contain relatively high concentrations (5%) of MtaBC isoenzymes 2 and 3 and of MtaA isoenzyme 4, as revealed by the N-terminal sequences of purified MtaBC and MtaA (see Materials and Methods). Consistently, the codon usage of the corresponding genes was that of highly expressed genes (Table 3). The MtaBC complex contained 1 mol of 5-hydroxybenzimidazolyl cobamide (factor III) per mol MtaC.
Alignments of the four deduced amino acid sequences encoded by mtaBC revealed that the isoenzymes MtaB3C3 and MtaB4C4 are more closely related (>99% sequence identity) than the other isoenzymes (<96%). All four isoenzymes are more closely related to each other than to any of the MtaBC isoenzymes in Methanosarcina species (<50%). This relationship indicates that the four MtaBC isoenzymes in M. stadtmanae are the result of three gene duplications, two of which occurred more recently. The finding that of the cytochrome-free methanogens only M. stadtmanae contains mtaBC homologues suggests that a set of mtaBC genes was acquired by lateral gene transfer from Methanosarcina species. However, this must have occurred a long time ago, as indicated by the low level of sequence identity of MtaBC from M. stadtmanae with MtaBC from Methanosarcina and by the different order of the genes in the mta operons of M. stadtmanae (mtaBC) and Methanosarcina (mtaCB). Alignment of the deduced amino acid sequences encoded by mtaA revealed a different picture. The four MtaA isoenzymes of M. stadtmanae are related to each other (<60%) almost as closely as they are related to the MtaA isoenzymes of Methanosarcina (<54%).
The possibility that in M. stadtmanae methyl-coenzyme M formation from methanol and coenzyme M could also proceed via methyl-H4MPT as an intermediate involving a methanol:H4MPT methyltransferase (MtxXAH) and a methyl-H4MPT:coenzyme M methyltransferase (MtrABCDEFGH) was considered. The genomes of Methanosarcina species harbor the mtxXAH genes, which have been proposed to code for a corrinoid-containing enzyme complex that catalyzes the formation of methyl-H4MPT from methanol and H4MPT and to be involved in methanol oxidation to CO2 (32). However, mtxXAH homologues are not found in the genome of M. stadtmanae. Consistently, in M. stadtmanae only 10% of the methyl group of methionine, which is biosynthetically derived from the methyl group of methyl-H4MPT, is synthesized from methanol. The other 90% is derived from C-2 of acetate via the hydroxymethyl group of serine or from C-1 of 3-hexulose-6-phosphate, as revealed by labeling experiments (62) (Fig. 2). The 10% labeling of the methyl group of methionine by methanol can be explained by the presence of a methyl-H4MPT:coenzyme M methyltransferase (MtrA-H). The genome of M. stadtmanae contains the mtrEDCBAFGH gene cluster (Msp0300 to Msp0307), which is predicted to code for the sodium ion-translocating methyl-tetrahydromethanopterin:coenzyme M methyltransferase complex, which like MtaABC is a corrinoid protein. So far, the mtr genes have been found in every methanogenic archaeon investigated but not in any other organism. During growth of methanogens on H2 and CO2 the Mtr complex catalyzes the exergonic transfer of the methyl group of methyl-H4MPT to coenzyme M (ΔGo′ = −30 kJ/mol), a step in CO2 reduction to methane, which is coupled to energy conservation. During growth of Methanosarcina on methanol the Mtr complex is involved in methanol oxidation to CO2, and the methyl transfer from methyl-coenzyme M to H4MPT is driven by the electrochemical sodium ion potential (29). Since M. stadtmanae cannot reduce CO2 to methane or oxidize methanol to CO2 and synthesizes the methyl group of methionine mainly from C-2 of acetate (Fig. 2), this methanogen probably does not really require the enzyme. Consistently, the specific activity of the Mtr complex in cell extracts of M. stadtmanae was found to be below the detection limit (98). However, there is evidence that in M. stadtmanae formaldehyde can be reduced with H2 to methane, which probably proceeds via methylene-H4MPT, methyl-H4MPT, and methyl-CoM as intermediates (51), indicating the presence of Mtr complex activity. Mtr complex might not be essential in M. stadtmanae. This is indicated by the recent finding that mtrABCDEFGH deletion mutants of M. barkeri can grow on methanol and H2 when the medium is supplemented with acetate (100).
Another pathway from methanol to methyl-coenzyme M that was considered starts with the oxidation of methanol to formaldehyde, which spontaneously reacts with H4MPT to form methylene-H4MPT. The latter compound is then reduced to methyl-H4MPT, from which the methyl group is transferred to coenzyme M. In the genome of M. stadtmanae, genes for F420-dependent methylene-H4MPT reductase (Mer) (Msp1128) and (as mentioned above) for methyl-H4MPT:coenzyme M methyltransferase (Mtr) are found. A gene for a formaldehyde-activating enzyme (Fae) (Msp1498) that catalyzes the formation of methylene-H4MPT from formaldehyde and H4MPT (Fig. 2) is also present in the genome. However, a CDS for a formaldehyde dehydrogenase is not apparent. The finding from labeling experiments that only 10% of the methyl group of methionine (62) is synthesized from methanol again indicates that, if present, this pathway has only minor importance. This is even more true as methanol is always contaminated by small amounts of formaldehyde generated from methanol by autoxidation, which could also explain the 10% labeling by methanol.
(ii) Methyl-coenzyme M reduction with coenzyme B to CH4 and CoM-S-S-CoB.
In all methanogens that have been investigated for this characteristic methane is generated by reduction of methyl-coenzyme M with coenzyme B (reaction 2). The reaction is catalyzed by methyl-coenzyme M reductase (Mcr or Mrt), which is composed of three subunits, subunits A, B, and G, each of which is present twice. Two moles of nickel porphinoid F430 per mol of heterohexamer is tightly but noncovalently bound. Five of the amino acid residues in McrA are post- or cotranslationally modified, and these amino acid positions are highly conserved. Some methanogens contain two methyl-coenzyme M isoenzymes, McrABG and MrtABG, which are formed under different growth conditions. The CDS for the three Mcr or Mrt subunits A, B, and G are organized in transcription units, which generally contain one or two additional open reading frames, subunits C and D, whose functions are unknown (95).
The genome of M. stadtmanae was found to harbor a transcription unit, mrtBDGA (Msp0318 to Msp0321), and separately an mcrC homologue (Msp0299). The mvhDGA cluster encoding the non-F420-reducing hydrogenase involved in reduction of methanol to methane with H2 is located directly upstream of the mrtBDGA cluster (see below). The sequence of mrtA is conserved with respect to five posttranslationally modified amino acids, glycine 447, glutamine 402, arginine 274, cysteine 454, and histidine 260. Genes encoding a second methyl-coenzyme M reductase were not found. Based on the deduced amino acid sequence, one of the three subunits (MrtB) is predicted to contain at least one hydrophopic segment that could form a transmembrane helix. Similar predictions have been made for the β subunits (McrB or MrtB) of methyl-coenzyme M reductases from other methanogens. The crystal structures of methyl-coenzyme M reductases from several methanogens have revealed that the hydrophobic segment of the β subunit is buried within the α2β2γ2 complex and that the surface of the complex is completely hydrophilic (22, 30). There is no indication from the biochemical properties of methyl-coenzyme M reductases that the enzyme could be membrane associated and involved in proton or sodium ion translocation.
(iii) CoM-S-S-CoB reduction to coenzyme M and coenzyme B.
In methanogens without cytochromes the heterodisulfide reductase complex is generally composed of the subunits HdrA, HdrB, and HdrC. HdrA is an iron-sulfur flavoprotein, and HdrB and HdrC are iron-sulfur proteins. The enzyme contains an active site iron-sulfur cluster located on subunit HdrB (19, 56). The genes coding for the three subunits are organized into two transcription units, hdrA and hdrCB (36). The physiological electron donor of HdrABC appears to be the iron-sulfur protein MvhD of the MvhADG complex (non-F420-reducing hydrogenase) (see below). HdrB exhibits sequence similarity to subunit GlpC of glycerol phosphate dehydrogenase from Escherichia coli (36) and to subunit SdhC of succinate dehydrogenases from archaea belonging to the order Sulfolobales (50), which are integral membrane proteins.
The genome of M. stadtmanae was found to contain two sets of hdrA, hdrB, and hdrC genes, one set organized in an hdrC2B2A2 transcription unit (Msp0125 to Msp0127) and the other set organized in two transcription units, hdrC1B1 (Msp1014 and Msp1013) and hdrA1 (Msp1476). The HdrABC complex purified from M. stadtmanae grown under laboratory conditions is the product of the hdrC1B1 and hdrA1 genes, as revealed by the N-terminal sequences (see Materials and Methods). The genome of M. stadtmanae lacks a CDS for HdrE (cytochrome b), which until now has been found only in Methanosarcina species. The hdrA2 gene in the hdrA2C2B2 cluster in M. stadtmanae is similar to an hdrA homologue found in the genome of Methanosarcina species since it is extended at the 3′ end by a sequence with similarity to mvhD on the protein level. The fusion of the two genes is considered an indication that HdrA and MvhD are also functionally associated in electron transfer from H2 to CoM-S-S-CoB (91). Another interesting aspect is that in Methanosarcina the order of the genes in the putative transcription unit is hdrACB, whereas in M. stadtmanae it is hdrC2B2A2.
(iv) Hydrogen activation for the reduction of CoM-S-S-CoB.
Methanogens without cytochromes, which with the exception of M. stadtmanae can grow on H2 and CO2 as sole energy sources, generally contain three different [NiFe] hydrogenases: a cytoplasmic F420-reducing hydrogenase (FrhABG), a cytoplasmic non-F420-reducing hydrogenase (MvhADG) (92), and a membrane-associated energy-converting hydrogenase (Eha and/or Ehb), which uses ferredoxin as an electron acceptor (33). So far, the energy-converting hydrogenase has been studied only in M. barkeri, in which the homologous enzyme (designated Ech) was shown to be involved in CO2 reduction to formylmethanofuran (energy metabolism), in reductive carboxylation reactions (e.g., pyruvate synthesis from acetyl-CoA), and in CO2 reduction to CO (CO2 assimilation) (60).
FrhABG is involved in methenyl-H4MPT reduction to methylene-H4MPT and in methylene-H4MPT reduction to methyl-H4MPT (CO2 reduction to methane), as well as in NADP reduction (biosynthesis). MvhADG appears to be involved only in CoM-S-S-CoB reduction. Some of the methanogens without cytochromes also contain an H2-forming methylene-H4MPT dehydrogenase (Hmd), which is a nickel- and iron-sulfur-cluster-free hydrogenase with a function in CO2 reduction to methane (54).
In the genome of M. stadtmanae CDS for three different hydrogenases are found: frhADGB (Msp1302 to Msp1305), mvhDGA (Msp0314 to Msp0316), and ehbABCDEFGHIJKLMNOPQ (Msp1457 to Msp1442 and Msp1436). There is also a second mvhD homologue (Msp0638), which is located at the 3′ end of two genes encoding a formate dehydrogenase (fdhAB; Msp0640 and Msp0639) and a third gene fused at the 5′ end to hdrA2 in the hdrB2C2A2 cluster. The genome lacks CDS for Eha and the nickel-free hydrogenase Hmd. As indicated below, in M. stadtmanae the Ehb and Frh hydrogenases probably have a purely anabolic function, Ehb in pyruvate synthesis from acetyl-CoA and Frh in NADP reduction (Fig. 2). Therefore, only the non-F420-reducing hydrogenase Mvh can be involved in methanol reduction to methane. Consistent with a function of Mvh in energy metabolism is the finding that the codon usage of the mvhDGA genes rather than that of the frhADGB and ehbA-Q genes is the codon usage of other highly expressed genes (Table 3).
The non-F420-reducing hydrogenase was purified from M. stadtmanae and identified as the mvhDGA gene product by N-terminal sequence comparisons (75). From the sequence of the mvhDGA genes it was deduced that MvhA is the [NiFe] center-harboring subunit and MvhD and MvhG are iron-sulfur proteins. There is no indication that in M. stadtmanae the non-F420-reducing hydrogenase is a membrane-associated enzyme (no predicted transmembrane helices) or is an extracytoplasmic enzyme (no 5′ twin-arginine motif).
In the genomes of most methanogens without cytochromes a mvhB gene is present at the 3′ end of the mvhDGA transcription unit. MvhB was shown to code for a polyferredoxin whose function is still not known (34, 74). A homologue of mvhB was not found in the genome of M. stadtmanae. An mvhB homologue is also not present in the genome of M. kandleri.
(v) ATP synthesis.
Methanogenic archaea generally contain a proton-translocating ATPase of the A1A0 type composed of 9 or 10 different subunits and encoded by an ahaHIKECFABD transcription unit (67). Such a gene cluster is also found in the genome of M. stadtmanae (Msp1141 to Msp1133). Based on codon usage the AhaA, AhaB, AhaF, AhaH, and AhaK subunits are predicted to be highly expressed (Table 3), which is consistent with a function of the ATPase in energy metabolism. From the primary structure it was deduced that AhaK is the hydrophobic proteolipid directly involved in proton translocation. AhaK is predicted to contain four transmembrane helices.
(vi) Energy conservation.
The genome analysis and biochemical studies revealed that in M. stadtmanae only five enzyme complexes are involved in methanol reduction to methane with H2, a reaction that is coupled to ADP phosphorylation, as shown by growth of the methanogen on H2 and methanol as sole energy sources. The five complexes are MtaABC, MrtABG, HdrABC, MvhADG, and AhaABCDEFHIK. Transmembrane prediction methods predict that three of these complexes (the Mta, Mvh, and Hdr complexes) are located in the cytoplasm, that the AhaCKI subunits of the A1Ao complex are membrane integrated, and that the MtrABG complex could be membrane associated via the MrtB subunit. However, as outlined above, the hydrophobic part of MrtB is buried within the enzyme complex rather than exposed to its surface, which is completely hydrophilic. Therefore, methyl-coenzyme M reductases have to be considered cytoplasmic enzymes. Conversely, HdrABC is most probably a membrane-associated enzyme, although none of its three subunits are predicted to contain transmembrane helices. The complex is proposed to be anchored via its HdrB subunit to the membrane, a conclusion based mainly on the observation that SdhC (CAA06782) (50), a protein with 31% sequence identity to HdrB, is a membrane protein and that transmembrane helices are not unequivocally predicted for this protein. In Methanothermobacter marburgensis HdrABC and MvhADG have been shown to form a tight complex partially associated with the cytoplasmic membrane (82, 92). In the complex HdrB harbors the active site for CoM-S-S-CoB reduction and MvhA harbors the active site for H2 oxidation. Electron transfer has been proposed to occur between the MvhD and HdrA subunits (89) based on the finding that in the genomes of Methanosarcina and M. stadtmanae there is an hdrA homologue that is fused to an mvhD homologue (see above). The proposed electron transfer from MvhD to HdrA remains to be shown experimentally. With cell suspensions of M. stadtmanae it has been shown that methanol reduction with H2 is coupled to the buildup of an electrochemical proton potential which drives the phosphorylation of ADP (87), most probably via the Aha complex. All the results together suggest that in M. stadtmanae the reduction of CoM-S-S-CoB with H2 is coupled principally with proton translocation, as shown in Fig. 3. In the topology model of the HdrABC/MvhADG complex the anchor function of HdrB is speculative.
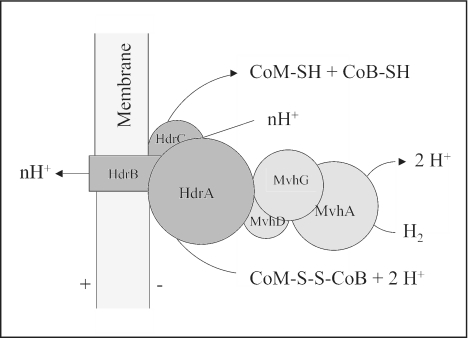
FIG. 3.
Topological model of the HdrABC/MvhADG complex catalyzing the reduction of CoM-S-S-CoB with H2 in a proton-translocating reaction in M. stadtmanae. In the model it is assumed that based on indirect evidence, HdrB is an integral membrane protein, although this is not reflected in its primary structure (see text).
The growth yield of M. stadtmanae on H2 and methanol in complex media was found to be approximately 4 g/mol methane (64), and the yield of M. barkeri on H2 and methanol was found to be 4.6 g/mol methane (68). Coupling of methanol reduction to methane with energy conservation thus proceeds in these two organisms with almost identical efficiencies.
(vii) 5-Hydroxybenzimidazolyl cobamide biosynthesis.
The corrinoid group is the prosthetic group of the methyltransferase (MtaABC) that catalyzes reaction 1. M. stadtmanae has an unusually high 5-hydroxybenzimidazolyl cobamide content, 4 nmol/mg protein; for comparison, M. marburgensis has a cobamide content of 0.3 nmol/mg protein. In the genome of M. stadtmanae all the genes required for synthesis of the corrin ring from eight glutamate residues (79) were found, which is consistent with the report that growth of the organism is not dependent on vitamin B12 (64). The finding that the genome of M. stadtmanae appears to lack a gene for a cobalamin-dependent ribonucleotide reductase was unexpected. Only a CDS for a glycyl radical ribonucleotide reductase (ribonucleotide reductase III) is present (Msp0254). In the genome of M. thermoautotrophicus genes for both a glycyl radical enzyme and a B12-dependent enzyme are found.
(viii) Coenzyme F430 biosynthesis.
The nickel porphinoid group is the prosthetic group of methyl-coenzyme M reductase that catalyzes reaction 2. M. stadtmanae contains approximately 0.3 nmol F430 per mg protein. The biosynthesis of F430 has not been elucidated yet. Biosynthesis branches off from the cobalamin biosynthesis pathway at the level of precorrin 2 (71). Since all methanogens synthesize F430 and only methanogens synthesize F430, the genes involved in F430 biosynthesis must be among the only approximately 40 CDS found in all methanogens and not in any other archaea (data not shown). These CDS include genes for the subunits of methyl-coenzyme M reductase (Mcr and/or Mrt), for methyl-H4MPT:coenzyme M methyltransferase (Mtr), and for several conserved hypothetical proteins. Some of the latter proteins could be involved in F430 biosynthesis and the posttranslational modification and activation of methyl-coenzyme M reductase. Conserved clusters of more than one CDS present in all methanogens could not be identified.
Inability of M. stadtmanae to use other methanogenic substrates.
Besides the genes encoding the enzymes that catalyze reactions 1 to 5, the genome of M. stadtmanae also contains CDS for all the enzymes required for the reduction of CO2 to methane and for the oxidation of methanol to CO2 (Fig. 2), as follows: formylmethanofuran dehydrogenase (FwdABCDG) (Msp0241 to Msp0245), which is a molybdopterin-dependent enzyme (FwdB has a putative molybdopterin cofactor binding motif); two formylmethanofuran:H4MPT formyltransferases (Ftr) (Msp0070 and Msp1502); methenyl-H4MPT cyclohydrolase (Mch) (Msp1238); F420-dependent methylene-H4MPT dehydrogenase (Mtd) (Msp0163); F420-dependent methylene-H4MPT reductase (Mer); and methyl-H4MPT:coenzyme M methyltransferase (MtrABCDEFGH) (see above). In cell extracts of M. stadtmanae only very low activities of these enzymes have been detected (between 20 and 40 mU/mg) (80, 98). As mentioned above, the genome lacks genes for the biosynthesis of molybdopterin (Table 2), indicating that M. stadtmanae cannot synthesize active formylmethanofuran dehydrogenase. Cell suspensions of the organism were shown to slowly oxidize [14C]formaldehyde to14CO2, most probably via methylene-H4MPT and formate (51), which is consistent with the absence of formylmethanofuran dehydrogenase activity. However, low specific activities (40 mU/mg) of this enzyme have been reported to be present in cell extracts of M. stadtmanae (80). Therefore, the possibility that M. stadtmanae might be able to take up molybdopterin present at trace levels in the yeast extract and/or rumen fluid, which is generally a component of the complex growth medium, has to be considered. It has been reported that methanofuran is absent in M. stadtmanae (98); however, the method employed was not sensitive enough to completely exclude the possibility that this coenzyme was present. In this respect it is of interest that in the genome of M. stadtmanae a CDS for a tyrosine decarboxylase (mfnA; Msp0329) involved in methanofuran biosynthesis (47) has been found. Nonetheless, together the findings can explain why the methanogen cannot reduce CO2 to methane or oxidize methanol to CO2.
It is interesting with respect to the function of formylmethanofuran dehydrogenase that the enzyme from Methylobacterium extorquens AM1, which lacks a molybdopterin binding site, catalyzes the hydrolysis rather than the dehydrogenation of formylmethanofuran (72). The function of formylmethanofuran dehydrogenase in M. stadtmanae could therefore be to catalyze the formation of formate, which is required for the synthesis of C-2 of purines and as an electron donor for ribonucleotide reduction (Fig. 2). It is also noteworthy that the sequences of the subunits of formylmethanofuran dehydrogenase from M. stadtmanae are most similar to those of the tungsten-containing isoenzyme present in many methanogens (95). There is one peculiarity: subunits A and G are most closely related to FwdA and FwdG from M. thermoautotrophicus, subunits C and D are most closely related to FwdC and FwdD from M. jannaschii and M. maripaludis, respectively, and subunit B, which harbors the molybdopterin binding site, is most closely related to FwdB from M. kandleri (99). The reason for this phylogenetic mixture is not apparent.
As mentioned above, the genome of M. stadtmanae lacks genes required for the synthesis of the nickel-containing carbon monoxide dehydrogenase/acetyl-CoA decarbonylase complex (Table 2), which in methanogens is either involved in methanogenesis from acetate or in acetyl-CoA synthesis from CO2 and a methyl group (95). This finding could explain why M. stadtmanae cannot use acetate as a methanogenic substrate and why this archaeon is dependent on acetate as a carbon source for growth.
Carbon assimilation.
M. stadtmanae is dependent on acetate and CO2 as main carbon sources for growth. Also, this organism appears to be auxotrophic for isoleucine or leucine and for thiamine (64). Labeling studies have revealed that the methyl group of methionine and positions 2 and 8 of the purines are mainly derived from C-2 of acetate (4, 10, 11, 51, 62) (Fig. 2). The labeling pattern in amino acids is consistent with assimilation of acetate via pyruvate involving reactions 6 to 9.
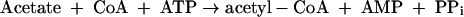 |
6 |
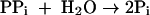 |
7 |
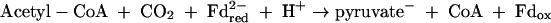 |
8 |
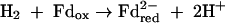 |
9 |
where Fd is a [4Fe-4S]2-containing ferredoxin assumed to have a redox potential identical to that of the acetyl-CoA + CO2/pyruvate couple (Eo′ = −500 mV). All other cell compounds are synthesized from acetyl-CoA and pyruvate.
(i) Pyruvate synthesis from acetate.
The genome of M. stadtmanae harbors CDS for two predicted acyl-CoA synthetases (AMP forming) (Msp0266 and Msp0447), one inorganic pyrophosphatase (ppa; Msp0625), one predicted acyl-CoA synthethase (ADP forming) (Msp0919), one pyruvate:ferredoxin oxidoreductase (PorABCD) (Msp0334 to Msp0337), five ferredoxins with [4Fe-4S]2 (Msp0147, Msp0236, Msp0237, Msp0768, and Msp0861), and one energy-converting hydrogenase (Ehb) (see above). The presence of these gene homologues is consistent with the hypothesis that reactions 6 to 9 are involved in pyruvate synthesis from acetate and CO2. Gene homologues for acetate kinase and phosphotransacetylase were not found.
The reductive carboxylation of acetyl-CoA to pyruvate (Eo′ = −500 mV) with H2 (Eo′ = −420 mV) as the electron donor (reactions 8 and 9) is an endergonic process, which can proceed only when it is pushed by the proton motive force or the sodium motive force. In some methanogens the reductive carboxylation of acetyl-CoA to pyruvate can be coupled to the exergonic oxidation of formylmethanofuran to CO2 (Eo′ = −530 mV) or the exergonic oxidation of CO to CO2 (Eo′ = −520 mV) (91). In M. stadtmanae this is not possible since the organism lacks an active formylmethanofuran dehydrogenase (Fmd) and a CO dehydrogenase (Cdh). However, the genome does contain CDS for an energy-converting hydrogenase, Ehb, which therefore is proposed to catalyze reaction 9. As deduced from the sequence, subunit N (Msp1444) of Ehb harbors the active site [NiFe] center. Interestingly, the ehbABCDEFGHIJKLMNOPQ cluster is interrupted between ehbP and ehbQ by two short open reading frames predicted to encode the iron transport proteins FeoA and FeoB (Msp1441 and Msp1440).
The energy-converting hydrogenase Ehb in M. stadtmanae is also required for other reductive carboxylations. The genome contains genes for the synthesis of 2-oxoglutarate:ferredoxin oxidoreductase (Kor) (korDABC; Msp1390 to Msp1387) in a conserved cluster together with the gene encoding subunit beta of succinyl-CoA synthetase (sucC; Msp1386), indicating that M. stadtmanae can generate 2-oxoglutarate by reductive carboxylation of succinyl-CoA. This organism also contains indolepyruvate:ferredoxin oxidoreductase (Ior) (iorAB; Msp1043 and Msp1042), indicating that it can assimilate indolacetate and phenylacetate by reductive carboxylation. Genes for 2-oxoisovalerate:ferredoxin oxidoreductase (Vor) present in the genomes of other methanogens were not found (94). Vor is involved in the assimilation of branched-chain fatty acids. Consistent with the absence of vor genes in the genome of M. stadtmanae is the finding that growth of this intestinal organism appears not to be dependent on the presence of branched-chain fatty acids (64).
(ii) Serine biosynthesis from pyruvate.
In the genome of M. stadtmanae CDS for pyruvate carboxylase (pycB; Msp1173), phosphoenolpyruvate synthase (ppsA; Msp0328), enolase (eno; Msp0862), two phosphoglycerate mutases (apgM1 or Msp0485 and apgM2 or Msp1299), phosphoglycerate dehydrogenase (serA; Msp1145), and phosphoserine phosphatase (Msp1096) were found, indicating that serine is synthesized from pyruvate via phosphoenolpyruvate, 2-phosphoglycerate, 3-phosphoglycerate, 3-phosphohydroxypyruvate, and 3-phosphoserine as intermediates (Fig. 2). A CDS for a phosphoserine transaminase appears to be absent. The phosphoserine phosphatase gene, however, contains a long 3′ extension, indicating that in M. stadtmanae phosphoserine phosphatase could be a bifunctional enzyme. CDS for pyruvate kinase, pyruvate phosphate dikinase, phosphoenolpyruvate carboxykinase, and phosphoenolpyruvate carboxylase were not detected.
(iii) C1 unit biosynthesis from serine.
A CDS for serine:H4MPT hydroxymethyl transferase (glyA; Msp1475) is present in the genome of M. stadtmanae, indicating that serine reacts with H4MPT, generating methylene-H4MPT and glycine (Fig. 2). H4MPT-dependent hydroxymethyltransferase activity was found in cell extracts of M. stadtmanae (52). Methylene-H4MPT is required for thymidylate synthesis, for synthesis of the methyl group of methionine via methyl-H4MPT, and for generation of formate. The latter compound could be formed via methenyl-H4MPT, formyl-H4MPT, and formylmethanofuran, as described previously for formate synthesis in M. extorquens (72). In M. stadtmanae formate is required for synthesis of C-2 of purines (10) and as an electron donor for the glycyl radical enzyme ribonucleotide reductase (20, 53). C-8 of purines is labeled by C-2 of acetate even in the presence of formate via an unknown pathway (10). The genome harbors genes for thymidylate synthase (thyA; Msp1237) and methionine synthase (metE; Msp1236), but a convincing CDS for the enzyme PurT, which in other methanogens catalyzes the ATP-dependent incorporation of formate into C-2 and C-8 of purines, was not found. The only possible homologue present is carbamoyl phosphate synthetase (carAB; Msp1201 and Msp1200). The genome also does not contain gene homologues for PurN and PurH, which could catalyze the transfer of the formyl group of N10-formyltetrahydrofolate to positions 2 and 8 of purines. This is in agreement with the findings that M. stadtmanae does not contain tetrahydrofolate and that for thermodynamic reasons N5-formyl-H4MPT cannot substitute for N10-formyltetrahydrofolate in the two formyltransferase reactions (8). The genome contains a CDS for a molybdopterin-dependent formate dehydrogenase. However, because of the lack of molybdopterin biosynthetic genes, this enzyme should not be functional, although low NAD(P)-dependent formate dehydrogenase activity has been reported for cell extracts of M. stadtmanae (51). Also, cell suspensions of the organism were shown to slowly oxidize [14C]formate to 14CO2 (51). This is another indication that M. stadtmanae might be able to take up molybdopterin present at trace levels in the yeast extract and/or rumen fluid present in the complex growth medium (see above). Growth of M. stadtmanae is not dependent on the presence of formate (64), indicating that this archaeon must somehow be able to synthesize this C1 unit.
Synthesis of C1 units from C-2 of acetate, as indicated by the labeling studies, does not necessarily have to proceed via C-3 of serine (Fig. 2). In methanogenic archaea 3-hexulose-6-phosphate is the precursor of pentose phosphates required for RNA and DNA biosynthesis (28). From hexulose-6-phosphate ribulose-5-phosphate and formaldehyde are formed in a reaction catalyzed by hexulose phosphate synthase (Hps). The formaldehyde is derived from C-1 of the hexulose phosphate, which in turn is derived from C-2 of acetate via C-3 of pyruvate, C-3 of dihydroxyacetone phosphate, C-1 of fructose bisphosphate, and C-1 of fructose-6-phosphate. The formaldehyde reacts with H4MPT to form methylene-H4MPT, a reaction catalyzed by the formaldehyde-activating enzyme (Fae) (Fig. 2). All the genes required for operation of this pathway are present in the genome of M. stadtmanae. Interestingly, as in other methanogens, the fae gene and the hps gene encoding hexulose phosphate synthase are fused (Msp1498).
(iv) F420-dependent enzymes.
In the genome of M. stadtmanae CDS for the following F420-dependent enzymes are present: F420-reducing hydrogenase (FrhABG); F420-dependent methylene-H4MPT dehydrogenase (Mtd); F420-dependent methylene-H4MPT reductase (Mer) (see above); F420H2:NADP oxidoreductase (Fno) (Msp0665); and F420H2 oxidase (FprA) (Msp0787) (Fig. 2). F420-reducing hydrogenase activity was not detectable in cell extracts of M. stadtmanae, indicating that the frhADBG genes were not expressed or were expressed at only low levels in the organism. The frhD gene is predicted to code for a protease involved in hydrogenase maturation. M. stadtmanae contains relatively low concentrations of F420 (0.16 nmol/mg protein, compared to 6 nmol/mg in M. marburgensis) (103), indicating that F420-dependent reactions are of minor importance quantitatively, which is in agreement with the finding that F420-dependent methylene-H4MPT dehydrogenase (Mtd) and F420-dependent methylene-H4MPT reductase (Mer) are required only for biosynthetic purposes. Likewise, F420H2:NADP oxidoreductase has only an anabolic function, the regeneration of NADPH (Fno) and F420H2 oxidase (FprA), and a function in the detoxification of O2, the reduction of O2 to 2H2O (81). Fno from M. stadtmanae has been purified and characterized (21).
(v) Thiamine and isoleucine/leucine auxotrophy.
In the genome of M. stadtmanae at least one CDS required for thiamine biosynthesis appears to be missing (Table 2). However, all of the genes for biosynthesis of leucine and isoleucine were found. The amino acid auxotrophy of this organism is not understood at this time. With respect to the dependence of growth on leucine or isoleucine, it is interesting that M. stadtmanae appears to lack a CDS for 2-oxoisovalerate:ferredoxin oxidoreductase (Vor), which in M. marburgensis has been shown to exhibit 2-oxoisohexanoate:ferredoxin oxidoreductase activity (94). Therefore, the organism should not be able (as a back-up) to synthesize the two branched-chain amino acids from 2-methylbutyrate and 3-methylbutyrate present in the volatile fatty acid fraction of the rumen fluid generally added to the growth medium of this intestinal inhabitant.
M. stadtmanae-specific genes.
Comparisons of whole-genome protein sequences revealed that the genome of M. stadtmanae contains 323 CDS without genes in the genomes of other archaea (data not shown). Of these, 73 CDS exhibit high levels of overall homology to genes encoding proteins of bacteria and eukaryotes (Table 4). These 73 CDS include 12 CDS which are predicted to code for a group of large proteins with a conserved N-terminal leader sequence and a repetitive structure (Fig. 4), 13 CDS which code for enzymes involved in the biosynthesis of glycosylated cell wall components in bacteria, and 5 CDS which are predicted to code for three subunits of a putative type III restriction-modification (R-M) system and for two subunits of a type I R-M system.
TABLE 4.
CDS in the genome of M. stadtmanae with close gene homologues only in nonarchaeal genomesa
| CDSb | Function | Length (bp) | Accession no. | Organism |
|---|---|---|---|---|
| Large CDS with a repetitive structure | ||||
| Msp0043 | Member of Asn/Thr-rich large protein family | 2,451 | ZP_00356360 | Chloroflexus aurantiacus |
| Msp0046 | Member of Asn/Thr-rich large protein family | 2,646 | CAF23662 | Parachlamydia sp. strain UWE25 |
| Msp0141 | Member of Asn/Thr-rich large protein family | 6,363 | BAB33965 | Escherichia coli O157:H7 |
| YP_189939 | Staphylococcus epidermidis RP62A | |||
| Msp0432 | Member of Asn/Thr-rich large protein family | 6,135 | NP_703779 | Plasmodium falciparum 3D7 (E)c |
| Msp0590 | Member of Asn/Thr-rich large protein family | 8,940 | CAF23662 | Parachlamydia sp. strain UWE25 |
| CAG36249 | Desulfotalea psychrophila LSv54 | |||
| Msp0713 | Member of Asn/Thr-rich large protein family | 6,456 | ZP_00356360 | Chloroflexus aurantiacus |
| Msp0762 | Member of Asn/Thr-rich large protein family | 10,071 | CAF23662 | Parachlamydia sp. strain UWE25 |
| NP_522741 | Ralstonia solanacearum GMI1000 | |||
| Msp0765 | Member of Asn/Thr-rich large protein family | 3,669 | ZP_00519573 | Solibacter usitatus Ellin6076 |
| CAF23662 | Parachlamydia sp. strain UWE25 | |||
| Msp0983 | Member of Asn/Thr-rich large protein family | 3,936 | ZP_00356360 | Chloroflexus aurantiacus |
| CAD71661 | Rhodopirellula baltica SH 1 | |||
| Msp1103 | Member of Asn/Thr-rich large protein family | 2,574 | ZP_00356360 | Chloroflexus aurantiacus |
| Msp1108 | Member of Asn/Thr-rich large protein family | 5,766 | CAF23662 | Parachlamydia sp. strain UWE25 |
| ZP_00525480 | Solibacter usitatus Ellin6076 | |||
| Msp1465 | Member of Asn/Thr-rich large protein family | 6,645 | AAG54838 | Escherichia coli O157:H7 strain EDL933 |
| CAG76164 | Erwinia carotovora subsp. atroseptica SCRI1043 | |||
| CDS involved in bacterial surface antigen biosynthesis | ||||
| Msp0039 | Predicted glycosyltransferase | 1,098 | AAK80269 | Clostridium acetobutylicum ATCC 824 |
| Msp0203 | Predicted glycosyltransferase | 1,959 | ZP_00327237 | Trichodesmium erythraeum IMS101 |
| ZP_00270340 | Rhodospirillum rubrum | |||
| Msp0207 | Predicted glycosyltransferase | 870 | AAR99614 | Geobacillus stearothermophilus |
| AAK80277 | Clostridium acetobutylicum ATCC 824 | |||
| Msp0212 | Predicted glycosyltransferase | 2,667 | AAP76661 | Helicobacter hepaticus ATCC 51449 |
| YP_130861 | Photobacterium profundum SS9 | |||
| Msp0215 | Predicted glycosyltransferase | 2,589 | YP_086549 | Bacillus cereus E33L |
| YP_022183 | Bacillus anthracis Ames Ancestor | |||
| Msp0220 | Predicted glycosyltransferase | 2,748 | NP_964416 | Lactobacillus johnsonii NCC 533 |
| NP_391310 | Bacillus subtilis subsp. subtilis 168 | |||
| Msp0441 | Predicted glycosyltransferase | 1,185 | ZP_00388322 | Streptococcus thermophilus LMD-9 |
| BAB80207 | Clostridium perfringens 13 | |||
| Msp0442 | Predicted glycosyltransferase | 1,077 | NP_816137 | Enterococcus faecalis V583 |
| BAB80207 | Clostridium perfringens 13 | |||
| Msp0495 | Predicted glycosyltransferase | 2,628 | CAB73858 | Campylobacter jejuni subsp. jejuni NCTC 11168 |
| ZP_00143779 | Fusobacterium nucleatum subsp. vincentii ATCC 49256 | |||
| Msp0500 | Predicted glycosyltransferase | 2,211 | ZP_00327237 | Trichodesmium erythraeum IMS101 |
| ZP_00270340 | Rhodospirillum rubrum | |||
| Msp0581 | Predicted UDP-N-acetylmuramyl tripeptide synthase | 1,344 | ZP_00504800 | Clostridium thermocellum ATCC 27405 |
| AAO35161 | Clostridium tetani E88 | |||
| Msp0921 | Putative poly(gamma-glutamate) biosynthesis protein, involved in capsule formation | 1,215 | YP_080918 | Bacillus licheniformis ATCC 14580 |
| NP_391469 | Bacillus subtilis subsp. subtilis 168 | |||
| Msp1540 | Partially conserved hypothetical protein, related to glycosyltransferase | 2,910 | ZP_00327237 | Trichodesmium erythraeum IMS101 |
| ZP_00270340 | Rhodospirillum rubrum | |||
| CDS for restriction-modification systems | ||||
| Msp0454 | Partially conserved hypothetical protein, related to type I restriction-modification system, restriction subunit | 1,350 | CAE10219 | Wolinella succinogenes |
| CAH07535 | Bacteroides fragilis NCTC 9343 | |||
| Msp0470 | Partially conserved hypothetical protein, related to type III restriction-modification system, methylation subunit | 1,182 | ZP_00504330 | Clostridium thermocellum ATCC 27405 |
| ZP_00531584 | Chlorobium phaeobacteroides BS1 | |||
| Msp0472 | Partially conserved hypothetical protein, related to type III restriction-modification system, methylation subunit | 768 | ZP_00504330 | Clostridium thermocellum ATCC 27405 |
| AAL94619 | Fusobacterium nucleatum subsp. nucleatum ATCC 25586 | |||
| Msp0473 | Type III restriction-modification system, restriction subunit | 2,988 | ZP_00504329 | Clostridium thermocellum ATCC 27405 |
| ZP_00143311 | Fusobacterium nucleatum subsp. vincentii ATCC 49256 | |||
| Msp0480 | Putative type I restriction-modification subunit, specificity subunit | 1,218 | YP_185367 | Staphylococcus aureus subsp. aureus COL |
| AAP11371 | Bacillus cereus ATCC 14579 | |||
| Remaining CDS | ||||
| Msp0059 | Conserved hypothetical proteinb | 618 | ZP_00288946 | Magnetococcus sp. strain MC-1 |
| Msp0064 | Predicted acetyltransferase | 609 | AAO75579 | Bacteroides thetaiotaomicron VPI-5482 |
| ZP_00423781 | Burkholderia vietnamiensis G4 | |||
| Msp0093 | Predicted flavoprotein | 1,242 | AAO35881 | Clostridium tetani E88 |
| ZP_00510807 | Clostridium thermocellum ATCC 27405 | |||
| Msp0128 | Predicted helicase | 2,196 | ZP_00357761 | Chloroflexus aurantiacus |
| Msp0140 | Predicted 3′-5′ exonuclease | 1,338 | BAB81107 | Clostridium perfringens 13 |
| NP_691978 | Oceanobacillus iheyensis HTE831 | |||
| Msp0408 | Putative nitroreductase protein | 507 | CAH08994 | Bacteroides fragilis NCTC 9343 |
| NP_661026 | Chlorobium tepidum TLS | |||
| Msp0528 | Putative gamma-glutamylcysteine synthetase | 1,404 | YP_015348 | Listeria monocytogenes 4b F2365 |
| NP_466292 | Listeria monocytogenes EGD-e | |||
| Msp0551 | Putative RNase HIII | 930 | NP_078232 | Ureaplasma parvum serovar 3 strain ATCC 700970 |
| Msp0580 | Predicted glutamine amidotransferase | 723 | AAO35160 | Clostridium tetani E88 |
| NP_621720 | Thermoanaerobacter tengcongensis MB4 | |||
| Msp0591 | Predicted carbonic anhydrase | 468 | AAT41924 | Fremyella diplosiphon |
| YP_113395 | Methylococcus capsulatus Bath | |||
| Msp0688 | Putative secreted RNAse barnase | 450 | AAB29635 | Bacillus circulans |
| YP_080806 | Bacillus licheniformis ATCC 14580 | |||
| Msp0697 | Predicted methyladenine glycosylase | 555 | CAB44429 | Bifidobacterium-longum biovar Longum |
| YP_053518 | Mesoplasma florum L1 | |||
| Msp0700 | Predicted phosphatase | 609 | CAH06545 | Bacteroides fragilis NCTC 9343 |
| AAO79289 | Bacteroides thetaiotaomicron VPI-5482 | |||
| Msp0760 | Predicted penicillin V acylase, choloylglycine hydrolase family | 981 | AAP20760 | Enterococcus faecium |
| NP_465591 | Listeria monocytogenes EGD-e | |||
| Msp0764 | Predicted nicotinate phosphoribosyltransferase | 1,416 | CAB65409 | Suberites domuncula (E) |
| AAH89296 | Xenopus laevis (E) | |||
| Msp0781 | d-Tyrosyl-tRNA(Tyr) deacylase (EC 3.1.-.-) | 447 | AAL94553 | Fusobacterium nucleatum subsp. nucleatum ATCC 25586 |
| NP_622822 | Thermoanaerobacter tengcongensis MB4 | |||
| Msp0797 | Predicted nitroreductase | 558 | AAL95419 | Fusobacterium nucleatum subsp. nucleatum ATCC 25586 |
| Msp0837 | Predicted glutamylcysteine synthetase | 1,413 | ZP_00404497 | Streptococcus pneumoniae TIGR4 |
| AAN59602 | Streptococcus mutans UA159 | |||
| Msp1165 | Predicted hydrolase | 891 | AAB68787 | Fervidobacterium islandicum |
| NP_952175 | Geobacter sulfurreducens PCA |
All CDS of M. stadtmanae encoding more than 100 amino acids were compared by BLAST analysis with all CDS of the genomes of all other archaea sequenced so far (<e−20). CDS without BLAST hits in this group were compared with the NCBI protein database (<e−20) (ftp.ncbi.nih.gov/BLAST/db/nr.tar.gz). All CDS with significant hits in this database are listed.
Additional conserved hypothetical proteins are encoded by Msp0219, Msp0332, Msp0419, Msp0421, Msp0465, Msp0467, Msp0468, Msp0469, Msp0529, Msp0530, Msp0534, Msp0679, Msp0681, Msp0734, Msp0803, Msp0845, Msp0941, Msp1113, Msp1235, Msp1344, Msp1385, Msp1474, Msp1489, and Msp1555.
E, eukaryote.
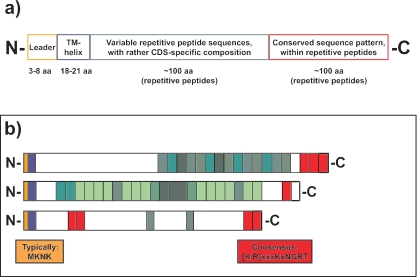
FIG. 4.
Schematic representation of the 37 putative proteins encoded by the group of large CDS with a repetitive structure in M. stadtmanae. (a) General composition shared by all proteins of this group, including an N-terminal leader sequence (yellow) followed by a predicted transmembrane (TM) helix (blue) and by a variable number of repeated peptide sequences (green and red). (b) Schematic composition of the three largest proteins of this group (Msp0762 protein, 3,357 amino acids [aa]; Msp0568 protein, 2,980 aa; Msp0597 protein, 2,469 aa). Two of these, the Msp0568 and Msp0597 proteins, are unique to M. stadtmanae; only the Msp0762 protein exhibits homology to bacterial proteins (Table 4). The short leader sequence is Lys and Asn rich, typically having the form MKNK. At least one copy of a short sequence motif is found at the C terminus of all proteins in this group [G(K/R)XXXKXNGRT]. Sequence variations of repetitive peptide sequences are indicated by green. Repetitive peptides of different large CDS do not necessarily have the same sequence.
(i) Large CDS with repetitive structure.
The 12 large CDS (>2,400 bp and up to 10,071 bp) identified in the whole-genome comparisons are members of a group of 37 CDS found in the genome of M. stadtmanae that share conserved structural features. With the exception of a single CDS (Msp1348) which exhibits partial homology to a CDS from the genome of M. thermautotrophicus, the 26 CDS in this group do not have close homologues either in the archaeal domain or in the two other domains. All 37 CDS have an N-terminal leader sequence, which is usually 3 to 8 amino acids long, followed by a predicted single transmembrane helix consisting of 18 to 21 amino acids and at least one copy of a conserved sequence pattern at the C terminus (Fig. 4). Repetitive peptide sequences that are about 100 amino acids long, whose amino acid compositions and numbers of repeats differ, seem to be a common structural feature of all proteins encoded by the large CDS. In these proteins the amino acids asparagine and threonine are overrepresented by a factor of two. Matches with known signal sequences could not be found. As the large CDS have no counterparts in other archaea sequenced so far but their products exhibit weak fragmentary similarity to proteins with unknown functions from different pathogenic bacteria, functions in the commensal lifestyle are assumed. With respect to the predominance of asparagine and threonine in the proteins encoded by the large CDS, it is interesting that the genome contains one CDS encoding a predicted protein composed of 91 amino acids with 14 tandem repeats having the form Asn-Asn-Ser-Ser (Msp0474).
(ii) CDS involved in bacterial surface antigen biosynthesis.
The genome of M. stadtmanae codes for 38 putative glycosyltransferases involved in the biosynthesis of glycosylated cell wall components. Among these glycosyltransferases are 11 proteins that exhibit high levels of homology to bacterial enzymes predicted to be involved in surface antigen biosynthesis. Furthermore, a predicted UDP-N-acetylmuramyl tripeptide synthase (Msp0581) and a putative poly(gamma-glutamate) biosynthesis protein involved in capsule formation in Bacillus species (Msp0921) indicate that many CDS identified as specific for M. stadtmanae among archaea have functions in the biosynthesis of glycosylated cell wall components. These components could represent an adaptation of this organism to the immunologically highly active intestinal growth habitat. The cell wall of M. stadtmanae is composed of pseudomurein as the major polymer. The pseudomurein appears to be unique with respect to the presence of serine. An S-layer was not found (42).
(iii) CDS for restriction-modification systems.
Quite unusual for archaea, the genome of M. stadtmanae harbors CDS for three putative subunits of a type III restriction-modification system (Msp0470, Msp0472, and Msp0473), which are widely distributed among bacteria, and two additional putative subunits of a type I R-M system (Msp0454 and Msp0480). The function of the R-M systems is to protect the organism from nonhost DNA. Additionally, an effect of DNA methylation on gene regulation in bacterial pathogens has been established (41).
Conclusion.
Analysis of the genome of M. stadtmanae in relation to the well-studied physiology and biochemistry of this organism allowed us to correlate the genotype with the phenotype of this methanogenic archaeon. Thus, the genetic data can explain why M. stadtmanae is restricted to H2 and methanol as energy sources and is dependent on acetate as a carbon source. Both the CDS found and the CDS not found compared to other methanogenic archaea helped to identify the proteins involved in methanol reduction to methane with H2, to predict how these proteins topologically interact, and to locate the site of energy conservation in subunit B of the HdrABC complex. For methanogens without cytochromes this has not been possible previously. In this respect our decision to determine the genome sequence of M. stadtmanae was rewarding. However, this was also true in other respects. The genome of the organism was found to harbor a number of CDS not present in the genome of any other archaeon sequenced so far. Many of these CDS are predicted to be involved in cell surface antigen synthesis. It is tempting to speculate that specific components of the archaeal cell wall are part of the adaptation of M. stadtmanae to its special habitat, the human large intestine. Lateral gene transfer between this archaeon and bacteria could be involved, as has been suggested for M. mazei and bacteria for another habitat (15). Such adaptations could result in phenotypic variability due to expression and glycosylation of special cell wall components. Similar mechanisms have been found in pathogenic bacteria that use them to avoid the human innate immune response. Large, tandem repeating units of surface proteins have been identified in group B streptococci and other gram-negative pathogens (61, 69). In the gram-negative bacterial pathogen Anaplasma marginale variation of cell wall structures is achieved by recombinatorial gene conversions between whole pseudogenes and between small segments within these genes (6). In analogy to A. marginale, short repetitive sequences within the large CDS of the M. stadtmanae genome could serve as homologous sites for segmental recombination. In M. stadtmanae proteins analogous to enzymes for the biosynthesis of cell wall components in gram-negative and gram-positive bacteria could serve as the machinery to glycosylate proteins encoded by the group of large CDS.
Acknowledgments
This work was supported by the Max Planck Society and by the Fonds der Chemischen Industrie (grants to R.K.T.) and by a grant from the Niedersächsisches Ministerium für Wissenschaft und Kultur (to G.G.).
We thank Rainer Merkl, Universität Regensburg, for providing the SIGI tool and for advice about using it and Mechthild Pohlschröder, University of Pennsylvania, for providing the TATFIND 1.2 search program.
Footnotes
Dedicated to Terry Miller.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 3.Baliga, N. S., R. Bonneau, M. T. Facciotti, M. Pan, G. Glusman, E. W. Deutsch, P. Shannon, Y. L. Chiu, R. R. Gan, P. L. Hung, S. V. Date, E. Marcotte, L. Hood, and W. V. Ng. 2004. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 14:2221-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bank, S., B. Yan, and T. L. Miller. 1996. Solid C13 CPMAS NMR spectroscopy studies of biosynthesis in whole cells of Methanosphaera stadtmanae. Solid State Nucl. Mag. Reson. 7:253-261. [DOI] [PubMed] [Google Scholar]
- 5.Bott, M. H., B. Eikmanns, and R. K. Thauer. 1985. Defective formation and/or utilization of carbon-monoxide in H2Co2 fermenting methanogens dependent on acetate as carbon source. Arch. Microbiol. 143:266-269. [Google Scholar]
- 6.Brayton, K. A., G. H. Palmer, A. Lundgren, J. Yi, and A. F. Barbet. 2002. Antigenic variation of Anaplasma marginale msp2 occurs by combinatorial gene conversion. Mol. Microbiol. 43:1151-1159. [DOI] [PubMed] [Google Scholar]
- 7.Brioukhanov, A., A. Netrusov, M. Sordel, R. K. Thauer, and S. Shima. 2000. Protection of Methanosarcina barkeri against oxidative stress: identification and characterization of an iron superoxide dismutase. Arch. Microbiol. 174:213-216. [DOI] [PubMed] [Google Scholar]
- 8.Buchenau, B., and R. K. Thauer. 2004. Tetrahydrofolate-specific enzymes in Methanosarcina barkeri and growth dependence of this methanogenic archaeon on folic acid or p-aminobenzoic acid. Arch. Microbiol. 182:313-325. [DOI] [PubMed] [Google Scholar]
- 9.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 10.Choquet, C. G., J. C. Richards, G. B. Patel, and G. D. Sprott. 1994. Purine and pyrimidine biosynthesis in methanogenic bacteria. Arch. Microbiol. 161:471-480. [Google Scholar]
- 11.Choquet, C. G., J. C. Richards, G. B. Patel, and G. D. Sprott. 1994. Ribose biosynthesis in methanogenic bacteria. Arch. Microbiol. 161:481-488. [Google Scholar]
- 12.Cohen, G. N., V. Barbe, D. Flament, M. Galperin, R. Heilig, O. Lecompte, O. Poch, D. Prieur, J. Querellou, R. Ripp, J. C. Thierry, J. Van der Oost, J. Weissenbach, Y. Zivanovic, and P. Forterre. 2003. An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Microbiol. 47:1495-1512. [DOI] [PubMed] [Google Scholar]
- 13.Daas, P. J. H., R. W. Wassenaar, P. Willemsen, R. J. Theunissen, J. T. Keltjens, C. vanderDrift, and G. D. Vogels. 1996. Purification and properties of an enzyme involved in the ATP-dependent activation of the methanol:2-mercaptoethanesulfonic acid methyltransferase reaction in Methanosarcina barkeri. J. Biol. Chem. 271:22339-22345. [DOI] [PubMed] [Google Scholar]
- 14.Deppenmeier, U. 2004. The membrane-bound electron transport system of Methanosarcina species. J. Bioenerg. Biomembr. 36:55-64. [DOI] [PubMed] [Google Scholar]
- 15.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Baumer, C. Jacobi, H. Bruggemann, T. Lienard, A. Christmann, M. Bomeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453-461. [PubMed] [Google Scholar]
- 16.Deppenmeier, U., T. Lienard, and G. Gottschalk. 1999. Novel reactions involved in energy conservation by methanogenic archaea. FEBS Lett. 457:291-297. [DOI] [PubMed] [Google Scholar]
- 17.Deppenmeier, U., V. Müller, and G. Gottschalk. 1996. Pathways of energy conservation in methanogenic archaea. Arch. Microbiol. 165:149-163. [Google Scholar]
- 18.Dongowski, G., A. Lorenz, and H. Anger. 2000. Degradation of pectins with different degrees of esterification by Bacteroides thetaiotaomicron isolated from human gut flora. Appl. Environ Microbiol. 66:1321-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duin, E. C., C. Bauer, B. Jaun, and R. Hedderich. 2003. Coenzyme M binds to a [4Fe-4S] cluster in the active site of heterodisulfide reductase as deduced from EPR studies with the [33S]coenzyme M-treated enzyme. FEBS Lett. 538:81-84. [DOI] [PubMed] [Google Scholar]
- 20.Eklund, H., U. Uhlin, M. Farnegardh, D. T. Logan, and P. Nordlund. 2001. Structure and function of the radical enzyme ribonucleotide reductase. Prog. Biophys. Mol. Biol. 77:177-268. [DOI] [PubMed] [Google Scholar]
- 21.Elias, D. A., D. F. Juck, K. A. Berry, and R. Sparling. 2000. Purification of the NADP+:F420 oxidoreductase of Methanosphaera stadtmanae. Can. J. Microbiol. 46:998-1003. [PubMed] [Google Scholar]
- 22.Ermler, U., W. Grabarse, S. Shima, M. Goubeaud, and R. K. Thauer. 1997. Crystal structure of methyl coenzyme M reductase: the key enzyme of biological methane formation. Science 278:1457-1462. [DOI] [PubMed] [Google Scholar]
- 23.Fischer, R., P. Gartner, A. Yeliseev, and R. K. Thauer. 1992. N5-Methyltetrahydromethanopterin-coenzyme-M methyltransferase in methanogenic archaebacteria is a membrane-protein. Arch. Microbiol. 158:208-217. [DOI] [PubMed] [Google Scholar]
- 24.Fitz-Gibbon, S. T., H. Ladner, U. J. Kim, K. O. Stetter, M. I. Simon, and J. H. Miller. 2002. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. USA 99:984-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Florin, T. H. J., G. Zhu, K. M. Kirk, and N. G. Martin. 2000. Shared and unique environmental factors determine the ecology of methanogens in humans and rats. Am. J. Gastroenterol. 95:2872-2879. [DOI] [PubMed] [Google Scholar]
- 26.Fütterer, O., A. Angelov, H. Liesegang, G. Gottschalk, C. Schleper, B. Schepers, C. Dock, G. Antranikian, and W. Liebl. 2004. Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proc. Natl. Acad. Sci. USA 101:9091-9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goenrich, M., R. K. Thauer, H. Yurimoto, and N. Kato. 2005. Formaldehyde activating enzyme (Fae) and hexulose-6-phosphate synthase (Hps) in Methanosarcina barkeri: a possible function in ribose-5-phosphate biosynthesis. Arch. Microbiol. 184:41-48. [DOI] [PubMed] [Google Scholar]
- 29.Gottschalk, G., and R. K. Thauer. 2001. The Na+-translocating methyltransferase complex from methanogenic archaea. Biochim. Biophys. Acta 1505:28-36. [DOI] [PubMed] [Google Scholar]
- 30.Grabarse, W. G., F. Mahlert, S. Shima, R. K. Thauer, and U. Ermler. 2000. Comparison of three methyl-coenzyme M reductases from phylogenetically distant organisms: unusual amino acid modification, conservation and adaptation. J. Mol. Biol. 303:329-344. [DOI] [PubMed] [Google Scholar]
- 31.Hamilton, P. T., and J. N. Reeve. 1985. Structure of genes and an insertion element in the methane producing archaebacterium Methanobrevibacter smithii. Mol. Gen. Genet. 200:47-59. [DOI] [PubMed] [Google Scholar]
- 32.Harms, U., and R. K. Thauer. 1997. Identification of the active site histidine in the corrinoid protein MtrA of the energy-conserving methyltransferase complex from Methanobacterium thermoautotrophicum. Eur. J. Biochem. 250:783-788. [DOI] [PubMed] [Google Scholar]
- 33.Hedderich, R. 2004. Energy-converting [NiFe] hydrogenases from archaea and extremophiles: ancestors of complex I. J. Bioenerg. Biomembr. 36:65-75. [DOI] [PubMed] [Google Scholar]
- 34.Hedderich, R., S. P. Albracht, D. Linder, J. Koch, and R. K. Thauer. 1992. Isolation and characterization of polyferredoxin from Methanobacterium thermoautotrophicum. The mvhB gene product of the methylviologen-reducing hydrogenase operon. FEBS Lett. 298:65-68. [DOI] [PubMed] [Google Scholar]
- 35.Hedderich, R., O. Klimmek, A. Kröger, R. Dirmeier, M. Keller, and K. O. Stetter. 1998. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol. Rev. 22:353-381. [Google Scholar]
- 36.Hedderich, R., J. Koch, D. Linder, and R. K. Thauer. 1994. The heterodisulfide reductase from Methanobacterium thermoautotrophicum contains sequence motifs characteristic of pyridine nucleotide-dependent thioredoxin reductases. Eur. J. Biochem. 225:253-261. [DOI] [PubMed] [Google Scholar]
- 37.Hellmer, J., R. Patzold, and C. Zeilinger. 2002. Identification of a pH regulated Na+/H+ antiporter of Methanococcus jannaschii. FEBS Lett. 527:245-249. [DOI] [PubMed] [Google Scholar]
- 38.Hendrickson, E. L., R. Kaul, Y. Zhou, D. Bovee, P. Chapman, J. Chung, E. Conway de Macario, J. A. Dodsworth, W. Gillett, D. E. Graham, M. Hackett,A. K. Haydock, A. Kang, M. L. Land, R. Levy, T. J. Lie, T. A. Major, B. C. Moore, I. Porat, A. Palmeiri, G. Rouse, C. Saenphimmachak, D. Soll, S. Van Dien, T. Wang, W. B. Whitman, Q. Xia, Y. Zhang, F. W. Larimer, M. V. Olson, and J. A. Leigh. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 186:6956-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Biol Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 40.Jensen, N. S., and E. Canale-Parola. 1986. Bacteroides pectinophilus sp. nov. and Bacteroides galacturonicus sp. nov.: two pectinolytic bacteria from the human intestinal tract. Appl. Environ. Microbiol. 52:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Julio, S. M., D. M. Heithoff, D. Provenzano, K. E. Klose, R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2001. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect. Immun. 69:7610-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kandler, O., and H. Konig. 1998. Cell wall polymers in Archaea (Archaebacteria). Cell. Mol. Life Sci. 54:305-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawarabayasi, Y., Y. Hino, H. Horikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Kato, T. Yoshizawa, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, S. Masuda, M. Yanagii, M. Nishimura, A. Yamagishi, T. Oshima, and H. Kikuchi. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Res. 8:123-140. [DOI] [PubMed] [Google Scholar]
- 44.Kawarabayasi, Y., Y. Hino, H. Horikawa, S. Yamazaki, Y. Haikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, H. Nakazawa, M. Takamiya, S. Masuda, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, H. Kikuchi, et al. 1999. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6:83-101. [DOI] [PubMed] [Google Scholar]
- 45.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, M. Sekine, S. Baba, H. Kosugi, A. Hosoyama, Y. Nagai, M. Sakai, K. Ogura, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Ohfuku, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, and H. Kikuchi. 1998. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5:55-76. [DOI] [PubMed] [Google Scholar]
- 46.Kawashima, T., N. Amano, H. Koike, S. Makino, S. Higuchi, Y. Kawashima-Ohya, K. Watanabe, M. Yamazaki, K. Kanehori, T. Kawamoto, T. Nunoshiba, Y. Yamamoto, H. Aramaki, K. Makino, and M. Suzuki. 2000. Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium. Proc. Natl. Acad. Sci. USA 97:14257-14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kezmarsky, N. D., H. Xu, D. E. Graham, and R. H. White. 2005. Identification and characterization of a l-tyrosine decarboxylase in Methanocaldococcus jannaschii. Biochim. Biophys. Acta 1722:175-182. [DOI] [PubMed] [Google Scholar]
- 48.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, and J. C. Venter. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 49.Krüer, M. 2003. Methanol:Coenzym-M-Methyltransferase: Zum Mechanismus der Substraktivierung und zur Funktion in Methanosphaera stadtmanae. Ph.D dissertation. Philipps-Universität, Marburg, Germany.
- 50.Lemos, R. S., C. M. Gomes, and M. Teixeira. 2001. Acidianus ambivalens complex II typifies a novel family of succinate dehydrogenases. Biochem. Biophys. Res. Commun. 281:141-150. [DOI] [PubMed] [Google Scholar]
- 51.Lin, Z., and R. Sparling. 1995. Oxidation-reduction of methanol, formaldehyde, serine, and formate in Methanosphaera stadtmanae using C14 short-term and long-term labeling. Can. J. Microbiol. 41:1048-1053. [Google Scholar]
- 52.Lin, Z. S., and R. Sparling. 1998. Investigation of serine hydroxymethyltransferase in methanogens. Can. J. Microbiol. 44:652-656. [DOI] [PubMed] [Google Scholar]
- 53.Logan, D. T., E. Mulliez, K. M. Larsson, S. Bodevin, M. Atta, P. E. Garnaud, B. M. Sjoberg, and M. Fontecave. 2003. A metal-binding site in the catalytic subunit of anaerobic ribonucleotide reductase. Proc. Natl. Acad. Sci. USA 100:3826-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyon, E. J., S. Shima, G. Buurman, S. Chowdhuri, A. Batschauer, K. Steinbach, and R. K. Thauer. 2004. UV-A/blue-light inactivation of the ‘metal-free’ hydrogenase (Hmd) from methanogenic archaea. Eur. J. Biochem. 271:195-204. [DOI] [PubMed] [Google Scholar]
- 55.Macdonald, T. T., and G. Monteleone. 2005. Immunity, inflammation, and allergy in the gut. Science 307:1920-1925. [DOI] [PubMed] [Google Scholar]
- 56.Madadi-Kahkesh, S., E. C. Duin, S. Heim, S. P. Albracht, M. K. Johnson, and R. Hedderich. 2001. A paramagnetic species with unique EPR characteristics in the active site of heterodisulfide reductase from methanogenic archaea. Eur. J. Biochem. 268:2566-2577. [DOI] [PubMed] [Google Scholar]
- 57.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mazmanian, S. K., C. H. Liu, A. O. Tzianabos, and D. L. Kasper. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107-118. [DOI] [PubMed] [Google Scholar]
- 59.Merkl, R. 2004. SIGI: score-based identification of genomic islands. BMC Bioinformatics 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meuer, J., H. C. Kuettner, J. K. Zhang, R. Hedderich, and W. W. Metcalf. 2002. Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc. Natl. Acad. Sci. USA 99:5632-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michel, J. L., L. C. Madoff, K. Olson, D. E. Kling, D. L. Kasper, and F. M. Ausubel. 1992. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc. Natl. Acad. Sci. USA 89:10060-10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller, T. L., X. Chen, B. Yan, and S. Bank. 1995. Solution 13C nuclear magnetic resonance spectroscopic analysis of the amino acids of Methanosphaera stadtmanae: biosynthesis and origin of one-carbon units from acetate and carbon dioxide. Appl. Environ. Microbiol. 61:1180-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller, T. L., and C. Z. Lin. 2002. Description of Methanobrevibacter gottschalkii sp. nov., Methanobrevibacter thaueri sp. nov., Methanobrevibacter woesei sp. nov. and Methanobrevibacter wolinii sp. nov. Int. J. Syst. Evol. Microbiol. 52:819-822. [DOI] [PubMed] [Google Scholar]
- 64.Miller, T. L., and M. J. Wolin. 1985. Methanosphaera stadtmaniae gen. nov., sp. nov.—a species that forms methane by reducing methanol with hydrogen. Arch. Microbiol. 141:116-122. [DOI] [PubMed] [Google Scholar]
- 65.Miller, T. L., and M. J. Wolin. 1983. Oxidation of hydrogen and reduction of methanol to methane is the sole energy source for a methanogen isolated from human feces. J. Bacteriol. 153:1051-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller, T. L., M. J. Wolin, E. C. Demacario, and A. J. L. Macario. 1982. Isolation of Methanobrevibacter smithii from human feces. Appl. Environ. Microbiol. 43:227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Müller, V. 2004. An exceptional variability in the motor of archael A1A0 ATPases: from multimeric to monomeric rotors comprising 6-13 ion binding sites. J. Bioenerg. Biomembr. 36:115-125. [DOI] [PubMed] [Google Scholar]
- 68.Müller, V., M. Blaut, and G. Gottschalk. 1986. Utilization of methanol plus hydrogen by Methanosarcina barkeri for methanogenesis and growth. Appl. Environ. Microbiol. 52:269-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pfaltz, A., A. Kobelt, R. Hüster, and R. K. Thauer. 1987. Biosynthesis of coenzyme F430 in methanogenic bacteria. Identification of 15,173-seco-F430-173-acid as an intermediate. Eur. J. Biochem. 170:459-467. [DOI] [PubMed] [Google Scholar]
- 72.Pomper, B. K., O. Saurel, A. Milon, and J. A. Vorholt. 2002. Generation of formate by the formyltransferase/hydrolase complex (Fhc) from Methylobacterium extorquens AM1. FEBS Lett. 523:133-137. [DOI] [PubMed] [Google Scholar]
- 73.Pritchett, M. A., and W. W. Metcalf. 2005. Genetic, physiological and biochemical characterization of multiple methanol methyltransferase isozymes in Methanosarcina acetivorans C2A. Mol. Microbiol. 56:1183-1194. [DOI] [PubMed] [Google Scholar]
- 74.Reeve, J. N., G. S. Beckler, D. S. Cram, P. T. Hamilton, J. W. Brown, J. A. Krzycki, A. F. Kolodziej, L. Alex, W. H. Orme-Johnson, and C. T. Walsh. 1989. A hydrogenase-linked gene in Methanobacterium thermoautotrophicum strain ΔH encodes a polyferredoxin. Proc. Natl. Acad. Sci. USA 86:3031-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reiß, R. 2002. Zum Verständnis des Energiestoffwechsels von Methanosphaera stadtmanae: Reinigung und biochemische Charakterisierung von Heterodisulfid-Reduktase und Hydrogenase. Diploma thesis. Phillips-Universität, Marburg, Germany.
- 76.Robb, F. T., D. L. Maeder, J. R. Brown, J. DiRuggiero, M. D. Stump, R. K. Yeh, R. B. Weiss, and D. M. Dunn. 2001. Genomic sequence of a hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 330:134-157. [DOI] [PubMed] [Google Scholar]
- 77.Ruepp, A., W. Graml, M. L. Santos-Martinez, K. K. Koretle, C. Volker, H. W. Mewes, D. Frishman, S. Stocker, A. N. Lupas, and W. Baumeister. 2000. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407:508-513. [DOI] [PubMed] [Google Scholar]
- 78.Sauer, K., and R. K. Thauer. 1999. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri—substitution of the corrinoid harbouring subunit MtaC by free cob(I)alamin. Eur. J. Biochem. 261:674-681. [DOI] [PubMed] [Google Scholar]
- 79.Sauer, K., and R. K. Thauer. 1999. The role of corrinoids in methanogenesis, p. 655-679. In R. Banerjee (ed.), Chemistry and biochemistry of B12. John Wiley & Sons, Inc., New York, N.Y.
- 80.Schwörer, B., and R. K. Thauer. 1991. Activities of formylmethanofuran dehydrogenase, methylenetetrahydromethanopterin dehydrogenase, methylenetetrahydromethanopterin reductase, and heterodisulfide reductase in methanogenic bacteria. Arch. Microbiol. 155:459-465. [Google Scholar]
- 81.Seedorf, H., A. Dreisbach, R. Hedderich, S. Shima, and R. K. Thauer. 2004. F420H2 oxidase (FprA) from Methanobrevibacter arboriphilus, a coenzyme F420-dependent enzyme involved in O2 detoxification. Arch. Microbiol. 182:126-137. [DOI] [PubMed] [Google Scholar]
- 82.Setzke, E., R. Hedderich, S. Heiden, and R. K. Thauer. 1994. H2:heterodisulfide oxidoreductase complex from Methanobacterium thermoautotrophicum: composition and properties. Eur. J. Biochem. 220:139-148. [DOI] [PubMed] [Google Scholar]
- 83.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Y. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. K. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shima, S., M. Sordel-Klippert, A. Brioukhanov, A. Netrusov, D. Linder, and R. K. Thauer. 2001. Characterization of a heme-dependent catalase from Methanobrevibacter arboriphilus. Appl. Environ. Microbiol. 67:3041-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Slesarev, A. I., K. V. Mezhevaya, K. S. Makarova, N. N. Polushin, O. V. Shcherbinina, V. V. Shakhova, G. I. Belova, L. Aravind, D. A. Natale, I. B. Rogozin, R. L. Tatusov, Y. I. Wolf, K. O. Stetter, A. G. Malykh, E. V. Koonin, and S. A. Kozyavkin. 2002. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. USA 99:4644-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sparling, R., M. Blaut, and G. Gottschalk. 1993. Bioenergetic studies of Methanosphaera stadtmanae, an obligate H2-methanol utilizing methanogen. Can. J. Microbiol. 39:742-748. [Google Scholar]
- 88.Sparling, R., L. T. Holth, and Z. S. Lin. 1993. Sodium-ion dependent active-transport of leucine in Methanosphaera stadtmanae. Can. J. Microbiol. 39:749-753. [Google Scholar]
- 89.Sprott, G. D., J. R. Brisson, C. J. Dicaire, A. K. Pelletier, L. A. Deschatelets, L. Krishnan, and G. B. Patel. 1999. A structural comparison of the total polar lipids from the human archaea Methanobrevibacter smithii and Methanosphaera stadtmanae and its relevance to the adjuvant activities of their liposomes. Biochim. Biophys. Acta 1440:275-288. [DOI] [PubMed] [Google Scholar]
- 90.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol 132:115-130. [DOI] [PubMed] [Google Scholar]
- 91.Stojanowic, A., and R. Hedderich. 2004. CO2 reduction to the level of formylmethanofuran in Methanosarcina barkeri is non-energy driven when CO is the electron donor. FEMS Microbiol. Lett. 235:163-167. [DOI] [PubMed] [Google Scholar]
- 92.Stojanowic, A., G. J. Mander, E. C. Duin, and R. Hedderich. 2003. Physiological role of the F420-non-reducing hydrogenase (Mvh) from Methanothermobacter marburgensis. Arch. Microbiol. 180:194-203. [DOI] [PubMed] [Google Scholar]
- 93.Tech, M., and R. Merkl. 2003. YACOP: enhanced gene prediction obtained by a combination of existing methods. In Silico Biol. 3:441-451. [PubMed] [Google Scholar]
- 94.Tersteegen, A., D. Linder, R. K. Thauer, and R. Hedderich. 1997. Structures and functions of four anabolic 2-oxoacid oxidoreductases in Methanobacterium thermoautotrophicum. Eur. J. Biochem. 244:862-868. [DOI] [PubMed] [Google Scholar]
- 95.Thauer, R. K. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144:2377-2406. [DOI] [PubMed] [Google Scholar]
- 96.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tietze, M., A. Beuchle, I. Lamla, N. Orth, M. Dehler, G. Greiner, and U. Beifuss. 2003. Redox potentials of methanophenazine and CoB-S-S-CoM, factors involved in electron transport in methanogenic archaea. Chembiochem 4:333-335. [DOI] [PubMed] [Google Scholar]
- 98.van de Wijngaard, W. M., J. Creemers, G. D. Vogels, and C. van der Drift. 1991. Methanogenic pathways in Methanosphaera stadtmanae. FEMS Microbiol. Lett. 64:207-211. [DOI] [PubMed] [Google Scholar]
- 99.Vorholt, J. A., M. Vaupel, and R. K. Thauer. 1997. A selenium-dependent and a selenium-independent formylmethanofuran dehydrogenase and their transcriptional regulation in the hyperthermophilic Methanopyrus kandleri. Mol. Microbiol. 23:1033-1042. [DOI] [PubMed] [Google Scholar]
- 100.Welander, P. V., and W. W. Metcalf. 2005. Loss of the mtr operon in Methanosarcina blocks growth on methanol, but not methanogenesis, and reveals an unknown methanogenic pathway. Proc. Natl. Acad. Sci. USA 102:10664-10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wennerhold, J. 2004. Heterodisulfid-Reduktasen des Methanothermobacter-Typs in Methanosarcina barkeri und Untersuchungen zur Rolle des H2:Heterodisulfid-Oxidoreduktase-Komplex bei der Energiekonservierung in Methanothermobacter marburgensis. Diploma thesis. Philipps-Universität, Marburg, Germany.
- 102.Wolfe, R. S. 1996. 1776-1996: Alessandro Volta's combustible air. 220 years after Volta's experiments, the microbial formation of methane approaches an understanding. ASM News 62:529-534. [Google Scholar]
- 103.Wong, D., Z. S. Lin, D. F. Juck, K. A. Terrick, and R. Sparling. 1994. Electron-transfer reactions for the reduction of NADP+ in Methanosphaera stadtmanae. FEMS Microbiol. Lett. 120:285-290. [Google Scholar]