Abstract
A mutation in the rsaL gene of Pseudomonas aeruginosa produces dramatically higher amounts of N-acyl homoserine lactone with respect to the wild type, highlighting the key role of this negative regulator in controlling quorum sensing (QS) in this opportunistic pathogen. The DNA binding site of the RsaL protein on the rsaL-lasI bidirectional promoter partially overlaps the binding site of the LasR protein, consistent with the hypothesis that RsaL and LasR could be in binding competition on this promoter. This is the first direct demonstration that RsaL acts as a QS negative regulator by binding to the lasI promoter.
Quorum sensing (QS) is a widespread bacterial intercellular communication system based on the production of signal molecules of which the extra cellular concentration is related to cell density. Individual cells sense the presence of the signal molecule, which allows the whole bacterial population to control gene expression in response to cell density. This form of regulation controls a diverse range of phenotypes, including various pathogenicity determinants, conjugation, biofilm formation, and production of antibiotics and secondary metabolites (16, 27, 31).
Several gram-negative bacteria use acylated homoserine lactones (acyl-HSL) as signal molecules in quorum sensing. The Pseudomonas aeruginosa QS system is one of the most extensively studied within this group of bacteria, reflecting the importance of this microorganism as an opportunistic pathogen of humans, other animals, and plants (10, 15). P. aeruginosa possesses two homologous QS systems encoded by the lasR/lasI and rhlR/rhlI gene pairs. The lasI and rhlI genes encode acyl-HSL synthase enzymes (LasI and RhlI) responsible for the synthesis of the N-3-oxo-dodecanoyl homoserine lactone (3OC12-HSL) and N-butyryl homoserine lactone (C4-HSL) signal molecules, respectively. The lasR and rhlR genes encode the regulators (i.e., LasR and RhlR) that respond to their cognate signals (i.e., 3OC12-HSL and C4-HSL) and activate transcription of lasI and rhlI, respectively, thus creating a positive feedback loop. Moreover, the two P. aeruginosa quorum-sensing systems are organized in a hierarchical manner, where the RhlR/RhlI system is subordinate to the LasR/LasI system, since expression of rhlR and rhlI is dependent upon LasR (18, 24).
In P. aeruginosa, QS is a major global regulatory system that has been estimated to control approximately 5% of the genes, including the most important virulence genes (21, 26). Indeed, P. aeruginosa quorum-sensing null mutants are severely impaired in virulence in all the infection model systems examined (reviewed in reference 24).
The P. aeruginosa QS system is intricately connected with other cellular global regulatory networks, since it is regulated by a number of regulatory factors, including Vfr (1), GacA (20), the LuxR homologues QscR and VqsR (3, 11), MvaT (5), the alternative sigma factors RpoS and RpoN (8, 29), PprB (6), RsmA (19), DksA (9), and RsaL (4). The additional regulation of QS by the above-mentioned factors most likely affects the timing of the response and increases the range of environmental and metabolic signals, in addition to cell-density, to which QS responds. The mode of action of these regulators in most cases is unknown, and with the exception of LasR (22), none have been shown to directly regulate lasI gene expression.
This study is focused on the RsaL protein encoded by the rsaL gene located between the lasR and lasI genes (Fig. 1). Overexpression of rsaL in P. aeruginosa PAO1 resulted in a 20-fold reduction of 3OC12-HSL production (4). However, the effect of an rsaL mutation on 3OC12-HSL synthesis in P. aeruginosa has never been investigated. Therefore, we compared 3OC12-HSL production in P. aeruginosa PAO1 and its rsaLderivative mutant. A P. aeruginosa rsaL transposon insertion mutant was supplied by the University of Washington Genome Center (www.genome.washington.edu/UWGC/pseudomonas) (Tables 1 and 2). The lactones produced by both strains were extracted and separated by thin-layer chromatography (TLC) and revealed by overlaying the TLC plate with a thin layer of Luria Bertani top agar seeded with Escherichia coli(pSB1075), a sensor strain for the detection of 3OC12-HSL and 3OC10-HSL (30). Results showed that 3OC12-HSL production is dramatically enhanced in the rsaL mutant (Fig. 2). Moreover, wild-type 3OC12-HSL levels were restored upon complementation of the rsaL mutant with the plasmid pPSRsaLPAO (Tables 1 and 2), expressing the rsaL gene, confirming that the mutated phenotype was due to rsaL inactivation and ruling out the occurrence of polar effects due to transposon insertion (Fig. 2). This result is consistent with what was recently observed in the rhizosphere-colonizing plant growth-promoting organism Pseudomonas putida WCS358, which has a QS genetic locus highly homologous to the las system of P. aeruginosa and organized in the same way (2). A P. putida WCS358 rsaL mutant discloses strong enhancement of the transcriptional activity of the ppuI gene (the lasI orthologue), producing dramatically higher levels of acyl-HSL with respect to the wild type (2).
FIG. 1.
Genetic organization of the P. aeruginosa las quorum sensing locus and detail of the rsaL-lasI bidirectional promoter (below). The lasR, rsaL, and lasI genes are depicted as arrows. Bent arrows indicate the rsaL- and lasI- divergent transcripts. The open box and the gray box indicate the LasR and RsaL binding sites, respectively. Numbers indicate the center of the LasR and RsaL binding sites with respect to the lasI transcription starting site (22).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics and plasmid construction | Reference or source |
|---|---|---|
| Strain | ||
| P. aeruginosa PAO1 | Wild type | American Type Culture Collection |
| P. aeruginosa PAO1 rsaL | rsaL::ISlacZ/hah; Tetr | University of Washington Genome Center |
| E. coli DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 (Nalr) relA1 Δ(lacIZYA-argF)U169 deoR(φ80dlacZΔM15) | Bethesda Research Laboratories |
| E. coli BL21(DE3)pLysS | High-stringency expression host; Chlr | Novagen |
| E. coli(pBS1075) | Acyl-HSL sensor strain; Ampr | 30 |
| Plasmid | ||
| pBluescript II KS+ | Cloning vector; Ampr | Stratagene |
| pDrive | TA-based cloning vector; Kanr, Ampr | QIAGEN |
| pET28b | Expression vector, Kanr | Novagen |
| pBBR1MCS-5 | Broad-host-range vector; Genr | 12 |
| pRsaLPAO6H | pET28b derivative in which a 255-bp PCR fragment, originated with primers P133FW and P134RV (Table 2), was cloned into the NcoI-HindIII sites of the vector; allows the overexpression in E. coli of a recombinant RsaL protein carrying a six-histidine tag at the C terminus | This study |
| pRsaLPAOWT | pET28b derivative in which a 258-bp PCR fragment originated with primers P133FW and P136RV (Table 2), was cloned into the NcoI-HindIII sites of the vector; allows the overexpression in E. coli of the wild-type RsaL protein | This study |
| pPSRsaLPAO | PCR fragment encompassing the wild-type rsaL gene was originated with primers P0FW and P0RV and plasmid pRsaLPAOWT as the template (Table 2) and was cloned into the KpnI-BamHI sites of pBBR1MCS-5 | This study |
| pPlasIS | pDrive derivative containing a 103-bp PCR fragment, originated with primers P193FW and P194RV (Table 2) | This study |
| pPlasI5′ | pBluescript II KS+ derivative in which a 313-bp PCR fragment, originated with primers P137FW and P138RV (Table 2), was cloned into the EcoRI-BamHI sites of the vector | This study |
| pPlasI3′ | pBluescript II KS+ derivative in which a 313-bp PCR fragment, originated with primers P139FW and P138RV (Table 2), was cloned into the XbaI-BamHI sites of the vector | This study |
TABLE 2.
Oligonucleotides used in this study
| Namea | Sequence (5′-3′)b | Positionc | Sited |
|---|---|---|---|
| P133FW | 5′-CATGCCATGGCTTCACACGAGAGAA-3′ | +1 | NcoI |
| P134RV | 5′-CCCAAGCTTCTCTCTGATCTTGCCTCTC-3′ | +240 | HindIII |
| P136RV | 5′-CCCAAGCTTTTACTCTCTGATCTTGCCTC-3′ | +243 | HindIII |
| P137FW | 5′-CGGAATTCGGTGGCCTTTGCCCGGA-3′ | +57 | EcoRI |
| P138RV | 5′-CGGGATCCCACTAACGTCCCAGCCTTT-3′ | −240 | BamHI |
| P139FW | 5′-GCTCTAGAGGTGGCCTTTGCCCGGA-3′ | +57 | XbaI |
| P193FW | 5′-CTTCGAGCCTAGCAAGGG-3′ | −24 | |
| P194RV | 5′-CTTCCTCCAAATAGGAAGCT-3′ | −126 | |
| P0FW | 5′-GGGTACCAATAATTTTGTTTAACTTTA-3′ | 329 | KpnI |
| P0RV | 5′-GGGATCCATTGCTCAGCGGTGGCAGC-3′ | 71 | BamHI |
All PCR was performed using Pfu polymerase (Stratagene) and P. aeruginosa genomic DNA as templates except PCR performed with P0FW and P0RV, for which the DNA template was plasmid pRsaLPAOWT (Table 1).
Introduced restriction sites are underlined.
Distance in base pairs from the rsaL ATG (for P0FW and P0RV, the position with respect to the pET28b sequence is given).
Restriction recognition sites.
FIG. 2.
TLC analysis of acyl-HSL produced by parent strain P. aeruginosa PAO1 and its rsaL-negative mutant derivative visualized by overlaying with acyl-HSL sensor strain E. coli(pSB1075) (30). Lane 1, synthetic 3OC12-HSL and 3OC10-HSL (0.8 μg); lane 2, PAO1; lane 3, PAO1 rsaL; lane 4, PAO1 rsaL carrying plasmid pPSRsaLPAO. Acyl-HSLs were extracted from spent supernatants, and a volume corresponding to 8 × 108 CFU was loaded for the TLC assay.
Therefore, RsaL plays a pivotal role in the hierarchy of regulators controlling quorum sensing, both in P. aeruginosa PAO1 and P. putida WCS358. This finding is also in agreement with a study in which the key role of RsaL in the repression of P. aeruginosa QS was inferred by mathematical modeling (7).
Genetic studies have shown that in E. coli the lasI promoter is not activated by the 3OC12-HSL/LasR complex when RsaL is also present (4). It was therefore suggested that RsaL could reduce 3OC12-HSL production by binding to the lasI promoter and inhibiting transcription. Since the repressive effect of RsaL was reduced upon the addition of exogenous 3OC12-HSL, it has also been speculated that RsaL and LasR could be in binding competition to the lasI promoter (4). However, another plausible explanation of the above genetic data could be that RsaL impairs binding of LasR to the lasI promoter by directly interacting with LasR and not with DNA. Therefore, in order to investigate the DNA binding properties of RsaL, we purified the recombinant protein RsaLPAO6H, which differs from the wild type for the presence of a six-histidine tag extension at the C terminus. The functionality of RsaLPAO6H with respect to the wild-type counterpart has been controlled by an in vivo assay system consisting of an E. coli strain expressing the lasR gene in trans and carrying a transcriptional fusion between the lacZ reporter gene and the lasI promoter. In this strain, the activity of the lasI promoter is activated upon the addition of exogenous 3OC12-HSL and can be measured quantitatively. In this assay system, introduction of plasmids pRsaLPAOWT and pRsaLPAO6H (Table 1) showed that the recombinant RsaLPAO6H protein can repress the activity of the lasI promoter to the same extent as the wild-type RsaL protein (data not shown).
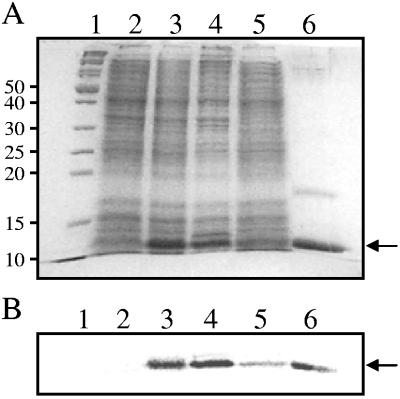
RsaLPAO6H was overexpressed in E. coli BL21(pLysS, pRsaLPAO6H) (Tables 1 and 2) and purified from the soluble cellular fraction by Ni2+ affinity under nondenaturing conditions according to the standard procedure suggested by the column manufacturer (Sigma-Aldrich, St. Louis, Mo.). The fractions containing highly purified RsaLPAO6H (>90% pure) were pooled, dialyzed against wash buffer (500 mM NaCl, 50% [vol/vol] glycerol, and 50 mM NaH2PO4 [pH 8.0]), and stored at −20°C. Key steps of the purification procedure are shown in Fig. 3.
FIG. 3.
Results of overexpression and purification of recombinant RsaLPAO6H are shown. (A) Analysis of protein samples from key steps of purification by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Lane 1, PageRuler protein ladder (Fermentas Inc.) (only molecular mass markers from 50 kDa to 10 kDa are indicated); lane 2, uninduced whole-cell extract; lane 3, induced whole-cell extract; lane 4, insoluble cell extract; lane 5, soluble cell extract; lane 6, pooled fractions eluted at 250 mM imidazole. The arrow indicates the purified RsaLPAO6H protein (10.8 kDa). (B) Western blot with anti-six-histidine antibodies of a sodium dodecyl sulfide-polyacrylamide gel identical to that shown in panel A.
The DNA binding properties of RsaLPAO6H were investigated by performing an electrophoretic mobility shift assay (EMSA), using a DNA probe encompassing the lasI promoter. The probe was obtained by 5′-end labeling of an EcoRI fragment derived from plasmid pPlasIS (Tables 1 and 2). The procedures for 5′-end labeling as well as for the EMSA have been previously described (13).
As shown in Fig. 4, incubation of the probe with increasing amounts of purified RsaLPAO6H leads to the formation of a complex endowed with lower electrophoretic mobility with respect to the probe alone. The minimum molar concentration of RsaLPAO6H able to shift almost all the probe is 8 nM, and since the protein preparation is >90% pure, it would contain a fairly low concentration of contaminants (≤0.8 nM), at least twofold lower than that of the DNA probe (2 nM). Therefore, the possibility that the observed complex may be due to coeluted proteins different from RsaLPAO6H can be ruled out. Moreover, the addition of an excess of unlabeled probe inhibited the formation of the complex, while the nonspecific competitor had no effect. The above results show that RsaLPAO6H is able to specifically bind the lasI promoter, demonstrating for the first time that RsaL is a DNA binding factor.
FIG. 4.
EMSA of RsaLPAO6H binding to the lasI promoter. The 32P-labeled DNA fragment contained nucleotides −83 to +20 with respect to the lasI transcriptional starting site. RsaLPAO6H concentrations (nM) are indicated below the lanes. The probe concentration for each sample was 2 nM. As specific and unspecific competitors, unlabeled probe (100 nM) and calf thymus DNA (1 μg) were added to lane 9 and 10, respectively. The arrow indicates the unshifted DNA probe.
In order to characterize the RsaL binding site, DNase I protection assays were performed on both strands of a DNA fragment encompassing the entire rsaL-lasI intergenic region (4, 23, 28) (Fig. 5A). Plasmids pPlasI3′ and pPlasI5′ were used to obtain the DNA probes, spanning from the first 19 codons of rsaL to the first 36 codons of lasI (Tables 1 and 2). The procedures for the DNase I protection assay have been described previously (14).
FIG. 5.
(A) DNase I footprints of RsaLPAO6H on the rsaL-lasI intergenic region. The plasmids pPlasI5′ (for labeling of the bottom strand) and pPlasI3′ (for labeling of the top strand) were utilized to generate EcoRI/SacI fragments used as probes (Tables 1 and 2). The probes were mixed with different amounts of RsaLPAO6H protein prior to DNase I digestion. Thick lines indicate the regions showing specific protection by RsaLPAO6H; arrows indicate hypersensitive sites. All numbering is in reference to the transcriptional starting site from the lasI promoter (23). M, Maxam and Gilbert sequencing reactions (A+G); lane 1, no RsaLPAO6H added; lanes 2 to 5, RsaLPAO6H added to a final concentration of 0.05, 0.5, 5, or 50 μM, respectively. (B) Sequence of the rsaL-lasI intergenic region. The lasI ATG starting codon is boldface and underlined, and the nucleotides complementary to the starting codon of rsaL (CAT) are boldface and double underlined. The lasI transcriptional starting site is boldface and capitalized (23). The sequence protected by RsaL in the DNase I protection assay is boldface and boxed, and hypersensitive sites are indicated by triangles. The sequence protected by LasR is gray shaded. The 5′-AAnTTATGnAA-3′ inverted repeats are indicated by arrows. The potential σ70-dependent −35 and −10 consensus sequences are indicated by dashed and solid thick lines, respectively.
The DNA region involved in RsaL binding spans at least from nucleotides −22 to −9 with respect to the lasI transcription starting point and is located approximately at the center of a palindromic region consisting of two 5′-AAnTTATGnAA-3′ inverted sequences interrupted by a single nucleotide (Fig. 5B). This dyad symmetry suggests that RsaL could bind DNA as a dimer. Even if the boundaries of the protection cannot be defined precisely due to the high AT content of this DNA region that makes it partly resistant to DNase I attack, the real RsaL-mediated protection could extend up to the extremes of the palindrome, as indicated by the presence of hypersensitive sites at the palindrome boundaries (Fig. 5A and B).
The RsaL binding site partially overlaps the −10 consensus for σ70 recognition (Fig. 5B), consistent with the proposed mechanism of action of RsaL as a transcriptional repressor (2, 4). Previous studies have shown that binding of LasR to a unique site located in the rsaL-lasI intergenic region activates the transcription of both rsaL- and lasI-divergent genes (22, 28). RsaL is implicated as well in the negative regulation of both rsaL and lasI transcription (4). Interestingly, the DNA region protected by LasR (22) is adjacent to the minimum RsaL-protected region and partially overlaps the palindromic sequence involved in RsaL binding (Fig. 5B). The close proximity of the LasR- and RsaL-protected sites supports the hypothesis that LasR and RsaL compete for binding to the rsaL-lasI bidirectional promoter. Therefore, it is likely that binding of RsaL to the rsaL-lasI promoter simultaneously inhibits lasI and rsaL transcription by impairing LasR binding. However, the possibility that LasR and RsaL could bind simultaneously to the lasI promoter cannot be ruled out, in which case, the RsaL-mediated repression of lasI transcription would lead to reduced synthesis of 3OC12-HSL and consequently to reduced expression of all the genes dependent on the LasR/3OC12-HSL complex for their transcription, including the rsaL gene itself.
Conclusions.
The P. aeruginosa QS system is finely regulated and integrated within the global cell regulatory network. Thanks to genetic and microarray studies, knowledge of the number of regulatory factors involved in P. aeruginosa QS regulation is increasing. However, the real impact of these regulators in determining the amount of acyl-HSL produced and the molecular mechanisms underlying their interaction with las and rhl genes in most cases remains unknown (10, 17).
This study shows the dramatic effect of the rsaL mutation in 3O-C12-HSL production, highlighting the importance of RsaL as a major negative regulator of QS in P. aeruginosa. Moreover, the demonstration that this protein is a DNA binding factor is of particular interest, considering that BLASTP and PSI-BLASTP (http://www.ncbi.nlm.nih.gov) analysis of the RsaL amino acid sequence did not show any significant homology with functionally characterized proteins, suggesting that RsaL could belong to a new class of transcriptional regulators. Interestingly, up to now, rsaL homologues have been reported only in the QS genetic loci of P. aeruginosa PAO1, P. putida WCS358, and P. putida IsoF (2, 4, 25).
To our knowledge, this is the first report of the direct binding of a QS regulator to the lasI promoter, with the exception of LasR (22).
The close proximity of the RsaL and LasR binding sites is in agreement with the previous hypothesis that these two proteins are in competition for binding to the rsaL-lasI bidirectional promoter (4). In this view, LasR would be able to trigger lasI transcription by outcompeting the transcriptional repressor RsaL at high cell densities, when the 3OC12-HSL levels reach a certain threshold, and/or if the levels of RsaL decrease in response to an unknown environmental/metabolic stimulus. In addition, an unknown signal molecule could bind RsaL and impair its binding to the DNA. The fact that rsaL expression itself is also dependent upon LasR adds further complexity to this regulatory mechanism. Thus, the interplay of RsaL and LasR on the lasI promoter, as well as the stimulus to which RsaL could respond, deserves further investigation.
Acknowledgments
We thank P. Williams and coworkers for providing synthetic acyl-HSL and the University of Washington Genome Center for supplying the rsaL mutant of P. aeruginosa.
This work was supported by grants from ISPESL (B/98-1/DIPIA/03) and from University “Roma Tre” (CLAR 2004). I.B. was supported by a fellowship from the Italian Cystic Fibrosis Research Foundation, Verona, Italy.
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. H. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertani, I., and V. Venturi. 2004. Regulation of the N-acyl homoserine lactone-dependent quorum-sensing system in rhizosphere Pseudomonas putida WCS358 and cross-talk with the stationary-phase RpoS sigma factor and the global regulator GacA. Appl. Environ. Microbiol. 70:5493-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chugani, S. A., M. Whiteley, K. M. Lee, D. D'Argenio, C. Manoil, and E. P. Greenberg. 2001. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Kievit, T., P. C. Seed, J. Nezezon, L. Passador, and B. H. Iglewski. 1999. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J. Bacteriol. 181:2175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diggle, S. P., K. Winzer, A. Lazdunski, P. Williams, and M. Camara. 2002. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 184:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong, Y. H., X. F. Zhang, H. M. Soo, E. P. Greenberg, and L. H. Zhang. 2005. The two-component response regulator PprB modulates quorum-sensing signal production and global gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 56:1287-1301. [DOI] [PubMed] [Google Scholar]
- 7.Fagerlind, M. G., S. A. Rice, P. Nilsson, M. Harlen, S. James, T. Charlton, and S. Kjelleberg. 2003. The role of regulators in the expression of quorum-sensing signals in Pseudomonas aeruginosa. J. Mol. Microbiol. Biotechnol. 6:88-100. [DOI] [PubMed] [Google Scholar]
- 8.Heurlier, K., V. Denervaud, G. Pessi, C. Reimmann, and D. Haas. 2003. Negative control of quorum sensing by RpoN (σ54) in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:2227-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jude, F., T. Kohler, P. Branny, K. Perron, M. P. Mayer, R. Comte, and C. van Delden. 2003. Posttranscriptional control of quorum-sensing-dependent virulence genes by DksA in Pseudomonas aeruginosa. J. Bacteriol. 185:3558-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juhas, M., L. Eberl, and B. Tummler. 2005. Quorum sensing: the power of cooperation in the world of Pseudomonas. Env. Microbiol. 7:459-471. [DOI] [PubMed] [Google Scholar]
- 11.Juhas, M., L. Wiehlmann, B. Huber, D. Jordan, J. Lauber, P. Salunkhe, A. S. Limpert, F. von Gotz, I. Steinmetz, L. Eberl, and B. Tummler. 2004. Global regulation of quorum sensing and virulence by VqsR in Pseudomonas aeruginosa. Microbiology 150:831-841. [DOI] [PubMed] [Google Scholar]
- 12.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 13.Leoni, L., P. Ascenzi, A. Bocedi, G. Rampioni, L. Castellini, and E. Zennaro. 2003. Styrene-catabolism regulation in Pseudomonas fluorescens ST: phosphorylation of StyR induces dimerization and cooperative DNA-binding. Biochem. Biophys. Res. Commun. 303:926-931. [DOI] [PubMed] [Google Scholar]
- 14.Leoni, L., G. Rampioni, V. Di Stefano, and E. Zennaro. 2005. Dual role of the response regulator StyR in styrene catabolism regulation. Appl. Environ. Microbiol. 71:5411-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyczac, J. B., C. L. Cannon, and G. B. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051-1060. [DOI] [PubMed] [Google Scholar]
- 16.Miller, M. B., and B. L. Bassler. 2001. Quorum-sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 17.Pearson, J. P. 2002. Early activation of quorum sensing. J. Bacteriol. 184:2569-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pesci, E. C., and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa, p. 259-273. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, D.C.
- 19.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. Holden, M. Camara, D. Haas, and P. Williams. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 21.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuster, M., M. L. Urbanowski, and E. P. Greenberg. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA 101:15833-15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seed, P. C., L. Passador, and B. H. Iglewski. 1995. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 177:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, R. S., and B. H. Iglewsky. 2003. Pseudomonas aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6:56-60. [DOI] [PubMed] [Google Scholar]
- 25.Steidle, A., M. Allesen-Holm, K. Riedel, G. Berg, M. Givskov, S. Molin, and L. Eberl. 2002. Identification and characterization of an N-acylhomoserine lactone-dependent quorum-sensing system in Pseudomonas putida strain IsoF. Appl. Environ Microbiol. 68:6371-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner, V. E., R. J. Gillis, and B. H. Iglewsky. 2004. Transcriptome analysis of quorum-sensing regulation and virulence factor expression in Pseudomonas aeruginosa. Vaccine 22(Suppl. 1):S15-S20. [DOI] [PubMed] [Google Scholar]
- 27.Whitehead, N. A., A. M. L. Barnard, H. Slater, N. J. L. Simpson, and G. P. C. Salmond. 2001. Quorum sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 28.Whiteley, M., and E. P. Greenberg. 2001. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J. Bacteriol. 183:5529-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. Jorgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163:185-192. [DOI] [PubMed] [Google Scholar]
- 31.Withers, H., S. Swift, and P. Williams. 2001. Quorum sensing as an integral component of gene regulatory networks in Gram-negative bacteria. Curr. Opin. Microbiol. 4:186-193. [DOI] [PubMed] [Google Scholar]