Abstract
Based on its genome sequence, the pathway of β-oxidative fatty acid degradation in Salmonella enterica serovar Typhimurium LT2 has been thought to be identical to the well-characterized Escherichia coli K-12 system. We report that wild-type strains of S. enterica grow on decanoic acid, whereas wild-type E. coli strains cannot. Mutant strains (carrying fadR) of both organisms in which the genes of fatty acid degradation (fad) are expressed constitutively are readily isolated. The S. enterica fadR strains grow more rapidly than the wild-type strains on decanoic acid and also grow well on octanoic and hexanoic acids (which do not support growth of wild-type strains). By contrast, E. coli fadR strains grow well on decanoic acid but grow only exceedingly slowly on octanoic acid and fail to grow at all on hexanoic acid. The two wild-type organisms also differed in the ability to grow on oleic acid when FadR was overexpressed. Under these superrepression conditions, E. coli failed to grow, whereas S. enterica grew well. Exchange of the wild-type fadR genes between the two organisms showed this to be a property of S. enterica rather than of the FadR proteins per se. This difference in growth was attributed to S. enterica having higher cytosolic levels of the inducing ligands, long-chain acyl coenzyme As (acyl-CoAs). The most striking results were the differences in the compositions of CoA metabolites of strains grown with octanoic acid or oleic acid. S. enterica cleanly converted all of the acid to acetyl-CoA, whereas E. coli accumulated high levels of intermediate-chain-length products. Exchange of homologous genes between the two organisms showed that the S. enterica FadE and FadBA enzymes were responsible for the greater efficiency of β-oxidation relative to that of E. coli.
In Escherichia coli, fatty acid metabolism is tightly regulated in order to allow a quick response to changes in the environment. Fatty acids are synthesized for incorporation into phospholipid membranes, and exogenously supplied fatty acids can be used as carbon and energy sources when coupled to the citric acid cycle via β-oxidation. Expression of the fad genes of the fatty acid degradation regulon is regulated at two levels. E. coli regards fatty acids as substrates of low status, and most other carbon sources are preferred growth substrates; thus, fad gene expression is under strong regulation by the global cyclic AMP-dependent catabolite repression system. The second specific regulatory mechanism is exerted by the FadR transcriptional factor, which plays a dual role in E. coli fatty acid metabolism. FadR specifically represses the transcription of each of the genes essential for fatty acid transport, activation, and β-oxidation, including fadL, fadD, fadE, fadBA, fadH, fadI and fadJ (6, 10, 11, 25, 30). FadR also acts as a transcriptional activator of the fabA and fabB genes, which encode key enzymes of unsaturated fatty acid biosynthesis (5, 21). FadR is also required to activate transcription of iclR, which encodes a repressor of the glyoxylate operon (19). Thus, FadR regulates both the conversion of fatty acids to acetyl-CoA and the utilization of this final product by the citric acid cycle.
Growth of wild-type E. coli strains in the presence of long-chain (>C12) fatty acids coordinately induces the fatty acid degradative (fad) enzymes (25, 32, 39). Fatty acids of medium (C7 to C11) or short (C4 to C6) chain lengths cannot induce synthesis of the fad enzymes in E. coli (2, 11, 15, 39). Due to this induction pattern, wild-type E. coli strains can utilize long-chain fatty acids, such as oleate (C18:1), but not medium-chain fatty acids (MCFAs), such as decanoate (C10), as a sole carbon and energy source. However, MCFAs can serve as growth substrates for fadR mutant strains that constitutively synthesize the fad enzymes (11, 31, 39). Such fadR mutant strains are readily obtained by selection for growth of wild-type cells on decanoate as the sole carbon source. In the case of short-chain fatty acids, growth of E. coli requires two degradative enzymes encoded by the atoD, atoA, and atoB genes (17, 18, 33) in addition to derepressed levels of the enzymes of the fad regulon. The three structural genes of the ato operon are cotranscribed, and transcription is positively regulated by the atoC gene product (23, 33). Mutants that grow on short-chain fatty acids can be readily isolated by plating fadR mutants on minimal medium containing butyrate as the sole carbon source (31). The resulting mutant colonies are constitutive for the ato enzymes and have been ascribed to mutations in the atoC regulatory gene (23, 31). However, from the genome sequence, AtoC seems very likely to be the response regulator of an AtoS-AtoC bacterial two-component system with atoS (located just upstream of atoC) encoding the sensor kinase. Hence, based on other such systems, atoS mutations might also result in constitutive expression.
The fatty acid degradation pathways of E. coli and Salmonella enterica serovar Typhimurium (hereafter called S. enterica) have long been thought to be essentially identical, based on comparisons of the genome sequences (11, 36). However, we found marked differences in the growth of wild-type strains of the two organisms on fatty acids, indicating that this assumption is not correct and that S. enterica is much more proficient at utilization of these carbon sources than is E. coli. In this paper we have characterized the differences in growth between E. coli and S. enterica on medium-chain fatty acids. We present the first direct evidence showing that, unlike E. coli, MCFAs weakly induce the β-oxidation enzymes of S. enterica. We also report that fatty acids are completely degraded to acetyl-CoA by the S. enterica β-oxidation enzymes, whereas the E. coli system gives strikingly incomplete degradation, which was shown to be largely due to differences in the fadE and fadBA gene products of the two organisms.
MATERIALS AND METHODS
Materials.
Labeled fatty acids and β-alanine were purchased from American Radiolabeled Chemicals (St. Louis, Mo.). Acyl-CoA standards, fatty acids, antibiotics, and most other chemicals were purchased from the Sigma Chemical Co. (St. Louis, Mo.). Bond Elut Jr. C18 columns were purchased from Chrom Tech (Apple Valley, Minn.). The goat anti-rabbit immunoglobulin G-horseradish peroxidase conjugate was obtained from Zymed Laboratories Inc. (San Francisco, Calif.).
Media and growth conditions.
Rich broth (RBO medium containing 10 g of tryptone, 1 g of yeast extract, and 5 g of NaCl per liter), 2× YT medium containing 16 g of tryptone, 5 g of NaCl, and 10 g of yeast extract per liter, and minimal medium M9 (28) supplemented with 1 mM MgSO4, 0.1 mM CaCl2, and 0.001% thiamine were used in growth phenotype studies on fatty acid plates. For CoA pool measurements, liquid cultures were grown in minimal medium E (28) supplemented with 0.001% thiamine and 0.1% vitamin free Casamino Acids (Difco) and β-alanine as previously indicated. The strains derived from E. coli UB1005 were also supplemented with 0.01% methionine. Acetate was used at a final concentration of 0.4% for a carbon source. Fatty acids were neutralized with KOH, solubilized with Tergitol NP-40, and used at final concentrations of 0.2% as the sole carbon source and at 5 mM for the induction experiments. Solid media contained 1.5% (wt/vol) Bactoagar (Difco, Milwaukee, Wis.). Antibiotics were used at the following concentrations (in mg/liter): sodium ampicillin, 100; kanamycin sulfate, 50; tetracycline HCl, 12; and chloramphenicol, 25. The phenotype of the various fad mutants was verified by testing for growth on minimal oleate plus acetate media and for no growth on media supplemented with oleate alone.
Bacterial strains and plasmids.
All bacterial strains are derivatives of E. coli K-12 or S. enterica LT2. The strains and plasmids used and generated in this study are listed in Table 1. Phage transduction and other basic genetic techniques were generally carried out as described by Miller (27). The phage λ Red-mediated recombination method of Datsenko and Wanner (14) was used to produce strains SI3, SI81, and SI158 from strain LT2. Strains SI3 and SI81 were constructed by amplification of the aminoglycoside 3′-phosphotransferase (kan) gene of plasmid pKD4 (14), using primers Sal-fadR-KO1 plus Sal-fadR-KO2 and Sal-panD-KO1 plus Sal-panD-KO2, respectively (Table 2). The appropriate PCR products were used to replace the entire coding sequences of fadR or panD in strain LT2 with the help of λ Red-encoding plasmid pKD46 (14). The correct constructs were verified by PCR, and the mutant genotypes were confirmed by the phenotypic observations that strain SI3 (ΔfadR) grew more rapidly than the wild-type strain on minimal media containing decanoate as the sole carbon and energy source, whereas strain SI81(ΔpanD) required β-alanine for growth on minimal media. Strain SI158 (ΔfadE) was constructed by amplification of the chloramphenicol acetyltransferase (cat) gene of plasmid pKD3 (14), using primers Sal-fadE-KO1 plus Sal-fadE-KO2. The resulting PCR product was used to replace the entire fadE coding sequence of strain LT2 as described above. Chloramphenicol-resistant colonies were checked by PCR for the expected insertion event, and the fadE genotype was confirmed by the inability of strain SI158 to grow on fatty acids. Strains SI194 and SI235 were obtained by P1 transduction of strains MC1061 and SI92, respectively, with a lysate grown on JWC266 and selection for kanamycin resistance. Strains SI199 and SI237 were generated by P1 transduction of strains SI196 and SI92, respectively, with a lysate grown on MFH8 and selection for tetracycline resistance. Strain SI226 was constructed by first removing the kanamycin resistance cassette from strain SI3, using the FLP plasmid pCP20 (9), followed by transduction with a P22 lysate grown on SI221, with selection for kanamycin resistance. Strain SI92 was obtained by P1 transduction of strain UB1005 with a lysate grown on strain NRD1, with selection for chloramphenicol resistance. Strains SI236 and SI245 were obtained by P1 transduction of strains SI92 and SI244, respectively, with a lysate grown on SI186 and selection for kanamycin resistance. Strain SI248 was produced by P1 transduction of strain SI245 with a lysate grown on SI241 and selection for chloramphenicol resistance. Strain SI239 was generated by first removing the kanamycin resistance cassette from strain SI81, using pCP20, followed by transduction with a P22 lysate grown on SI3 and selection for kanamycin resistance.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strain | ||
| UB1005 | metB1 relA1 spoT1 gyrA216 | 1 |
| MC1061 | araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL (Strr) hsdR2 mrcA mcrB1 | 7 |
| LT2 | Salmonella enterica serovar Typhimurium | S. Maloy |
| MFH8 | UB1005 fadR::Tn10 | 21 |
| MFH9 | UB1005 fadR::Tn5 | 21 |
| SI3 | LT2 fadR::kan | This study |
| JWC266 | BW25113 fadE::kan | 4 |
| SI194 | MC1061 fadE::kan | This study |
| SI158 | LT2 fadE::cat | This study |
| SI196 | MC1061 fadE-lacZ | This study |
| SI221 | LT2 fadE-lacZ | This study |
| SI199 | MC1061 fadE-lacZ fadR::Tn10 | This study |
| SI226 | LT2 fadR fadE-lacZ | This study |
| SI184 | MFH9 attPP22::(S. enterica fadR cat) | This study |
| SI217 | SI3 attPP22::(E. coli fadR cat) | This study |
| SI231 | MFH9 attPP22::(E. coli fadR cat) | This study |
| NRD1 | DY330 panD::cat | N. Delay |
| SI92 | UB1005 panD::cat | This study |
| SI81 | LT2 panD::kan | This study |
| SI186 | MG1655 fadBA::kan | 22 |
| SI235 | UB1005 panD::cat fadE::kan | This study |
| SI236 | UB1005 panD::cat fadBA::kan | This study |
| SI237 | UB1005 panD::cat fadR::Tn10 | This study |
| SI239 | LT2 panD fadR::kan | This study |
| SI244 | UB1005 panD fadE attPP22::(fadE; bla) | This study |
| SI241 | UB1005 panD fadBA attPP22::(fadBA; cat) | This study |
| SI245 | SI244 fadBA::kan | This study |
| SI248 | SI245 attPP22::(fadBA; cat) | This study |
| BL21(DE3)/ pLysS | ompT hsdSB(rB− mB−) gal dcm | 37 |
| Plasmid | ||
| pACYC177 | General cloning vector | 8 |
| pCR2.1 | General cloning vector | Invitrogen |
| pET16b | T7 promoter-based expression vector | Novagen; 37 |
| pKD3 | bla FRT cat FRT PS1 PS2 oriR6K | 14 |
| pKD4 | bla FRT ahp FRT PS1 PS2 oriR6K | 14 |
| pKD46 | bla pBAD gam bet exo pSC101 ori(Ts) | 14 |
| pCP20 | bla cat cI857 PRflp pSC101 ori(Ts) | 9 |
| pCD22PSK | R6Kγori attP22 kana | 34 |
| pINT-A22 | Plac-int(P22) bla repA(Ts)b | 34 |
| pXINT-A22 | Plac-xis/int(P22) bla repA(Ts)c | 34 |
| pAH81 | bla oriR attP21 | 20 |
| pAH121 | int(P21) bla repA(Ts) | 20 |
| pRK34 | pET16b carrying E. coli K-12 fadD | 29 |
| pSH11 | pACYC177 carrying E. coli K-12 fadR | 22 |
| pSH12 | pACYC177 carrying LT2 fadR | 22 |
| pSH53 | pET16b carrying LT2 fadD | This study |
| pSH61 | pCD22PSK carrying E. coli fadR | This study |
| pSH62 | pCD22PSK carrying S. enterica fadR | This study |
| pSH68 | pCD22PSK carrying S. enterica fadBA | This study |
| pSH72 | pAH81 carrying S. enterica fadE | This study |
Plasmid vector for integration at P22 attachment sites.
Temperature-sensitive helper plasmid for pCD-vector integration.
Temperature-sensitive helper plasmid for pCD-vector excision.
TABLE 2.
PCR primers used
| Primer | Sequence (5′-3′) |
|---|---|
| Sal-fadR-KO1 | ATGGTCATTAAGGCGCAGAGCCCGGCGGGT TTCGCGGAA |
| GAGTATGTGTAGGCTGGAGCTGCTTC | |
| Sal-fadR-KO2 | TTATCGTCCCTGAATCGCTAAATCGCCGGG CAGATTTTTC |
| TGCACATATGAATATCCTCCTTAG | |
| Sal-panD-KO1 | CACAAGCGCCTGCTTAGCCAAGGTAAACGA CAGGGTAAA |
| GAAGTTTGTGTAGGCTGGAGCTGCTTC | |
| Sal-panD-KO2 | ACCATGTCGCGGCTGATCAGCAACCAGCCG CAGGGATAA |
| GGACTACATATGAATATCCTCCTTAG | |
| Sal-fadE-KO1 | CATCACAAGTGGTCAGACCTCCTACAAGCA AGGGAGCTT |
| TTCGTTTGTGTAGGCTGGAGCTGCTTC | |
| Sal-fadE-KO2 | GCGAAAGCGGGGCAGGAAAGACACGAGACT GATTATGC |
| GGCTTCCATATGAATATCCTCCTTAG | |
| Sal-fadBA-1 | CTGCAGTCTGCCGAGCGTGATCACATC |
| Sal-fadBA-2 | ATCGATTGCCGCTGCGCTGCAATGCGA |
| Sal-fadE-1 | CTGCAGGTACAGGCGAGAAGCATAAGC |
| Sal-fadE-2 | ATCGATGGCAGGAAAGACACGAGACTG |
| Sal-fadD-1 | GCCATATGTTGAAGAAGGTTTGGCTTAACCG |
| Sal-fadD-2 | CCGGATCCTCAGGCTTTATTGTCTACTTTGC |
| CAT-N | CAGGAGCTAAGGATCCTAAAATGGAG |
| CAT-C | GGGCACCAAGATCTGCCTTAAAAAAA |
| att-R | TCCGAATTCGTGCGTAATAAATGAC |
| att-L | TCCGAATTCATGAGGTTGTACATAAGTGA |
To construct plasmid pSH53, the fadD gene of strain LT2 was amplified via PCR using Easy-A high-fidelity polymerase (Stratagene) under standard PCR conditions, using primer set Sal-fadD-1 plus Sal-fadD-2. The resultant 1.7-kb PCR product was purified via a QIAGEN spin column and cloned into pCR2.1 (Invitrogen). The insert of the resultant plasmid was sequenced to verify the fidelity of the cloned PCR product. The sequenced plasmid was digested with NdeI and BamHI (sites within the primers) and ligated to pET16b cut with the same enzymes to give in-frame fusion of the coding sequence of S. enterica FadD to the amino terminal His tag encoded by the vector to give pSH53.
Construction of fadE-lacZ transcriptional fusions.
Strains SI196 and SI221 were constructed using a λ Red- and FLP-mediated site-specific recombination method (16). The kanamycin or chloramphenicol resistance cassettes of strains SI194 and SI158 were removed by FLP recombinase as described above to leave behind a single FLP recombinase target (FRT) site. The FRT sites was then used for site-specific integration of a lac fusion plasmid, pCE70 (16), containing an FRT site upstream of the promoterless lacZY genes, a kanamycin resistance gene, and the R6K origin of replication. Transformants were plated on RBG Kan X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at 37°C, which resulted in the stable integration of the fusion plasmid due to the loss of pCP20 and produced the fadE-lacZ transcriptional fusion strains SI196 and SI221.
CRIM plasmid integration.
Plasmids pSH61 and pSH62 were obtained by subcloning the BamHI-XhoI and HindIII-XhoI fragments of pSH11 and pSH12, respectively, into pCD22PSK (34). Strain MFH9, carrying the CRIM helper plasmid pINT-A22 (34), was transformed with pSH61 or pSH62, with selection for resistance to both ampicillin and chloramphenicol at 30°C. The plasmids specifically integrated into the bacteriophage P22 attachment site attP22 of E. coli strain MFH9. The resulting integrants SI184 (from pSH62) and SI231 (from pSH61) were purified by restreaking them twice onto RBO-chloramphenicol plates at 42°C. Ampicillin-sensitive colonies were identified, and the constructs were verified by PCR, using primer sets CAT-N plus CAT-C and att-R plus att-L. The helper plasmid pXINT-A22 (34) was used to recover the integrated plasmids. Strain SI217 was generated by integration of plasmid pSH61 into the bacteriophage P22 attachment site attA of S. enterica strain SI3, essentially as described above. To construct strains SI241 and SI244, the fadBA and fadE genes of strain LT2 were amplified via PCR using Easy-A high-fidelity polymerase with primer sets Sal-fadBA-1 plus Sal-fadBA-2 and Sal-fadE-1 plus Sal-fadE-2, respectively. The resultant 3.6-kb fadBA PCR product and the fadE 2.7-kb PCR product were purified via QIAGEN spin columns and cloned into pCR2.1. The inserts of the resultant plasmids were sequenced to verify the fidelity of the cloned PCR products. The fadBA and fadE inserts were subcloned into pCD22PSK and pAH81, using BamHI-PstI and EcoRI digests, producing plasmids pSH68 and pSH72, respectively. Strain SI241 was constructed by first removing the kanamycin and chloramphenicol resistance cassettes from strain SI236 as described above and then integrating plasmid pSH68 into the attP22 attachment site as described above. To generate strain SI244, the kanamycin and chloramphenicol resistance cassettes were first removed from strain SI235 as described above. The resulting strain carrying the CRIM helper plasmid pAH121 (20) was transformed with pSH72, with selection for ampicillin resistance at 37°C. Plasmid pSH72 specifically integrated into bacteriophage P21 attachment site attP21, producing strain SI244. The construct was confirmed by PCR as previously described (20).
β-Galactosidase assays.
Overnight cultures were grown in RBO medium or RBO medium supplemented with fatty acid to 5 mM. The overnight cultures were subcultured into the same medium and shaken at 37°C. When the cultures reached mid-log phase, the cells were pelleted and washed twice with RBO medium containing 0.5% Tergitol NP-40 and three times with RBO medium to remove fatty acids and detergent. After the final rinse, the cells were resuspended to 4 × 108 cells/ml and assayed for β-galactosidase activity after chloroform/sodium dodecyl sulfate lysis as described by Miller (27). The cell debris in the assay mix was removed by centrifugation before absorbance was read at 420 nm. All cultures contained Tergitol NP-40 and received washing treatment without regard to fatty acid supplementation.
Western blotting.
Western blotting of cell extracts of E. coli and S. enterica strains was performed by growing overnight cultures in RBO medium. These cultures were diluted 1:100 into the same medium and grown until mid-log phase. Crude extracts were loaded on an equal protein basis and separated on 12% resolving, 5% stacking polyacrylamide gel using a Mini-Protean II apparatus (Bio-Rad). The separated proteins were then electrophoretically transferred to an Immobilon-P membrane (Millipore) for 60 min at 90 V. The membrane was incubated first in TBS buffer (20 mM Tris-HCl [pH 7.5] and 150 mM NaCl) containing 0.05% Tween 20 and 5% nonfat milk powder for 1 h at room temperature with gentle shaking. Following blocking, the membrane was probed with rabbit polyclonal antibody (raised against E. coli His6-FadR protein) diluted 1:1,000 in the antibody buffer (0.25% Triton X-100 and 2% nonfat milk powder in TBS buffer) for an additional hour. After being rinsed four times with wash buffer (0.05% Tween 20 in TBS buffer), the membrane was incubated with a secondary antibody conjugated with horseradish peroxidase (Amersham) diluted 1:20,000 in antibody buffer for 1 h at room temperature. The membrane was washed as described above, and the FadR proteins were visualized by incubation of the membrane in ECL plus chemiluminescent detection reagents (Amersham Biosciences) and imaged on ECL Hyperfilm (Amersham Biosciences).
Expression and purification of His6-FadD proteins.
The His6-FadD proteins of E. coli and S. enterica were overexpressed in E. coli BL21(λDE3)/pLysS, harboring plasmids pRK34 (29) and pSH53, respectively, using the T7 polymerase-dependent expression system. Cultures (1 liter) were grown in LB broth supplemented with ampicillin and chloramphenicol at 37°C until an optical density of about 0.5 was reached, and then 1 mM isopropyl-β-d-thiogalactopyranoside was added, followed by incubation for 3 h. The cells were harvested by centrifugation at 4,000 × g for 20 min. The pellets were resuspended (at a fourfold concentration) in ice-cold 20 mM Tris-HCl (pH 8.0) and centrifuged for 5 min at 4,000 × g. The pellets were stored at −80°C until further use. All further purification steps were performed at 5°C. For purification of the His-tagged proteins, frozen cell pellets were thawed on ice for 15 min prior to the addition of 20 ml of buffer A (50 mM sodium phosphate [pH 8.0], 300 mM NaCl, and 10 mM imidazole). Lysozyme (1 mg/ml) was added, and after 30 min of incubation on ice the cells were disrupted via sonication. The lysates were centrifuged at 10,000 × g for 30 min, and 3 ml of Ni2+-nitrilotriacetic acid resin (QIAGEN, Valencia, Calif.) was added to the resultant supernatant. The slurry was rotated slowly at 5°C for 60 min, and the resin was loaded into a 5-ml column and washed with five volumes of buffer B (50 mM sodium phosphate [pH 8.0], 300 mM NaCl, and 20 mM imidazole). The His6-tagged protein was eluted by 1 ml of buffer C {50 mM sodium TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] [pH 8.0] and 300 mM NaCl} containing 250 mM imidazole. The elution was repeated four times. The purity of the samples was monitored via sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Fractions containing the protein of interest were pooled and dialyzed against two changes of buffer C for 12 h at 5°C. Following dialysis, 2-mercaptoethanol and glycerol were added to final concentrations of 10 mM and 10%, respectively, prior to storage at −80°C.
Assay of acyl-CoA synthetase activity.
Wild-type E. coli and S. enterica strains were grown overnight in RBO supplemented with 5 mM decanoate. The cells were harvested by centrifugation, washed, and resuspended in Tris buffer (100 mM Tris-HCl [pH 7.5], 10 mM 2-mercaptoethanol, 2 mM EDTA, and 1% Triton X-100). Cells were lysed by sonication on ice, with four 10 s bursts and 10 s cooling intervals. Acyl-CoA synthetase activities were determined in crude cell extracts or using the purified His-tagged FadD proteins of E. coli and S. enterica. The specific activities were calculated by measuring the formation of radiolabeled acyl-CoAs from 1-14C-labeled fatty acids, using the assay of Kameda and Nunn (24). Unless otherwise stated, reaction mixtures contained 200 mM Tris-HCl (pH 7.5), 50 μM ATP, 8 mM MgCl2, 2 mM EDTA, 20 mM NaF, 0.1% Triton X-100, 0.5 mM CoA, 10 μM fatty acids, and cell extract in a total volume of 200 μl. The reactions were incubated at room temperature for 15 min and then spun at the top speed of a microcentrifuge for 1 min. Reactions were initiated by the addition of CoA and were terminated by the addition of 1 ml of isopropyl alcohol, n-heptane, and 1 M H2SO4 (40:10:1, by volume). The residual radioactive free fatty acid was removed by extraction with n-heptane. The aqueous fraction was then counted in a Beckman-Coulter LS6500 scintillation counter to measure synthesis of fatty acyl-CoA. In the negative-control experiments, CoA was omitted. The protein concentrations of the enzyme extracts and purified enzyme samples were determined using the Bradford assay (3), with bovine serum albumin as a standard.
CoA pool analyses.
To label the CoA pools, strains carrying deletions of the panD gene (which are auxotrophic for β-alanine) were used. The strains were labeled with β-[3-3H]alanine as described in the figure legends. CoA derivatives were extracted, and the compositions of CoA pools were determined by reversed-phase high-pressure liquid chromatography (HPLC), using a protocol modified from that of Roughan (35). Samples (1 ml) of cultures were transferred to a microcentrifuge tube and trichloroacetic acid (TCA) was added to a final concentration of 6%. After mixing, the tubes were placed in an ice slurry for 3 min and spun at the top speed of a microcentrifuge for 3 min at 4°C. The supernatant was saved, and the pellet was resuspended in 1 ml of 1% TCA. The tubes were centrifuged for 3 min at maximum microcentrifuge speed at 4°C, and the supernatants were saved. The combined supernatants were loaded on a Bond Elut Jr. C18 cartridge previously equilibrated with 2 ml of methanol and then with 8 ml of 1 mM HCl. After sample loading, the cartridge was washed with 6 ml of 1 mM HCl and the CoA species were eluted with 3 ml of 0.1 M ammonium acetate in 65% ethanol. The eluant was dried in a vacuum centrifuge at room temperature, and the residue was resuspended in 0.5 ml of 50 mM ammonium acetate (pH 5.0). For HPLC analysis, appropriate CoA standards (Sigma) were added to the sample immediately prior to the injection of samples of 100 μl. The HPLC analysis was performed on a Waters C18 column (38). Solvent A was 50 mM ammonium acetate (pH 5.0) and solvent B was acetonitrile. The column was developed at a flow rate of 1 ml/min. The starting conditions of 98.8% A and 1.2% B were maintained for 5 min, followed by a 50-min gradient from 1.2% to 8% B, a 10-min gradient from 8% to 40% B, and a final 50-min gradient from 40% to 60% B. Radioactivity was monitored by an in-line Beckman 110B Radioisotope detector with a flow-through scintillation counter. The identities of the tritium-labeled CoA species were determined by comparison with the retention times of the internal standards.
RESULTS
Differential growth of E. coli and S. enterica on medium-chain fatty acids.
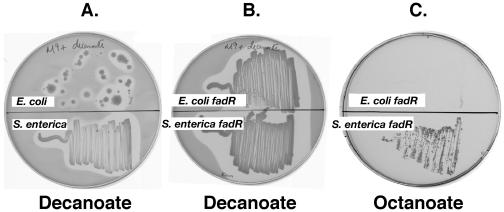
Different growth phenotypes were observed for wild-type E. coli strain UB1005, wild-type S. enterica serovar Typhimurium strain LT2, and their fadR derivatives on M9 minimal medium containing fatty acids of different chain lengths as sole carbon and energy sources (Table 3). The fatty acids tested included decanoate (C10:0), octanoate (C8:0) and hexanoate (C6:0), with the long-chain acid oleate (C18:1) as a control. As previously observed (11, 31, 39), wild-type E. coli strain UB1005 was unable to grow on decanoic acid (C10), although several fadR mutant colonies arose on the plate, whereas S. enterica wild-type strain LT2 grew slowly on this acid (Fig. 1A). Moreover, the E. coli fadR strain MFH9, which has constitutive levels of fad enzymes, grew well on decanoate (Fig. 1B) but failed to grow on hexanoic acid (C6) (data not shown) and grew only very poorly on octanoic acid (C8) (Fig. 1C), whereas the S. enterica fadR strain SI3 demonstrated much better growth on both acids (Fig. 1A to C and Table 3). Note that Spector and coworkers (36) mentioned that, during selections for fadR mutants of S. enterica LT2, the mutant strains were overgrown by the background of wild-type cells present on the selection plates. However, this observation of the growth of wild-type S. enterica on decanoate does not seem to have been further investigated.
TABLE 3.
Growth of E. coli and S. enterica strains on fatty acids of various chain lengths
| Strain | Growth on indicated carbon sourceb
|
||||
|---|---|---|---|---|---|
| C18:1 | C10 | C8 | C6 | C2 | |
| E. coli UB1005 | 5+ | −a | − | − | 5+ |
| S. enterica LT2 | 5+ | 2+ | − | − | 5+ |
| E. coli UB1005 fadR | 5+ | 5+ | − | − | 5+ |
| S. enterica LT2 fadR | 5+ | 5+ | 2+ | 1+ | 5+ |
Isolated colonies (characteristically fadR mutants) appeared following extended incubation (>4 days) at 37°C.
Growth of pinpoint colonies was visible after extended incubation (>5 days) at 37°C. The cultures were grown at 37°C on plates of M9 medium supplemented with oleate (C18:1), decanoate (C10), octanoate (C8), hexanoate (C6), or acetate (C2). After 7 days of incubation, growth was scored as positive (+) if pinpoint colonies were present or as negative (−) if no colonies were seen. If large colonies appeared after 2 days of incubation, growth was given a score of 5+, and if smaller colonies appeared, growth was given a lower score proportional to the size of the colonies (e.g., 2+).
FIG. 1.
Growth of E. coli and S. enterica on medium-chain-length fatty acids. Plate A, growth of wild-type E. coli strain UB1005 and wild-type S. enterica strain LT2 on decanoate. Plates B and C, growth of strains MFH9 (UB1005 fadR::Tn5) and SI3 (LT2 fadR::kan) on decanoate and octanoate, respectively. M9 minimal medium plates were supplemented with 0.2% of the specified fatty acid. Plates A and B were incubated for 7 and 3 days, respectively, whereas plate C was incubated for 5 days. The clear zones around the areas of growth are due to consumption of decanoate, which renders the medium turbid.
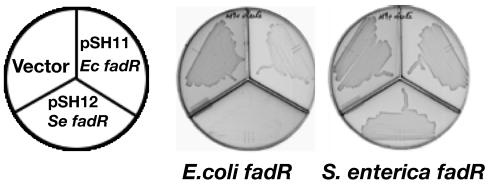
Although both the wild-type strains UB1005 and LT2 efficiently utilized oleate (C18:1) as the sole carbon and energy source, increased levels of E. coli or S. enterica FadR produced from the multicopy plasmid resulted in strong inhibition of the growth of E. coli fadR strain MFH9 on oleate (22). In contrast, the S. enterica fadR strain SI3 grew well on oleate in the presence of a plasmid encoding either E. coli FadR or S. enterica FadR (Fig. 2). The differences in growth could be a result of one or a combination of factors. These are (i) differential levels of fad gene repression, which in turn could be due to differences in the FadR expression levels or the differing affinities for the fad regulon operators; (ii) differences in the interactions of acyl-CoA ligands of various chain lengths with FadR and their efficiency in dissociating the FadR-DNA complex; and (iii) differences in acyl-CoA pool levels, which would not only regulate FadR activity but also serve as substrates for β-oxidation.
FIG. 2.
Growth of E. coli and S. enterica fadR derivatives on oleate in response to a plasmid carrying a fadR gene. Growth of strains MFH9 (UB1005 fadR) and SI3 (LT2 fadR), harboring either pACYC177-derived plasmids pSH11 (carrying E. coli fadR) or pSH12 (carrying S. enterica fadR) or the empty vector. The M9 minimal medium plates were supplemented with 0.2% oleic acid and incubated for 3 days at 37°C.
Comparison of fadE transcriptional regulation.
To quantitate the efficiency of FadR repression as well as the efficiency of induction by various chain lengths of fatty acids in E. coli and S. enterica, we analyzed fadE transcriptional regulation (FadE catalyzes the first step of the β-oxidation cycle), using fadE-lacZ transcriptional fusion strains. These strains were constructed using homologous recombination catalyzed by the λ Red proteins, followed by removal of the antibiotic cassette by FLP-mediated site-specific recombination (16) (see Materials and Methods). As expected, the β-galactosidase activities of E. coli strain SI196 (MC1061 fadE-lacZ) and S. enterica strain SI221 (LT2 fadE-lacZ) were similarly low in noninducing medium (RBO) and high in the inducing medium (RBO supplemented with oleate). In contrast, the MCFAs, decanoate and octanoate, failed to induce β-galactosidase activity in E. coli strain SI196, whereas partial inductions of about 2.5- and 2-fold were observed for these acids, respectively, in S. enterica strain SI221 (Table 4). The short-chain fatty acid, hexanoate, failed to induce either strain. These results suggested that the weak growth observed in the case of wild-type S. enterica on decanoate might be due to weak induction of the β-oxidation enzymes.
TABLE 4.
Transcriptional regulation of fadE-lacZ fusion derivatives of wild-type strains of E. coli and S. enterica in response to various-chain-length fatty acids
| Strain | β-Galactosidase activity in RB medium and indicated fatty acida
|
||||
|---|---|---|---|---|---|
| RB | RB-C18:1 | RB-C10 | RB-C8 | RB-C6 | |
| E. coli SI19 | 104 ± 11 | 998 ± 90 | 124 ± 13 | 108 ± 10 | 101 ± 9 |
| S. enterica SI221 | 125 ± 14 | 1,049 ± 121 | 301 ± 25 | 242 ± 19 | 131 ± 13 |
β-Galactosidase activities are expressed as the amount of o-nitrophenol produced per min per mg of protein and are the averages of three separate experiments, each assayed in duplicate. The fadE-lacZ strains were E. coli SI196, which is a derivative of MC1061, and S. enterica SI221, which is a derivative of LT2. The RB medium was supplemented with fatty acids to 5 mM.
β-Galactosidase activities were also determined for E. coli strain SI199 (fadR fadE-lacZ) and S. enterica strain SI226 (fadR fadE-lacZ) in response to plasmid-encoded overproduction of FadR (Table 5). Note that the presence of E. coli fadR on pACYC177 (the vector we used) gave about twofold-stronger repression of the fad regulon than was seen in a wild-type cell, presumably due to greater occupancy of the fad operators by the increased number of FadR molecules. Strains E. coli SI199 and S. enterica SI226, harboring either the E. coli or S. enterica fadR plasmids, showed similar β-galactosidase activities under noninducing conditions, and these levels were about 20-fold less than those seen in the strains carrying the empty vector, pACYC177 (Table 5). In the presence of oleate, β-galactosidase activities remained repressed by about 3-fold in E. coli strain SI199, carrying the E. coli plasmid, and 8-fold in E. coli strain SI199, carrying the S. enterica fadR plasmid, whereas only about 1.5-fold repression was observed in S. enterica strain SI226, carrying the E. coli plasmid, and 3-fold repression was observed in S. enterica strain SI226, carrying the S. enterica fadR plasmid (Table 5). These results are consistent with the growth phenotypes on oleate plates of E. coli and S. enterica carrying a FadR-encoding plasmid and led us to examine the reason for the higher induction of fadE promoter activity observed in S. enterica in the presence of oleate or decanoate.
TABLE 5.
Transcriptional regulation of fadE-lacZ fusions by expression of plasmid-encoded FadR in fadR strains of E. coli and S. enterica
| Strain | Plasmid present | Plasmid-encoded FadR | β-Galactosidase activitya
|
|
|---|---|---|---|---|
| RB | RB-C18:1 | |||
| E. coli SI199 (MC1061 fadR fadE-lacZ) | None | None | 1121 ± 115 | 1109 ± 96 |
| pACYC177 | None | 1057 ± 94 | 1083 ± 87 | |
| pSH11 | E. coli | 58 ± 13 | 371 ± 52 | |
| pSH12 | S. enterica | 53 ± 11 | 141 ± 17 | |
| S. enterica SI226 (LT2 fadR fadE-lacZ) | None | None | 1095 ± 97 | 1066 ± 104 |
| pACYC177 | None | 1081 ± 105 | 1057 ± 89 | |
| pSH11 | E. coli | 64 ± 10 | 701 ± 68 | |
| pSH12 | S. enterica | 60 ± 12 | 382 ± 47 | |
β-Galactosidase activities are expressed as the amount of o-nitrophenol produced per min per mg of protein and are the averages of three separate experiments, each assayed in duplicate. RB-oleate is RB medium supplemented with 5 mM oleate (C18:1).
Exchange of fadR genes between E. coli and S. enterica.
The E. coli and S. enterica FadR proteins differ in only 7 of 293 residues, but interestingly, six of the mismatches are in the C-terminal half, where acyl-CoA binding occurs. We tested the hypothesis that decanoyl-CoA might dissociate the complex of DNA with S. enterica FadR more efficiently than E. coli FadR, resulting in weak induction, which would allow slow growth. To test this hypothesis, the CRIM plasmid pSH62, carrying the S. enterica fadR together with its promoter, was integrated into the bacteriophage P22 attachment site (attP22) of the E. coli fadR strain MFH9. The resulting strain, SI184, which contained a single copy of the fadR gene (that of S. enterica) failed to grow on decanoate. We also found that integration of the CRIM plasmid pSH61, carrying the E. coli fadR gene, into the bacteriophage P22 attachment site allowed growth of a S. enterica fadR strain on decanoate but blocked growth of an E. coli fadR strain on this medium. The levels of FadR expression in the two organisms have been assayed by Western blotting analyses and found to be essentially identical (22). These results indicated that growth differences between E. coli and S. enterica on decanoate were not due to differences in the interactions of decanoyl-CoA with the two FadR proteins or to differential operator interactions.
Comparison of Acyl-CoA production in E. coli and S. enterica.
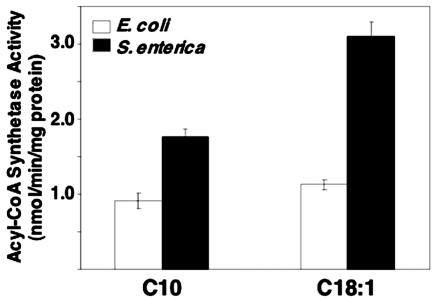
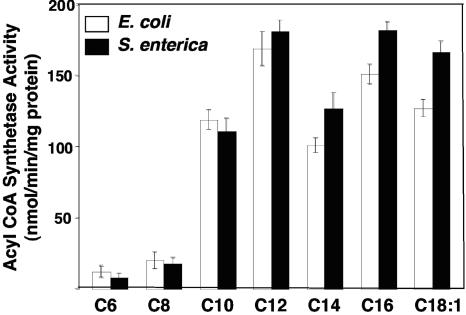
Given that the FadR proteins of the two organisms had no physiologically important functional differences, we next asked if the levels of intracellular inducer accumulated by the two organisms were similar, since higher inducer levels could explain the growth of S. enterica. We first attempted to use strains defective in β-oxidation to determine the level of acyl-CoAs accumulated within the cells. To detect the acyl-CoAs produced, panD bacterial strains were labeled with the CoA precursor, β-[3-3H]alanine, and the extracted CoA pools were analyzed by reversed-phase HPLC as described in Materials and Methods (panD encodes aspartate 1-decarboxylase, the enzyme responsible for β-alanine synthesis) (see reference 13). Unfortunately, our attempts were futile due to the fact that strains SI235 (panD fadE) and SI236 (panD fadBA) grown in acetate medium supplemented with oleate or decanoate accumulated no significant levels of oleoyl-CoA or decanoyl-CoA (data not shown). As previously reported (25, 26), these strains are severely deficient in fatty acid uptake, presumably because fatty acid transport is tightly coupled to its catabolism. We therefore measured the activities of acyl-CoA synthetase, the enzyme that converts free fatty acids to their respective acyl-CoA thioesters, in the two bacteria. Acyl-CoA synthetase activities in crude cell extracts of the wild-type strains E. coli UB1005 and S. enterica LT2 grown on RBO in the presence of 5 mM decanoate were monitored, using either 1-14C-labeled decanoic or oleic acid as the substrate. The S. enterica strain had acyl-CoA synthetase activities about two- and threefold higher than parallel cultures of E. coli when decanoic and oleic acid, respectively, were used as substrates (Fig. 3). The results suggested that the weak induction of the S. enterica β-oxidation enzymes in cells grown on decanoate was probably due to higher decanoyl-CoA levels produced, perhaps in concert with the modest increase in FadR binding of this ligand. However, it remained possible that the increased acyl-CoA synthetase activity was a property of the S. enterica FadD protein rather than its levels of expression. We therefore purified the E. coli and S. enterica FadD proteins to homogeneity and determined their specific activities with fatty acids of various chain lengths. No significant differences in the specific activities of the two proteins were found for any fatty acid tested (Fig. 4). These results indicate that the higher acyl-CoA synthetase activity of crude extracts of S. enterica strains relative to those of E. coli must be attributed to differences in fadD regulation that result in a greater number of active FadD molecules in S. enterica.
FIG. 3.
Fatty acyl-CoA synthetase activities in crude extracts of wild-type E. coli and S. enterica. Enzyme activities were calculated for crude extracts of strains UB1005 (wild-type E. coli) and LT2 (wild-type S. enterica) using [1-14C]decanoic acid (C10:0) or [1-14C]oleic acid (C18:1) as a substrate and grown in RBO medium supplemented with 5 mM decanoate. The amount of labeled acyl-CoA species formed in 15 min at room temperature was determined as described in Materials and Methods. The values represent the mean ± the standard deviation of the results of three independent experiments.
FIG. 4.
Fatty acyl chain length specificities of the purified His6-FadD proteins of E. coli and S. enterica. The specific activities (nmol of product formed/min/mg protein) were measured by assaying the formation of 1-14C-fatty acyl-CoAs from 1-14C-labeled fatty acid substrates as described in Materials and Methods. A standard 200-μl reaction containing 0.6 to 5 μg of enzyme was used. Values represent means ± standard deviations (n = 4). C18:1 is oleic acid.
Compositions of the CoA Pools of E. coli and S. enterica.
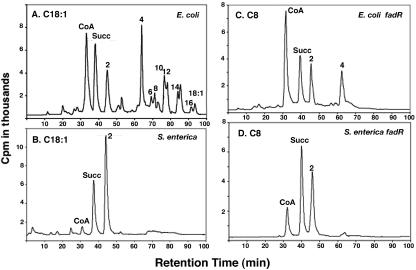
Although increased acyl-CoA synthetase activity could account for the differences in growth of the two organisms on decanoate, it seemed unlikely to explain the much poorer growth on the medium-chain fatty acids, octanoate and hexanoate, because even an E. coli fadR strain having constitutive expression of the fad regulon shows only very slow growth on octanoate (and no detectable growth on hexanoate) despite high-level expression of acyl-CoA synthetase and the other Fad enzymes. The levels of transcription of the Fad regulon genes are similar in the fadR derivatives of the two organisms (Table 4). Hence, the ability of S. enterica fadR strains to grow well on these short-chain acids must be due to a factor other than enzyme levels. To address this question, we determined the CoA pool compositions of cultures of E. coli strain SI237 (UB1005 panD fadR) and S. enterica strain SI239 (LT2 panD fadR) grown on acetate supplemented with either octanoate (C8) or hexanoate (C6) and compared them with the pools seen in cultures grown on oleate. The unexpected result was that S. enterica was much more efficient than E. coli in degrading fatty acyl-CoAs (Fig. 5). Specifically, E. coli cultures grown on any of the fatty acids tested accumulated high levels of an intermediate having the retention time of butyryl-CoA (C4). In contrast, S. enterica showed complete degradation of oleate, octanoate, and hexanoate to acetyl-CoA. These results explain the failure of E. coli fadR strains to grow on hexanoate and their feeble growth on octanoate plus the ability of S. enterica fadR strains to show significant growth on these acids. Hexanoic and octanoic acids are very poor acyl-CoA synthetase substrates (Fig. 4). Thus, the small amount of hexanoyl-CoA or octanoyl-CoA generated in E. coli apparently undergoes only one or two cycles of β-oxidation before it cannot be further metabolized, whereas in S. enterica, both hexanoyl-CoA and octanoyl-CoA are completely degraded to acetyl-CoA (C2) and thus yield more acetyl-CoA per acyl-CoA molecule initially formed.
FIG. 5.
CoA metabolite pools of E. coli (A and C) and S. enterica (B and D). For panels A and B, overnight cultures of E. coli strain SI92 (UB1005 panD) (A) and S. enterica strain SI81 (LT2 panD) (B) were grown in minimal medium E supplemented with 1.5 μM β-alanine and 0.2% oleic acid. The cells were harvested, washed, and resuspended in minimal medium E alone and subcultured at a ratio of 1:100 in minimal medium E supplemented with 0.2% oleic acid but lacking β-alanine. After 6 h of β-alanine starvation, 0.5 μM β-[3-3H]alanine was added and cells were allowed to grow until they reached an optical density at 600 nm of 0.5. For panels C and D, fadR derivatives of the same strains were grown in minimal medium E supplemented with 0.4% acetate and 1.5 μM β-alanine. The cultures were harvested and washed to remove β-alanine and then diluted in the same growth medium lacking β-alanine. Dilutions were made such that after 6 h of β-alanine starvation the culture had reached mid-exponential phase. Octanoic acid (5 mM) and β-[3-3H]alanine (0.5 μM) were added to both cultures, followed by incubation for an additional 75 min. The cells were then harvested and extracted, and the distribution of the label among the CoA species was determined by reversed-phase HPLC as described in Materials and Methods. The identities of the tritium-labeled CoA species were determined by comparisons with the retention times of the standards. The numbers used to label the peaks correspond to the carbon chain lengths of acyl-CoA species: acetyl-CoA (C2), butyryl-CoA (C4), hexanoyl-CoA (C6), octanoyl-CoA (C8), decanoyl-CoA (C10), lauroyl-CoA (C12), myristoyl-CoA (C14), palmitoyl-CoA (C16), and oleoyl-CoA (C18:1). However, note that the separation depends on the hydrophobic surface area and that the acyl chain substitutions (e.g., double bonds, hydroxyl group) of the intermediates of degradation alter the hydrophobicity of the chains. Therefore, long-chain acyl-CoAs could be about one carbon atom longer than they appear. For short-chain acyl-CoAs, the identifications are valid, since substitutions would result in shifts to very early retention times. Succ, succinyl-CoA.
The FadBA and FadE proteins of S. enterica are largely responsible for the more efficient degradation.
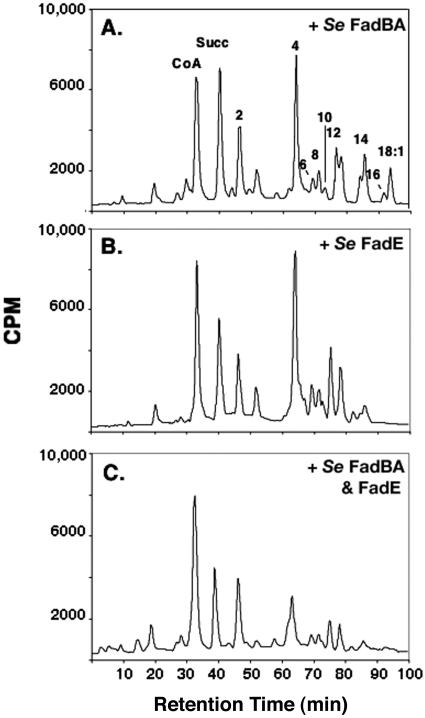
Given the lack of large differences in the expression levels of the fad regulon enzymes between the two organisms, it seemed that the differing efficiencies of β-oxidation must be due to the enzymes that catalyze the steps of the degradative cycle, FadE and FadBA. To evaluate the contributions of these major β-oxidation enzymes, we deleted the fadE or fadBA genes of a S. enterica fadR strain. Neither of these mutant strains grew on octanoate or hexanoate, indicating that there were no other enzymes available that could replace the functions of fadE or fadBA (data not shown). We then constructed an E. coli strain in which the chromosomal copies of fadBA were replaced by a single copy of the fadBA homologues of S. enterica (see Materials and Methods) and analyzed the CoA pool of a panD derivative of this strain grown on oleate. The pool composition of this strain indicated no improvement in efficiency in degrading long- and some medium-chain fatty acid intermediates (Fig. 6A). However, a similar replacement of the E. coli fadE gene with those of S. enterica gave a marked improvement in degradative efficiency (Fig. 6B). When both fadE and fadBA chromosomal copies were replaced by the respective homologues from S. enterica, the levels of the acyl-CoA intermediate with the retention time of butyryl-CoA (C4) was also reduced significantly (Fig. 6C). These data indicate that the function of FadE and FadBA of S. enterica is a major factor in the efficient utilization of various-chain-length fatty acids.
FIG. 6.
Efficiency of oleate utilization in E. coli strains expressing S. enterica β-oxidation enzymes. The CoA pools of E. coli strains SI241 (chromosomal fadBA replaced by S. enterica fadBA) (A), SI244 (chromosomal fadE replaced by S. enterica fadE) (B), and SI248 (chromosomal fadE and fadBA genes replaced by the S. enterica homologues) (C) grown on oleate were obtained and analyzed as described in the Fig. 5 legend. Designations are the same as those in Fig. 5.
DISCUSSION
Our initial hypothesis to explain the differing growth phenotypes of E. coli and S. enterica on medium-chain fatty acids was that, unlike E. coli FadR, S. enterica FadR would bind medium-chain fatty acyl-CoAs and dissociate from the fad operon operators. The resulting induction would then allow utilization of the fatty acid carbon chains. However, in our prior work, isothermal titration calorimetry showed that S. enterica FadR bound decanoyl-CoA only modestly better than did E. coli FadR (22). This difference seemed unlikely to explain the differences in growth on this fatty acid, and this hypothesis was subsequently eliminated by exchanging the FadR genes between the two species. An E. coli strain whose chromosomal fadR copy had been replaced with the S. enterica fadR gene failed to grow on decanoate. Moreover, an S. enterica fadR strain containing a single copy of the E. coli fadR gene remained able to grow on decanoate. Finally, the levels of FadR are virtually identical in the two organisms (22), and similarly weak fadE induction by decanoate were seen in S. enterica fadE-lacZ transcriptional fusion strains that carried single copies of either the E. coli or S. enterica fadR genes (data not shown).
Given that the fad gene operators and fad induction amplitudes in the two organisms were virtually identical, we then turned to the remaining player in Fad operon regulation, the acyl-CoA ligand. Our hypothesis was that upon fatty acid supplementation the acyl-CoA pools may be higher in S. enterica than in E. coli. If so, this could explain the weak MCFA induction of β-oxidation enzymes in S. enterica and the finding that FadR overexpression inhibits growth of E. coli, but not that of S. enterica, on oleate. Indeed, we observed two- to threefold-higher acyl-CoA synthetase activity in S. enterica than in E. coli, with either decanoic or oleic acid as a substrate (Fig. 3). However, since the purified His6-FadD proteins of the two organisms did not show significant differences in activity toward medium- or long-chain fatty acids (Fig. 4), the increased acyl-CoA synthetase activity must be due to differences in FadD expression. Note that, although the FadR binding sites of the E. coli and S. enterica fadD promoter regions are the most divergent of the fad operators in the two organisms, these promoters cannot be highly repressed because induction of the fad regulon requires the acyl-CoAs produced by FadD (12).
The most significant finding of this work is that S. enterica β-oxidation enzymes utilize fatty acids much more efficiently than do those of E. coli. Complete degradation of oleoyl-CoA was observed when the CoA pools of S. enterica grown on oleate as the carbon source were analyzed. In contrast, short- and medium-chain acyl-CoA intermediates accumulated in E. coli cells grown on oleate. In agreement with our results, it was long ago reported that wild-type E. coli performs incomplete oxidative degradation of oleate (25). When the rates of radioactive CO2 production from oleate labeled at various positions within the acyl chain were compared to that of the carboxyl carbon, the rate of release of carbon atoms 10 and 18 was only 0.4 and 0.33, respectively (25). This incomplete oxidation is metabolically expensive and becomes more so as the acyl chain length is shortened because each acyl chain requires activation in an ATP-requiring reaction in order to enter the first β-oxidation cycle, whereas subsequent cycles of oxidation require no additional activation. Thus, it is more favorable energetically to produce a given amount of acetyl-CoA by complete degradation of a long acyl chain than by partial degradation of multiple acyl chains of whatever chain length. Thus, S. enterica extracts all of the acetyl-CoA equivalents from fatty acids of all chain lengths, whereas E. coli extracts fewer equivalents from medium- and long-chain fatty acids and almost none from short-chain acids. In E. coli FadR strains, the breakpoint lies between the C8 and C10 acids. These strains grow well on decanoate but only very feebly on octanoate. The fact that octanoic acid is a very poor substrate for acyl-CoA synthetase whereas decanoyl-CoA is a good substrate (about sixfold better than octanoate), coupled with the lesser yield of acetyl-CoA from the few octanoate molecules that become activated renders octanoic acid a very poor carbon source for E. coli. However, in S. enterica FadR strains, although octanoic acid activation remains a problem, the organism extracts a full measure of acetyl-CoA from all activated octanoic acid molecules and growth proceeds, albeit slowly. The same arguments apply more strongly to growth on hexanoate but to a much lesser extent to decanoate, where E. coli can grow well despite only partial oxidation of the carbon chain. Note that the accumulation of material having the retention time of butyryl-CoA in E. coli fadR mutants was not foreseen, since we had supposed that induction of the ato operon by accumulated acetoacetyl-CoA would provide the thiolase necessary for complete conversion to acetyl-CoA. Therefore, it seems that if acetoacetyl-CoA accumulates, its concentration is insufficient to induce the ato operon. In contrast, we might have expected accumulation of butyryl-CoA in S. enterica, since this organism lacks the ato operon, which appears to be due to deletion of the ato region together with several downstream genes of unknown function. However, despite this deletion and the lack of genes elsewhere in the genome that encode Ato protein homologues, S. enterica does not accumulate butyryl-CoA. Indeed, we have found that both wild-type and fadR strains of S. enterica LT2 fail to grow on butyrate and do not give rise to mutants able to utilize this carbon source, such as those that occur readily in E. coli fadR strains. The ato system seems designed to degrade exogenous acetoacetate, suggesting that, unlike E. coli, S. enterica does not encounter acetoacetate in its environment. Note that E. coli has recently been shown to contain homologues of FadB and FadA, called FadJ and FadI, whose expression is controlled by FadR (6). These proteins are responsible for the residual aerobic growth of E. coli fadBA mutants on oleate and play a role in the anaerobic fatty acid degradation that suggests that their preferred substrates are medium- and short-chain acyl-CoAs (6). However, since fadBA fadR mutants of S. enterica are unable to grow on hexanoate or octanoate, the close homologues of the fadIJ genes present in S. enterica do not play an important role in utilization of these acids.
The finding that the FadBA and FadE proteins of S. enterica are much more efficient at complete oxidation of fatty acids than their E. coli homologues is surprising, given the high sequence identities (>91%) of the proteins from the two organisms and the similar ecological niches occupied. Our results indicate that no single step of the β-oxidation cycle is responsible for the greater efficiency of the S. enterica system. The high levels of amino acid sequence conservation suggests that identification of the residues responsible for the differing efficiencies should be straightforward, although there are an appreciable number of active sites involved.
Acknowledgments
This work was supported by NIH grant AI15650 from the National Institute of Allergy and Infectious Diseases.
We thank an anonymous reviewer for very helpful comments on the original submitted manuscript.
REFERENCES
- 1.Bachman, B. J. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36:524-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, P. N., and C. C. DiRusso. 1994. Molecular and biochemical analyses of fatty acid transport, metabolism, and gene regulation in Escherichia coli. Biochim. Biophys. Acta 1210:123-145. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, J. W., and J. E. Cronan, Jr. 2002. The enigmatic Escherichia coli fadE gene is yafH. J. Bacteriol. 184:3759-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, J. W., and J. E. Cronan, Jr. 2001. Escherichia coli FadR positively regulates transcription of the fabB fatty acid biosynthetic gene. J. Bacteriol. 183:5982-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, J. W., R. M. Morgan-Kiss, and J. E. Cronan, Jr. 2003. A new Escherichia coli metabolic competency: growth on fatty acids by a novel anaerobic beta-oxidation pathway. Mol. Microbiol. 47:793-805. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 8.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 10.Clark, D. 1981. Regulation of fatty acid degradation in Escherichia coli: analysis by operon fusion. J. Bacteriol. 148:521-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, D. P., and J. E. Cronan, Jr. 1996. Two-carbon compounds and fatty acids as carbon sources, p. 343-357. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.). Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 12.Cronan, J. E., Jr. 1997. In vivo evidence that acyl coenzyme A regulates DNA binding by the Escherichia coli FadR global transcription factor. J. Bacteriol. 179:1819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cronan, J. E., Jr. 1980. β-Alanine synthesis in Escherichia coli. J. Bacteriol. 141:1291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiRusso, C. C., T. L. Heimert, and A. K. Metzger. 1992. Characterization of FadR, a global transcriptional regulator of fatty acid metabolism in Escherichia coli. Interaction with the fadB promoter is prevented by long chain fatty acyl coenzyme A. J. Biol. Chem. 267:8685-8691. [PubMed] [Google Scholar]
- 16.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 17.Frerman, F. E. 1973. The role of acetyl coenzyme A: butyrate coenzyme A in the transferase uptake of butyrate by isolated membrane vesicles of Escherichia coli. Arch. Biochem. Biophys. 159:444-452. [DOI] [PubMed] [Google Scholar]
- 18.Frerman, F. E., and W. Bennett. 1973. Studies on the uptake of fatty acids by Escherichia coli. Arch. Biochem. Biophys. 159:434-443. [DOI] [PubMed] [Google Scholar]
- 19.Gui, L., A. Sunnarborg, and D. C. LaPorte. 1996. Regulated expression of a repressor protein: FadR activates iclR. J. Bacteriol. 178:4704-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henry, M. F., and J. E. Cronan, Jr. 1992. A new mechanism of transcriptional regulation: release of an activator triggered by small molecule binding. Cell 70:671-679. [DOI] [PubMed] [Google Scholar]
- 22.Iram, S. H., and J. E. Cronan. 2005. Unexpected functional diversity among FadR fatty acid transcriptional regulatory proteins. J. Biol. Chem. 280:32148-32156. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins, L. S., and W. D. Nunn. 1987. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J. Bacteriol. 169:42-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kameda, K., and W. D. Nunn. 1981. Purification and characterization of acyl coenzyme A synthetase from Escherichia coli. J. Biol. Chem. 256:5702-5707. [PubMed] [Google Scholar]
- 25.Klein, K., R. Steinberg, B. Fiethen, and P. Overath. 1971. Fatty acid degradation in Escherichia coli. An inducible system for the uptake of fatty acids and further characterization of old mutants. Eur. J. Biochem. 19:442-450. [DOI] [PubMed] [Google Scholar]
- 26.Maloy, S. R., C. L. Ginsburgh, R. W. Simons, and W. D. Nunn. 1981. Transport of long and medium chain fatty acids by Escherichia coli K12. J. Biol. Chem. 256:3735-3742. [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Morgan-Kiss, R. M., and J. E. Cronan. 2004. The Escherichia coli fadK (ydiD) gene encodes an anerobically regulated short chain acyl-CoA synthetase. J. Biol. Chem. 279:37324-37333. [DOI] [PubMed] [Google Scholar]
- 30.Nunn, W. D. 1986. A molecular view of fatty acid catabolism in Escherichia coli. Microbiol. Rev. 50:179-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overath, P., G. Pauli, and H. U. Schairer. 1969. Fatty acid degradation in Escherichia coli. An inducible acyl-CoA synthetase, the mapping of old-mutations, and the isolation of regulatory mutants. Eur. J. Biochem. 7:559-574. [PubMed] [Google Scholar]
- 32.Overath, P., and E. M. Raufuss. 1967. The induction of the enzymes of fatty acid degradation in Escherichia coli. Biochem. Biophys. Res. Commun. 29:28-33. [DOI] [PubMed] [Google Scholar]
- 33.Pauli, G., and P. Overath. 1972. ato Operon: a highly inducible system for acetoacetate and butyrate degradation in Escherichia coli. Eur. J. Biochem. 29:553-562. [DOI] [PubMed] [Google Scholar]
- 34.Platt, R., D. L. Reynolds, and G. J. Phillips. 2003. Development of a novel method of lytic phage delivery by use of a bacteriophage P22 site-specific recombination system. FEMS Microbiol. Lett. 223:259-265. [DOI] [PubMed] [Google Scholar]
- 35.Roughan, G. 1994. A semi-preparative enzymic synthesis of malonyl-CoA from [14C]acetate and 14CO2: labelling in the 1, 2 or 3 position. Biochem. J. 300:355-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spector, M. P., C. C. DiRusso, M. J. Pallen, F. Garcia del Portillo, G. Dougan, and B. B. Finlay. 1999. The medium-long-chain fatty acyl-CoA dehydrogenase (fadF) gene of Salmonella typhimurium is a phase 1 starvation-stress response (SSR) locus. Microbiology 145:15-31. [DOI] [PubMed] [Google Scholar]
- 37.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 38.Subrahmanyam, S., and J. E. Cronan, Jr. 1998. Overproduction of a functional fatty acid biosynthetic enzyme blocks fatty acid synthesis in Escherichia coli. J Bacteriol. 180:4596-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weeks, G., M. Shapiro, R. O. Burns, and S. J. Wakil. 1969. Control of fatty acid metabolism. I. Induction of the enzymes of fatty acid oxidation in Escherichia coli. J. Bacteriol. 97:827-836. [DOI] [PMC free article] [PubMed] [Google Scholar]