Abstract
The core human U6 promoter consists of a proximal sequence element (PSE) located upstream of a TATA box. The PSE is recognized by the snRNA-activating protein complex (SNAPc), which consists of five types of subunits, SNAP190, SNAP50, SNAP45, SNAP43, and SNAP19. The TATA box is recognized by TATA box binding protein (TBP). In addition, basal U6 transcription requires the SANT domain protein Bdp1 and the transcription factor IIB-related factor Brf2. SNAPc and mini-SNAPc, which consists of just SNAP43, SNAP50, and the N-terminal third of SNAP190, bind cooperatively with TBP to the core U6 promoter. By generating complexes smaller than mini-SNAPc, we have identified a 50-amino-acid region within SNAP190 that is (i) required for cooperative binding with TBP in the context of mini-SNAPc and (ii) sufficient for cooperative binding with TBP when fused to a heterologous DNA binding domain. We show that derivatives of mini-SNAPc lacking this region are active for transcription and that with such complexes, TBP can still be recruited to the U6 promoter through cooperative interactions with Brf2. Our results identify complexes smaller than mini-SNAPc that are transcriptionally active and show that there are at least two redundant mechanisms to stably recruit TBP to the U6 transcription initiation complex.
An important event during activation of transcription is the establishment of a stable transcription initiation complex. For RNA polymerase II, transcription initiation complexes appear to be partially disassembled at each round of transcription, losing transcription factor IIB (TFIIB), RNA polymerase II, and TFIIF as well as, in some cases, TFIIE and TFIIH (23, 24, 33, 34). Other factors including TFIIA, TATA box binding protein (TBP), some of the TFIID TBP-associated factors (TAFs), and, in a crude extract containing mediator, TFIIH, TFIIE, and mediator itself, are left behind and form a scaffold for reinitiation of transcription (33). In the case of RNA polymerase III transcription, initiation complexes preformed on 5S- and tRNA-type promoters can direct several rounds of transcription in the presence of concentrations of sarcosyl that inhibit formation of new initiation complexes, suggesting that the complexes are stable for several rounds of transcription (8, 9).
The human U6 snRNA promoter is a type 3 RNA polymerase III promoter characterized by gene-external promoter elements, which can be divided into enhancer and core regions. The enhancer region, referred to as the distal sequence element, is located about 200 bp upstream of the transcription start site and contains binding sites for the POU domain protein Oct-1 and the zinc finger protein STAF (see reference 7 for a review). Both of these factors can activate U6 transcription and can be localized to the U6 promoter region in vivo by chromatin immunoprecipitation experiments (14, 20-22, 26, 28, 35).
The U6 core promoter region consists of a proximal sequence element (PSE) and a TATA box located about 50 and 25 bp, respectively, upstream of the transcription start site. The PSE recruits the snRNA-activating protein complex (SNAPc), a complex consisting of five types of subunits, SNAP190, SNAP50, SNAP45, SNAP43, and SNAP19 (reference 5 and references therein). Of these subunits, the largest (SNAP190) contains an unusual Myb domain with a half Myb repeat (Rh) followed by four full-length Myb repeats (Ra, Rb, Rc, and Rd), of which the last two are required for DNA binding of the complex (19, 32). The TATA box recruits TBP (7). TBP is one component of the TFIIIB activity, which for 5S- and tRNA-type promoters also contains the SANT domain protein Bdp1 (previously called human B') and the TFIIB-related factor Brf1 (4, 16, 25, 30). Basal transcription from the human U6 promoter requires a specialized TFIIIB activity in which the Brf1 protein is replaced by another TFIIB-related factor known as Brf2 (previously called BRFU or TFIIIB50 [25, 29]; for a universal nomenclature of TFIIIB subunits, see reference 31).
All of the U6 promoter elements are required for stable assembly of a U6 transcription initiation complex (11), which presumably depends on a number of protein-protein interactions among members of the complex. One such interaction mediates cooperative binding of the transcription activator Oct-1 and the basal factor SNAPc and involves a glutamate residue at position 7 within the Oct-1 POU domain and a lysine at position 900 within SNAP190 (2, 17). In the natural U6 promoter, this protein-protein contact is mediated by a positioned nucleosome that brings the octamer sequence in close proximity to the PSE (35). A subcomplex of SNAPc called mini-SNAPc, which consists of SNAP43, SNAP50, and the N-terminal third of SNAP190, cannot bind cooperatively with Oct-1 because it lacks the SNAP190 region containing the critical lysine residue at position 900. Surprisingly, however, mini-SNAPc binds more efficiently to a PSE than the full-length SNAPc, suggesting that the full-length complex contains a built-in damper that down-regulates its binding to DNA and that is deleted in mini-SNAPc (19).
In addition to the interaction between the activator Oct-1 and SNAPc, protein-protein contacts are likely to occur among core promoter binding factors, because they also display cooperative binding. For example, TBP and Brf2 bind cooperatively to a TATA box (1), and SNAPc and TBP bind cooperatively to probes containing both a PSE and a TATA box (18). We have shown that mini-SNAPc, although unable to bind cooperatively with the Oct-1 POU domain, can bind cooperatively with TBP (19). Because mini-SNAPc binds relatively efficiently to DNA on its own, cooperative binding in this case results, in effect, in recruitment of TBP to the DNA (19).
Here we have assembled complexes that were smaller than mini-SNAPc (stmSNAPc) and tested their ability to recruit TBP to the DNA. The results identify a 50-amino-acid (aa) region within the N-terminal part of SNAP190 that is required for cooperative binding of mini-SNAPc with TBP. When fused to the RcRd repeats of SNAP190 or the GAL4 DNA binding domain, this region is sufficient to recruit TBP to a TATA box. By using stmSNAPcs lacking the TBP-recruiting region, we show that there are at least two redundant mechanisms to recruit TBP to U6 transcription initiation complexes. Our results also identify a stmSNAPc that is 192 aa smaller than mini-SNAPc and yet is still capable of directing basal RNA polymerase III transcription.
MATERIALS AND METHODS
Protein expression and purification.
The various versions of the SNAP190, SNAP43, and SNAP50 proteins were expressed in Escherichia coli with the T7 system of Studier and colleagues (27). In some cases, both SNAP43 and SNAP50 were expressed from a single plasmid. All SNAP190 derivatives were tagged at their C terminus with six histidines. The lysates from cells expressing SNAP43, SNAP50, and SNAP190 were then mixed and incubated with rotation for 2 h at 4°C, and the SNAP complexes were purified by affinity chromatography on nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (Qiagen). TBP and the RcRd portion of SNAP190 were expressed as glutathione S-transferase (GST) fusion proteins in E. coli and purified by affinity chromatography on glutathione agarose beads. The proteins were then either eluted with reduced glutathione (G-TBP) or cleaved from the GST moiety with thrombin. Brf2 carrying a His tag at its C terminus was expressed in E. coli and purified by affinity chromatography on Ni-NTA agarose beads. The GAL4 DNA binding domain (GAL4 DBD-HT) and the (1-90)-GAL4 DNA binding domain fusion protein [(1-90)GAL4 DBD-HT], both carrying a His tag at their C termini, were expressed in E. coli with the T7 system and were purified by affinity chromatography on Ni-NTA agarose beads.
Electrophoretic mobility shift assays (EMSAs).
The binding reactions were performed in a total volume of 20 μl containing 80 mM KCl, 20 mM HEPES (pH 7.9), 5 mM MgCl2, 0.2 mM EDTA, 10% glycerol, 1 mM dithiothreitol (DTT), 0.07% Tween 20, 0.5 μg of poly(dG-dC) · (dG-dC), 0.25 μg of pUC118, and 20 μg of fetal bovine serum. The reaction mixtures were incubated for 20 min at 4°C before addition of the radiolabeled probe, followed by a 30-min incubation at 30°C. The binding reactions were fractionated on a 5% polyacrylamide gel (39:1 acrylamide-bisacrylamide) in TGEM buffer (50 mM Tris base, 380 mM glycine, 2 mM EDTA, 5 mM MgCl2). The probes used in Fig. 5 had the sequence 5′ CATTTCTCT ATCGATAGGT ACCGGAGGAC TGTCCTCCGC GGAGGACTGT CCTCCGCGGA GGACTGTCCT CCGGCTGCATATAAGCAGCT GCTTTTTCTC GAGTACTGG 3′, in which the three G17 GAL4 binding sites (3) are underlined and the TATA box is indicated in bold type. The italicized AT in the TATA box was changed to GG in the probe carrying a mutated TATA box.
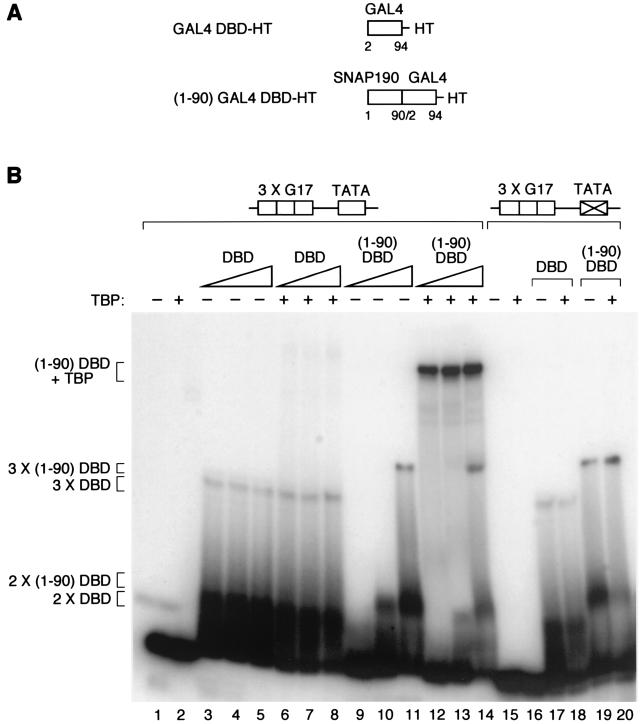
FIG. 5.
SNAP190 aa 1 to 90 can recruit TBP to the DNA when fused to a heterologous DNA binding domain. (A) The structures of the various fusion proteins are illustrated. HT, His Tag. (B) EMSA performed with probes containing three tandem copies of the G17 GAL4 DNA binding site (3 X G17) (3) inserted 5 bp upstream of the wild-type (lanes 1 to 14) or mutated (lanes 15 to 20) HIV-1 TATA box (see Materials and Methods for the sequences of the probes), and the proteins indicated above the lanes. The presence (+) or absence (−) of TBP and the amounts (indicated by the thickness of the triangle) of the proteins containing the GAL4 DNA binding domain (DBD) are shown over the gel. DBD, GAL4 DBD-HT; (1-90)DBD, (1-90)GAL4 DBD-HT (see panel A). Both the GAL4 DBD-HT and (1-90)GAL4 DBD-HT proteins were titrated over a threefold range. In lanes 17 and 18, the same amount of GAL4 DBD-HT was used as in lane 5. In lanes 19 and 20, the same amount of (1-90)GAL4 DBD-HT was used as in lane 11. The proteins present in the various protein-DNA complexes are indicated to the left of the panels.
Transcription.
To deplete SNAPc from whole-cell extracts (15), 1 ml of extract was incubated with 0.5 ml of protein A agarose beads covalently linked to anti-SNAP190 antibodies (CS696, directed against aa 1 to 9 of SNAP190 [19]) and 0.5 ml of beads covalently linked to anti-SNAP43 antibodies (CS48, directed against aa 355 to 368 of SNAP43 [6]). In vitro transcription reactions were performed as described previously (10) with the following modifications. The transcription reactions were performed in a total volume of 40 μl containing 100 ng of the pU6/Hae/RA.2 template, 500 ng of poly(dG-dC) · (dG-dC), and 8 μl of whole-cell extract diluted 1:2 as a result of the treatment with the antibody beads and were complemented with 12.5 ng of recombinant TBP and 2 μl of ATP mix (0.3 M ATP, 10 μg of phosphocreatine kinase per ml, and 10 mM creatine kinase). The RNAs were analyzed by RNase T1 protection as described previously (12).
RESULTS
The SNAP190 region from aa 34 to 83 is required for cooperative binding with TBP in the context of mini-SNAPc.
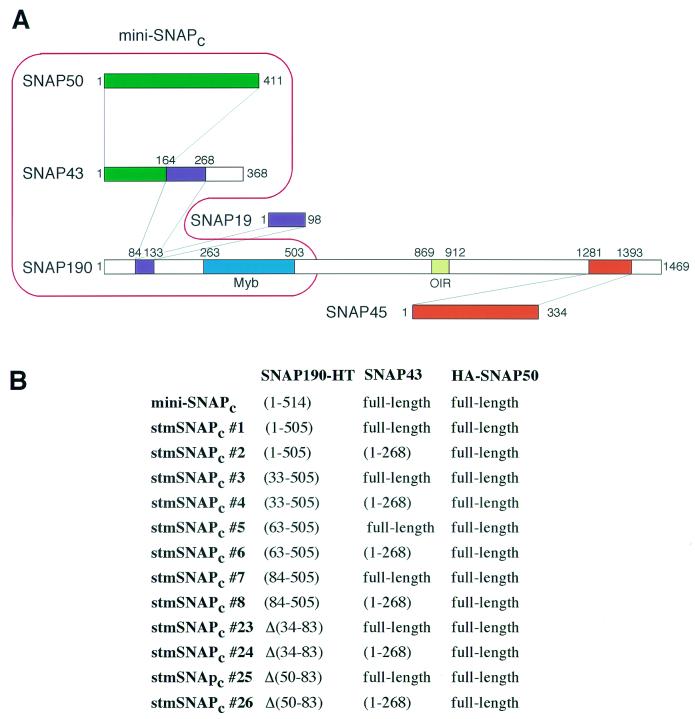
To determine which region of SNAPc might be required for cooperative binding with TBP, we generated subcomplexes of SNAPc that can bind to the PSE on their own and monitored their ability to recruit TBP in gel shift assays. Figure 1 shows a map of subunit-subunit interactions within the full-length SNAPc that we established previously (13). Mini-SNAPc, which efficiently recruits TBP, consists of SNAP50, SNAP43, and the N-terminal 514 aa of SNAP190 including the SNAP190 Myb domain (19). As shown in Fig. 1A, within the mini-SNAPc subunits, neither the first 83 aa of SNAP190 nor the SNAP43 region extending from aa 269 to 368 are required for subunit-subunit interactions. Indeed, we previously showed that stmSNAPcs that bind to the PSE can be assembled (13). We assembled a series of such complexes, which are listed in Fig. 1B. StmSNAPc#1 is identical to mini-SNAPc except that the SNAP190 fragment present is slightly shorter at the C terminus, ending after aa 505. All other stmSNAPcs contain a SNAP190 truncated after aa 505, and in addition stmSNAPc#3, stmSNAPc#5, and stmSNAPc#7 lack increasing amounts of SNAP190 N-terminal sequences up to aa 83; stmSNAPc#23 is missing SNAP190 aa 34 to 83, and stmSNAPc#25 is missing SNAP190 aa 50 to 83. Each of these SNAP190 deletions was also used to assemble complexes in which SNAP43 is truncated at aa 268 (stmSNAPc#2, -#4, -#6, -#8, -#24, and -#26).
FIG. 1.
Composition of various SNAPc subcomplexes. (A) Map of the known subunit-subunit interactions within SNAPc. Within SNAP190, aa 84 to 133 associate with SNAP19 and aa 164 to 268 of SNAP43, aa 263 to 503 correspond to the Myb domain with the Rh, Ra, Rb, Rc, and Rd repeats, aa 869 to 912 correspond to the Oct-1-interacting region (OIR), and aa 1281 to 1393 associate with SNAP45. Within SNAP43, aa 1 to 164 associate with SNAP50. Mini-SNAPc contains SNAP190 aa 1 to 514, SNAP43, and SNAP50, as outlined in red. (B) The composition of each stmSNAPc is indicated. All stmSNAPcs including stmSNAPc#23-26 contain a SNAP190 protein truncated after aa 505.
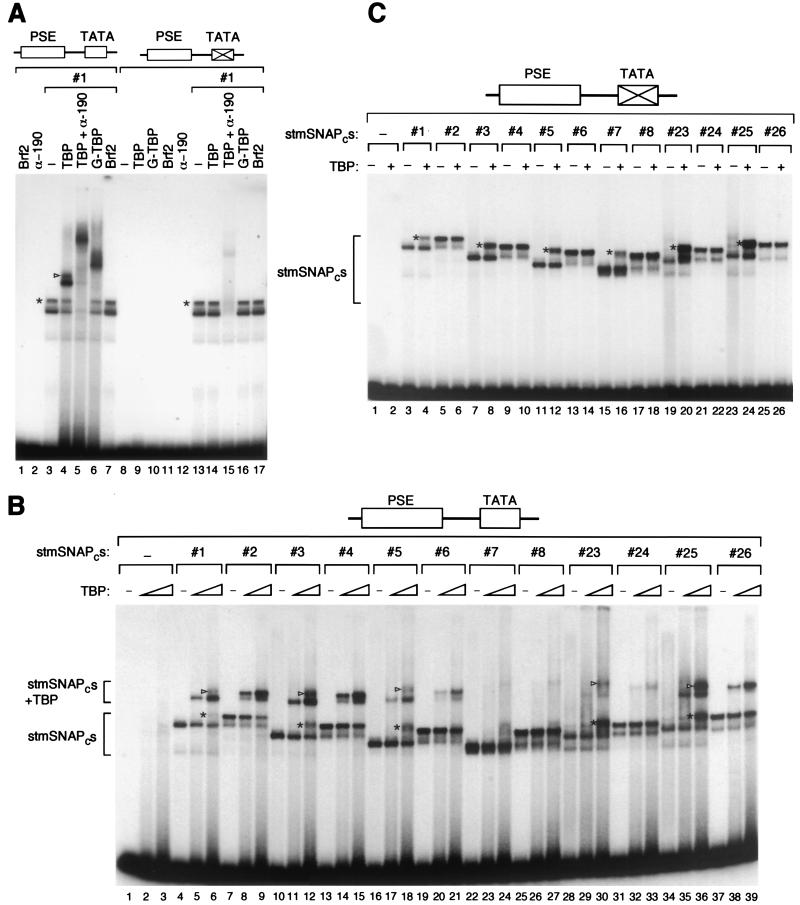
We first characterized the complexes observed upon addition of stmSNAPc#1 and TBP to a probe containing the high-affinity mouse U6 PSE and the human U6 TATA box. As shown in Fig. 2A, binding of stmSNAPc#1 to the PSE gave a doublet of complexes (the slower-migrating complex is labeled with an asterisk) (lane 3). The relative amounts of these two complexes changed under different conditions and from experiment to experiment, with the lower complex usually being prominent with stmSNAPcs containing full-length SNAP43, and the upper complex usually being prominent with stmSNAPcs containing a truncated SNAP43 lacking the C-terminal 100 aa (see below). We do not know how these two complexes differ, but one possibility is that they have different conformations. Addition of Brf2 to these complexes had no effect (Fig. 2A, lanes 7 and 17). In contrast, addition of TBP, which on its own was unable to bind efficiently to the probe (Fig. 2B), gave rise to a doublet of slower-migrating complexes (lane 4). These complexes contained both stmSNAPc#1, because they were supershifted by addition of an anti-SNAP190 antibody (lane 5), and TBP, because their migration was retarded when TBP was replaced by a larger GST-TBP fusion protein (lane 6). As expected, none of the TBP-containing complexes were observed on probes containing a mutated TATA box (lanes 8 to 17). However, the complexes containing just stmSNAPc#1 were still formed on these probes, as expected (lanes 13 to 17) and could be supershifted by addition of the anti-SNAP190 antibody (lane 15). Thus, on probes containing both a PSE and a TATA box, TBP can be recruited to the TATA box by stmSNAPc#1.
FIG. 2.
A 50-aa region of SNAP190 is required for cooperative binding of the complex with TBP. (A) EMSA performed with the proteins indicated above the lanes and a probe containing the wild-type mouse U6 PSE and either the wild-type or a mutated human U6 TATA box as indicated above the lanes. In panels A to C, the upper complex in the doublet observed with stmSNAPc#1 (#1) is labeled with an asterisk to the left of the complex, and the upper complex of the doublet observed with stmSNAPc#1 and TBP is labeled with an arrowhead to the left of the complex. α-190, anti-SNAP190 antibody; −, only complex #1 was added. (B) EMSA performed with the proteins indicated above the lanes and a probe containing the wild-type mouse U6 PSE and the human U6 TATA box. The absence (−) or presence of the stmSNAPc proteins (indicated by the stmSNAPc number) is indicated over the lanes. The absence (−) and amount (indicated by the height of the triangle) of TBP is indicated over the gel. TBP was titrated over a threefold range. The protein-DNA complexes containing the stmSNAPcs only or both the stmSNAPcs and TBP are indicated. (C) EMSA performed with the proteins indicated above the lanes and a probe containing a wild-type mouse U6 PSE and a mutated TATA box. The presence or absence (−) and the amount (indicated by the height of the triangle over the lanes) of TBP are indicated over the lanes.
To compare the abilities of the different stmSNAPcs to recruit TBP, we first performed a titration experiment to determine the amount of each complex required to obtain equal binding to the high-affinity mouse U6 PSE. As shown in Fig. 2B, we were able to titrate each complex so as to obtain roughly equal amounts of protein-DNA complex (compare lanes 4, 7, 10, 13, 16, 19, 22, 25, 28, 31, 34, and 37). (Note that in this case, the stmSNAPcs containing full-length SNAP43, such as stmSNAPc#1, gave rise only to the lower complex of the doublet observed in Fig. 2A.) Little or no detectable binding was obtained with increasing amounts of full-length TBP on its own (lanes 2 and 3). In sharp contrast, when both stmSNAPc#1 and TBP were incubated with the probe, the complex containing both stmSNAPc#1 and TBP appeared, as before (complex labeled stmSNAPcs + TBP [lanes 5 and 6]). (Note that at high concentrations of TBP, the upper band of the doublet appeared [lane 6, see also lanes 12, 18, 30, and 36, complex labeled with an asterisk], and as before, this complex was supershifted similarly to the corresponding main stmSNAPc-PSE complex with TBP [see lanes 6, 12, 18, 30, and 36, complex labeled with an arrowhead]).
Deletion of the N-terminal 32 aa of SNAP190 had no effect on TBP recruitment (stmSNAPc#3; Fig. 2B, lanes 11 and 12); however, deletion of the first 62 aa weakened TBP recruitment (stmSNAPc#5; lanes 17 and 18), and deletion of the first 83 aa resulted in a barely detectable stmSNAPc-TBP complex (stmSNAPc#7; lanes 23 and 24). This suggested that a region required for cooperative binding with TBP lay between SNAP190 aa 63 (or slightly N-terminal of aa 63) and aa 84. Surprisingly, however, an internal deletion encompassing aa 50 to 83 of SNAP190 had little effect (stmSNAPc#25; lanes 35 and 36), and only a larger internal deletion from aa 34 to 83 reduced cooperative binding significantly (stmSNAPc#23; lanes 29 and 30). Combination of the SNAP190 deletions with deletion of the last 100 aa of SNAP43 had little additional effect on cooperative binding with TBP (compare each odd-numbered complex with the next even-numbered complex). Together, these results indicate that in the context of mini-SNAPc, the SNAP190 region extending from aa 34 to 83 contains redundant sequences required for cooperative binding with TBP.
When we tested whether cooperative binding is dependent on a wild-type PSE and TATA box, we did not detect binding of the stmSNAPcs to probes containing a mutant PSE, consistent with our previous results (13), even upon addition of TBP (data not shown). The results with a probe containing a wild-type mouse U6 PSE and a mutant TATA box for all of the stmSNAPcs are shown in Fig. 2C. As before for stmSNAPc#1 (Fig. 2A), we did not observe any of the stmSNAPc-TBP complexes in the absence of the TATA box, indicating that cooperative binding is dependent on both an intact PSE and an intact TATA box, as observed previously (18). However, for all stmSNAPcs containing full-length SNAP43, we did observe formation of the upper complex of the doublet upon addition of TBP (Fig. 2C, lanes 4, 8, 12, 16, 20, and 24, complex labeled with an asterisk). As shown above, the mobility of this complex was not affected by replacing TBP with a larger GST-TBP fusion protein or with Brf2 (Fig. 2A, compare lanes 16 and 17), again suggesting that it corresponds to another conformation of the stmSNAPcs.
The N-terminal 90 aa of SNAP190 are sufficient for cooperative binding with TBP when fused to the SNAP190 RcRd Myb repeats.
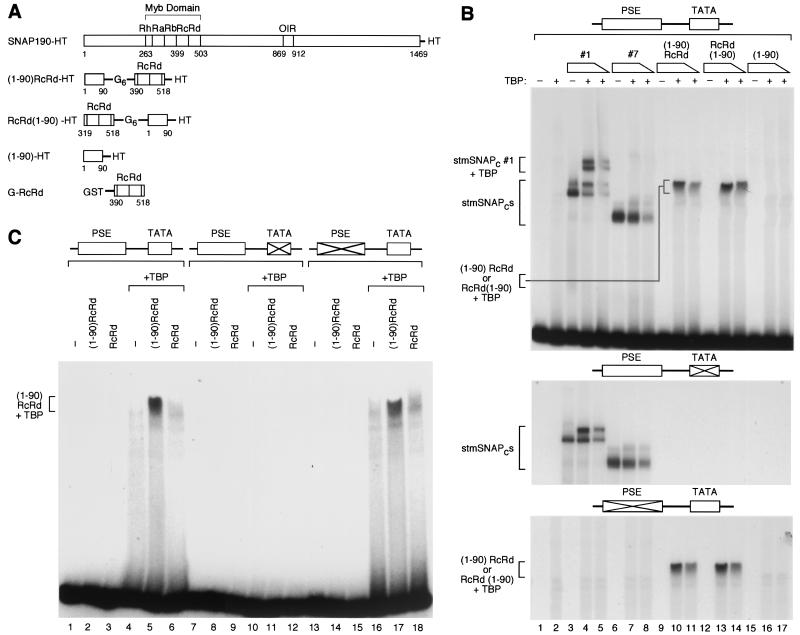
The above results suggest that in a complex consisting of SNAP43 (or the first 268 aa of SNAP43), SNAP50, and the N-terminal third of SNAP190, the N terminus of SNAP190 is required for cooperative binding with TBP. To determine whether this SNAP190 region might recruit TBP in the absence of other SNAPc subunits, we fused it via a six-glycine linker to either the N [(1-90)RcRd] or C [RcRd(1-90)] terminus of the RcRd repeats of SNAP190 (Fig. 3A). The SNAP190 RcRd repeats can bind to the PSE on their own, as visualized with a gel lacking MgCl2, although with reduced specificity (32; M. Wong and N. Hernandez, unpublished results). As illustrated in Fig. 3A, we also generated a protein consisting of just the N-terminal 90 aa of SNAP190. All the constructs carried a His tag (HT) for easy purification from E. coli lysates.
FIG. 3.
When fused to the SNAP190 RcRd Myb repeats, SNAP190 aa 1 to 90 are sufficient to recruit TBP to a TATA box. (A) The structures of the various fusion proteins are illustrated. HT, His tag; G6, a run of six glycines. (B) EMSA performed with probes containing either wild-type or mutated mouse U6 PSE and wild-type or mutated human U6 TATA box, as indicated above the panels, and the proteins indicated above the lanes. The absence (−) or presence (+) of TBP is indicated over the gel. TBP was titrated over a threefold range. The proteins present in the various protein-DNA complexes are indicated to the left of the panels. (C) EMSA performed with the probes and proteins indicated [−, absence of (1-90)RcRd and RcRd proteins] above the lanes. The proteins present in the protein-DNA complex are indicated to the left.
The results are shown in Fig. 3B. As before, stmSNAPc#1, but not stmSNAPc#7, efficiently recruited TBP to the DNA (lanes 1 to 8), and this effect was dependent on both an intact TATA box (second panel) and an intact PSE (third panel). Neither the (1-90)RcRd nor the RcRd (1-90) fusion proteins bound to DNA on their own (lanes 9 and 12). However, upon addition of TBP, increasing amounts of both fusion proteins gave rise to increasing amounts of a protein-DNA complex [lanes 10, 11, 13, and 14, complexes labeled (1-90)RcRd or RcRd(1-90) + TBP]. These complexes were dependent on an intact TATA box (second panel) but surprisingly not on an intact PSE (third panel). We then checked whether the SNAP190 region from aa 1 to 90 was required for this effect. As shown in Fig. 3C, neither the (1-90)RcRd protein nor the RcRd protein bound efficiently to DNA on their own (lanes 2 and 3), although it should be noted that the same preparation of RcRd protein bound to DNA, as visualized with a gel lacking MgCl2, consistent with our previous results (data not shown) (32). Importantly, in the presence of TBP, the (1-90)RcRd fusion protein, but not the RcRd protein, gave a prominent complex (compare lane 5 to lane 6), and this complex was dependent, as before, on an intact TATA box (lane 11) but not on an intact PSE (lane 17). Thus, the prominent complex is dependent on the presence of the first 90 aa of SNAP190 in the fusion protein.
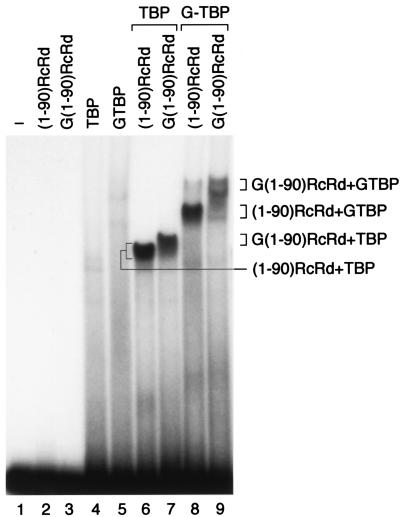
Since the prominent complex observed with TBP and the (1-90)RcRd fusion protein did not depend on a intact PSE, we wondered whether it indeed contained both TBP and the SNAP190-derived fusion proteins. As shown in Fig. 4, ( 1-90)RcRd and TBP each bound poorly to the probe alone, as did GST fusion derivatives of each protein [G(1-90)RcRd and GTBP, lanes 2 to 5]. When (1-90)RcRd was combined with TBP, however, a protein-DNA complex was observed, as before (lane 6). The complex migrated with increasingly slower mobility upon replacement of (i) (1-90)RcRd with the larger protein G(1-90)RcRd (lane 7), (ii) TBP with the larger protein GTBP (lane 8), and (iii) both (1-90)RcRd and TBP with the larger GST fusion proteins (lane 9). It is not clear why fusion of the GST moiety to the (1-90)RcRd protein affected migration of the complex less than fusion of the GST moiety to TBP (lanes 7 and 8). Nevertheless, the observation that fusion of the GST moiety to either protein does retard migration of the complex confirms that the complex does indeed contain both components. Together, these results suggest that a fusion protein consisting of the first 90 aa of SNAP190 and the RcRd DNA binding domain is capable of recruiting TBP to the DNA. The observation that this recruitment is not dependent on an intact PSE suggests that the (1-90)RcRd and RcRd(1-90) proteins bind with low specificity to DNA.
FIG. 4.
The complex observed in the presence of TBP and the (1-90)RcRd-HT fusion protein contains both TBP and (1-90)RcRd-HT. EMSA performed with the proteins indicated above the lanes and a probe containing the wild-type mouse U6 PSE and the human U6 TATA box. G(1-90)RcRd and GTBP are GST fusion proteins. The proteins present in each protein-DNA complex are indicated to the right. In lane 1 no proteins were added to the probe.
The N-terminal 90 aa of SNAP190 are sufficient for cooperative binding with TBP when fused to the GAL4 DNA binding domain.
We also expressed two proteins, shown in Fig. 5A, consisting of either the GAL4 DNA binding domain alone (GAL4 DBD-HT, aa 2 to 94 of GAL4) or the first 90 aa of SNAP190 fused to the GAL4 DNA binding domain [(1-90)GAL4 DBD-HT] with, in each case, a His tag (HT) at the C terminus. We tested the abilities of these two proteins to recruit TBP to probes carrying three tandem GAL4 DNA binding sites (G17 site [3]) upstream of either a wild-type or mutated TATA box derived from the human immunodeficiency virus type 1 (HIV-1) promoter (see Materials and Methods for the sequences of these probe). The presence of three GAL4 binding sites allowed us, in effect, to test three different spacings between the GAL4 site and the TATA box in a single probe. As shown in Fig. 5B, TBP on its own bound very weakly to the probe, as expected, such that the complex is not visible at this exposure of the gel (lane 2). In contrast, both the GAL4 DBD-HT and the (1-90)GAL4 DBD-HT proteins [labeled DBD and (1-90)DBD, respectively, in Fig. 5B] bound to DNA on their own (lanes 3 to 5 and 9 to 11). The GAL DBD-HT protein formed at least two complexes even at the lowest concentration used (lane 3, complexes labeled 3xDBD and 2xDBD), while the (1-90)GAL4 DBD protein formed at least two complexes only at the highest concentration used [lane 11, complexes labeled 3x (1-90)DBD and 2x (1-90)DBD]. These complexes probably correspond to occupancy of two and three of the three GAL4 DNA binding sites on the probe (a weak, smaller complex probably corresponding to occupancy of a single site can be seen just above the probe signal on low exposures of the gel). Very strikingly, in the presence of TBP, a new, prominent complex was obtained with the (1-90)GAL4 DBD-HT but not the DBD-HT protein [compare lanes 12 to 14 with lanes 6 to 8, complex labeled (1-90)DBD + TBP]. This complex migrated more slowly than either the TBP-TATA box complex (which is visible in lane 2 on very long exposures of the gel) or the 3x (1-90)DBD complex (lane 11), was dependent on the presence of a wild-type TATA box (compare lanes 12 to 14 with lane 20), and probably corresponds, therefore, to a complex containing both TBP and the (1-90)GAL4 DBD-HT protein. Thus, the first 90 aa of SNAP190 are capable of recruiting TBP to the DNA when fused to at least two types of DNA binding domain, the RcRd repeats from SNAP190 or the DNA binding domain of the Saccharomyces cerevisiae GAL4 protein. This indicates that the first 90 aa of SNAP190 are sufficient to recruit TBP to a TATA box when fused to a heterologous DNA binding domain.
The stmSNAPcs are competent for U6 transcription.
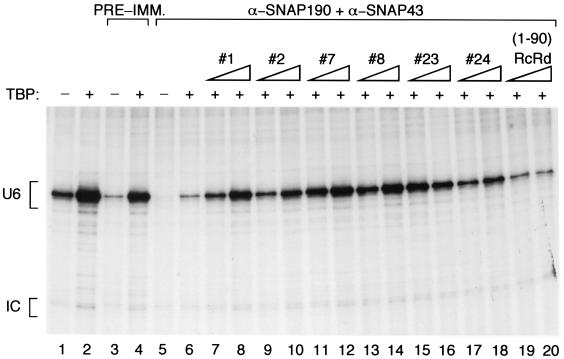
We depleted a transcription extract of endogenous SNAPc with a mixture of anti-SNAP190 and anti-SNAP43 antibodies and tested the abilities of the various stmSNAPcs to restore transcription in the depleted extract. As shown in Fig. 6, addition of TBP to the untreated extract stimulated transcription, indicating that TBP was limiting (lane 2). Upon treatment of the extract with preimmune antibody beads, transcription was diminished but less so than upon depletion with anti-SNAP190 and anti-SNAP43 beads (compare lanes 3 and 5). Upon addition of TBP, transcription from the extract treated with preimmune beads was restored to near-wild-type levels, whereas transcription from the extract depleted with anti-SNAP190 and anti-SNAP43 beads was stimulated only slightly (compare lanes 4 and 6). To test the stmSNAPcs, we complemented the depleted extract with a constant amount of TBP and increasing amounts of the various stmSNAPcs. The amounts of the various stmSNAPcs were normalized as in Fig. 2B to give rise to equal binding to the mouse U6 PSE. As shown in Fig. 6, lanes 7 to 18, the various stmSNAPcs restored transcription to different levels, but there was no correlation between the ability of an stmSNAPc to restore transcription and its ability to recruit TBP. Thus, stmSNAPc#7, stmSNAPc#8, stmSNAPc#23, and stmSNAPc#24, which cannot recruit TBP efficiently (Fig. 2), were as active or more active than stmSNAPc#1 and stmSNAPc#2, which recruit TBP efficiently. Addition of the (1-90)RcRd protein did not stimulate transcription above the levels observed with addition of just TBP, indicating that this protein is inactive for transcription (compare lanes 19 and 20 to lane 6). Together, these results indicate that the ability of the stmSNAPcs to recruit TBP to the TATA box is not essential for transcription. Further, they show that stmSNAPc#8, which contains SNAP190 aa 84 to 505, SNAP43 aa 1 to 268, and SNAP50, contains all the information required to direct basal levels of RNA polymerase III transcription. On the other hand, the (1-90)RcRd SNAP190 derivative on its own is not capable of directing transcription, indicating that it is lacking essential parts present in stmSNAPc#8.
FIG. 6.
The transcription activities of different stmSNAPcs does not correlate with their abilities to recruit TBP. A whole-cell extract was either left untreated (lanes 1 and 2) or treated with preimmune (PRE-IMM.) antibody beads (lanes 3 and 4) or with a mixture of anti-SNAP190 (α-SNAP190) and anti-SNAP43 (α-SNAP43) antibody beads (lanes 5 to 20). The extracts were then complemented with the proteins indicated above the lanes. The presence (+) or absence (−) of TBP and the amount of the stmSNAPc protein (indicated by the height of the triangle) are indicated over the lanes. The extracts were programmed with the plasmid pU6/Hae/RA.2, which carries the human U6 promoter. The titrations of the stmSNAPcs and the (1-90)RcRd protein were over a threefold range. The transcripts were analyzed by RNase T1 protection. The signals corresponding to correctly initiated U6 RNA (U6) or to a control RNA (IC) included in the reaction mixtures to monitor RNA handling and recovery are indicated to the left of the gel.
TBP can be recruited to the DNA through cooperative binding with Brf2.
Cooperative binding of SNAPc and TBP to the U6 promoter achieves both recruitment of TBP and recruitment of SNAPc. The stmSNAPcs, however, lack the damper of DNA binding present in the full-length SNAPc. In our experiments, we adjusted their concentrations so as to obtain in each case efficient binding to the PSE. Thus, under these conditions, cooperative binding mainly serves to recruit TBP. The observation that stmSNAPcs unable to recruit TBP efficiently are nevertheless active for transcription suggests, then, that there is another route to recruit TBP to the U6 promoter. Indeed, Brf2 has been shown to bind cooperatively with TBP (1). We asked, therefore, whether Brf2 could recruit TBP to the U6 promoter in the presence of stmSNAPcs. The results are shown in Fig. 7 .
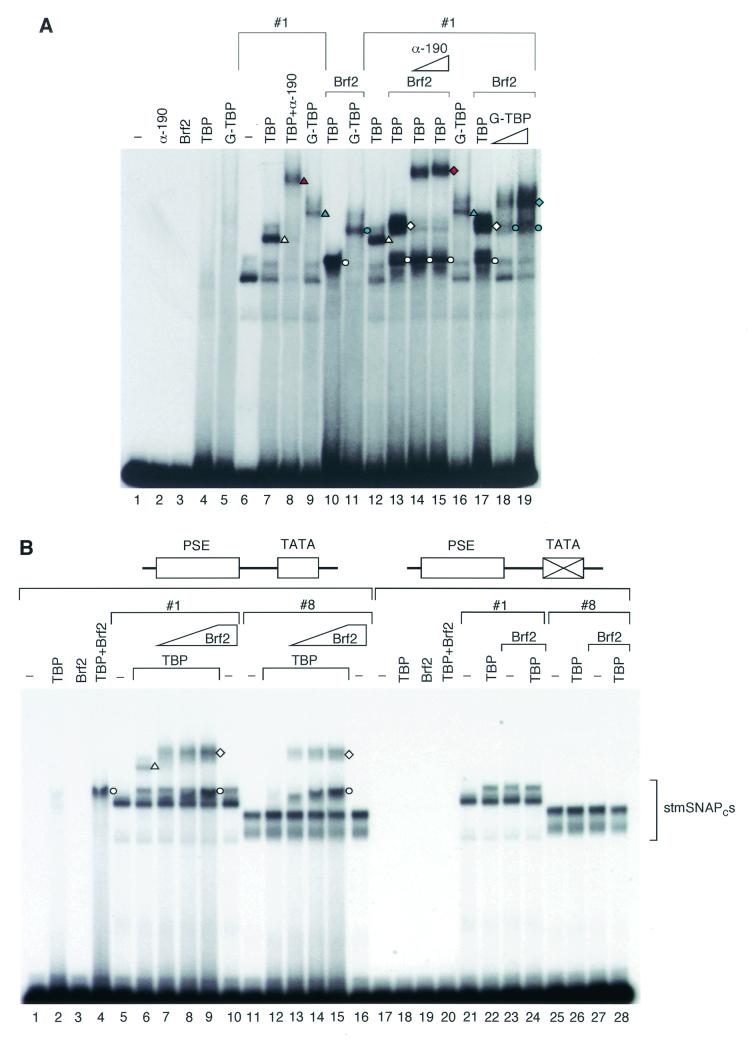
FIG. 7.
TBP can be recruited to the U6 transcription initiation complex in the absence of SNAP190 aa 1 to 83. EMSA performed with the proteins indicated above the lanes and a probe containing the wild-type mouse U6 PSE and the human U6 TATA box. The amounts (indicated by the height of the triangle) of an anti-SNAP190 antibody (α-190) (CS696) and G-TBP, a GST-TBP fusion protein, are indicated over the lanes.The symbols to the right of different protein-DNA complexes indicate complexes containing proteins as follows: white circles, TBP and Brf2; blue circles, G-TBP and Brf2; white triangles, stmSNAPc#1 and TBP; blue triangles, stmSNAPc#1 and G-TBP; red triangle, stmSNAPc#1, TBP, and anti-SNAP190; white diamonds, stmSNAPc#1, TBP, and Brf2; blue diamonds, stmSNAPc#1, G-TBP, and Brf2; red diamonds, stmSNAPc#1, TBP, Brf2, and anti-SNAP190. (B) EMSA performed with probes containing the wild-type mouse U6 PSE and either a wild-type or mutant human U6 TATA box as indicated above the lanes, and the proteins indicated above the lanes. The Brf2 titrations were over a threefold range. The symbols at the right of different protein-DNA complexes indicate complexes containing proteins as follows: white circles, TBP and Brf2; white triangles, stmSNAPc#1 and TBP; white diamonds, stmSNAPc#1 or stmSNAPc#8, TBP, and Brf2.
We first characterized the complexes obtained with various combinations of Brf2, stmSNAPc#1, and TBP, as shown in Fig. 7A. As before, TBP alone did not bind efficiently to the probe, nor did a GST-TBP fusion protein, Brf2, and an antibody directed against SNAP190 (lanes 2 to 5). StmSNAPc#1 alone bound to the probe (lane 6) and was able to recruit TBP, as expected (lane 7, complex labeled with a white triangle). The stmSNAPc#1-TBP complex was retarded in its migration by an antibody directed against SNAP190 (lane 8, red triangle) or by replacement of TBP by the larger GST-TBP fusion protein (lane 9, blue triangle), consistent with it containing both stmSNAPc#1 and TBP. Brf2 and TBP, each of which alone binds very poorly, if at all, to the probe (lanes 3 and 4) gave a strong complex when added together (lane 10, white circle). This complex migrated more slowly when TBP was replaced with the larger GST-TBP fusion protein (lane 11, blue circle), indicating that it does indeed contain TBP. Thus, as observed before (1), TBP and Brf2 bind cooperatively.
When Brf2 was added to the stmSNAPc#1-TBP complex, a new complex of lower mobility was obtained (Fig. 7A, lane 13, white diamond). The mobility of this complex was retarded by increasing amounts of anti-SNAP190 antibody (lanes 14 and 15, red diamond) and by replacement of TBP with the larger GST-TBP fusion protein (lanes 18 and 19, blue diamond), consistent with it containing stmSNAPc#1 and TBP. Moreover, complexes formed in the presence of Brf2 consistently migrated slightly more slowly than the corresponding complexes lacking Brf2, confirming that these complexes indeed contain Brf2 [compare (i) the stmSNAPc#1-TBP-Brf2 complex in lane 13 (white diamond) with the stmSNAPc#1-TBP complex in lane 12 (white triangle); (ii) the stmSNAPc#1-TBP-Brf2-α-190 complex in lanes 14 and 15 (red diamond) to the stmSNAPc#1-TBP-α-190 complex in lane 8 (red triangle); (iii) the stmSNAPc#1-G-TBP-Brf2 complex in lanes 18 and 19 (blue diamond) to the stmSNAPc#1-G-TBP complex in lane 16 (blue triangle)]. Together, these results indicate that TBP binds cooperatively not only with stmSNAPc#1 but also with Brf2. Further, a complex containing all three components is efficiently obtained.
We next compared stmSNAPc#1, which can recruit TBP, with stmSNAPc#8, which is deficient in TBP recruitment, for assembly into complexes containing TBP and Brf2. As shown in Fig. 7B, neither TBP nor Brf2 bound efficiently to DNA on their own, but together, they formed a strong complex, as before (lane 4, white circle). Both stmSNAPc#1 and stmSNAPc#8 bound efficiently to the mouse U6 PSE (lanes 5 and 11) but only stmSNAPc#1 recruited TBP to the DNA (compare lanes 6 and 12, white triangle). Furthermore, we did not observe cooperative binding of Brf2 and any of the two stmSNAPcs (lanes 10 and 16). Upon addition of increasing amounts of Brf2, increasing amounts of the stmSNAPc#1-TBP-Brf2 complex were obtained (lanes 7 to 9, white diamond). Remarkably, addition of Brf2 to the binding reaction mixture containing stmSNAPc#8 and TBP also resulted in efficient formation of a complex containing the three components, indicating that TBP can be recruited through Brf2 (lanes 13 to 15, white diamond). In addition, a complex containing just TBP and Brf2 was observed in the presence of both stmSNAPcs (for example, lane 15, white circle), although with stmSNAPc#1 the complex was difficult to visualize because it comigrated with the complex that probably corresponds to an altered conformation of stmSNAPc#1 (compare lane 9, white circle, with the binding reaction mixture lacking Brf2 in lane 6). All the complexes containing TBP and Brf2 were dependent on the presence of a TATA box in the probe (lanes 18 to 28).
DISCUSSION
We have shown before that TBP and SNAPc bind cooperatively to the U6 promoter. Here we have identified a region in mini-SNAPc that is required for recruitment of TBP. Thus, as shown in Fig. 8, we now know of five functional regions within SNAP190. Two of these are involved in association with other SNAPc subunits; the region extending from aa 84 to 133 associates with SNAP43 and SNAP19, and the region extending from aa 1281 to 1393 associates with SNAP45 (13). The Myb domain extends from aa 263 to 503 and contains the two Myb repeats Rc and Rd that are required for DNA binding (19, 32). Two other regions are involved in cooperative binding with other members of the U6 initiation complex: (i) the Oct-1-interacting region (OIR) from aa 869 to 912, which allows cooperative binding with Oct-1 through direct protein-protein contacts (2, 17); and (ii) the TBP recruitment region (TRR1) from aa 34 to 83, which is required for cooperative binding with TBP in the context of mini-SNAPc. The TRR1 is also sufficient to recruit TBP when fused to a heterologous DNA binding domain.
FIG. 8.
Known functional regions of SNAP190. The known functional regions of SNAP190 follow: SNAP190 aa 34 to 83, TBP recruitment region 1 (TRR1); SNAP190 aa 84 to138, SNAP19- and SNAP43-interacting region; SNAP190 aa 263 to 503, Myb domain; SNAP190 aa 869 to 912, Oct-1-interacting region (OIR); and SNAP190 aa 1281 to 1393, SNAP45-interacting region.
With the full-length SNAPc and with probes containing the low-affinity human U6 PSE, cooperative binding of SNAPc and TBP effectively results in recruitment of both proteins to the DNA. In such a context, the cooperative interaction between the two proteins is necessary for efficient transcription from the human U6 core promoter (18). With mini-SNAPc on probes containing the high-affinity mouse U6 PSE, cooperative binding results mainly in the recruitment of TBP to the DNA because mini-SNAPc binds relatively efficiently on its own (18, 19). Here we show that derivatives of mini-SNAPc unable to recruit TBP efficiently can still direct efficient transcription. We also show that in the presence of Brf2, trimeric complexes containing mini-SNAPc, Brf2, and TBP can efficiently be formed on the U6 core promoter, even with mini-SNAPc derivatives unable to recruit TBP. This is because, at the concentrations used, the mini-SNAPc derivatives can bind to DNA on their own, and Brf2 and TBP recruit each other to the DNA. These observations indicate that there are redundant mechanisms to recruit TBP to the U6 transcription initiation complex. There are also redundant mechanisms to recruit SNAPc to the DNA, since it can be recruited by cooperative binding with TBP or with Oct-1. Such redundant mechanisms may ensure efficient formation of the transcription initiation complex on chromatin templates in vivo. Furthermore, they probably ensure that once the U6 transcription initiation complex is formed, it remains stable for several rounds of transcription.
It will be very interesting to determine whether SNAPcs that still contain the damper of DNA binding but are unable to bind cooperatively with TBP are inactive for transcription from the core U6 promoter lacking the Oct-1 binding site. We do not know, however, whether deletion of the SNAP190 TRR1 will be sufficient to debilitate recruitment of TBP by the full-length SNAPc. Indeed, although this region is required for efficient TBP recruitment in the context of stmSNAPc#1 (and is sufficient to recruit TBP when fused to the RcRd repeats), it is quite possible that the full-length complex contains other regions, either in the C-terminal two thirds of SNAP190 or in SNAP45 or in SNAP19, that act redundantly with the SNAP190 TRR1 region.
The smallest stmSNAPc we tested in transcription is stmSNAPc#8, which lacks the last 100 aa of SNAP43 as well as the first 83 aa and the last 9 aa of the SNAP190 truncation present in mini-SNAPc. Remarkably, this complex is active for basal RNA polymerase III transcription from the human U6 promoter, indicating that it contains all the information required to recruit, ultimately, RNA polymerase III. We do not know whether RNA polymerase III directly contacts SNAPc, but our results suggest that any essential contact would involve subunits or part of subunits present in stmSNAPc#8, the smallest functional SNAPc subcomplex we have assembled so far.
Acknowledgments
We thank Winship Herr and Ashish Saxena for comments on the manuscript, L. Schramm for reagents, J. Duffy and P. Renna for artwork and photography.
This work was funded in part by NIH grant GM38810. N.H. and B.M. are supported by the Howard Hughes Medical Institute.
REFERENCES
- 1.Cabart, P., and S. Murphy. 2001. BRFU, a TFIIB-like factor, is directly recruited to the TATA-box of polymerase III small nuclear RNA gene promoters through its interaction with TATA-binding protein. J. Biol. Chem. 276:43056-43064. [DOI] [PubMed] [Google Scholar]
- 2.Ford, E., M. Strubin, and N. Hernandez. 1998. The Oct-1 POU domain activates snRNA gene transcription by contacting a region in the SNAPc largest subunit that bears sequence similarities to the Oct-1 coactivator OBF-1. Genes Dev. 12:3528-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giniger, E., and M. Ptashne. 1988. Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc. Natl. Acad. Sci. USA 85:382-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henry, R. W., E. Ford, R. Mital, V. Mittal, and N. Hernandez. 1998. Crossing the line between RNA polymerases: transcription of human snRNA genes by RNA polymerases II and III. Cold Spring Harbor Symp. Quant. Biol. 63:111-120. [DOI] [PubMed] [Google Scholar]
- 5.Henry, R. W., V. Mittal, B. Ma, R. Kobayashi, and N. Hernandez. 1998. SNAP19 mediates the assembly of a functional core promoter complex (SNAPc) shared by RNA polymerases II and III. Genes Dev. 12:2664-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry, R. W., C. L. Sadowski, R. Kobayashi, and N. Hernandez. 1995. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerases II and III. Nature 374:653-657. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez, N. 2001. snRNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 276:26733-26736. [DOI] [PubMed] [Google Scholar]
- 8.Kassavetis, G. A., B. R. Braun, L. H. Nguyen, and E. P. Geiduschek. 1990. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 60:235-245. [DOI] [PubMed] [Google Scholar]
- 9.Kovelman, R., and R. G. Roeder. 1990. Sarkosyl defines three intermediate steps in transcription initiation by RNA polymerase III: application to stimulation of transcription by E1A. Genes Dev. 4:646-658. [DOI] [PubMed] [Google Scholar]
- 10.Kuhlman, T. C., H. Cho, D. Reinberg, and N. Hernandez. 1999. The general transcription factors IIA, IIB, IIF, and IIE are required for RNA polymerase II transcription from the human U1 snRNA promoter. Mol. Cell. Biol. 19:2130-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunkel, G. R., and D. A. Danzeiser. 1992. Formation of a template committed complex on the promoter of a gene for the U6 small nuclear RNA from the human requires multiple sequence elements, including the distal region. J. Biol. Chem. 267:14250-14258. [PubMed] [Google Scholar]
- 12.Lobo, S., and N. Hernandez. 1989. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell 58:55-67. [DOI] [PubMed] [Google Scholar]
- 13.Ma, B., and N. Hernandez. 2001. A map of protein-protein contacts within the small nuclear RNA-activating protein complex SNAPc. J. Biol. Chem. 276:5027-5035. [DOI] [PubMed] [Google Scholar]
- 14.Mach, C. M., B. W. Hargrove, and G. R. Kunkel. 2002. The small RNA gene activator protein, SphI postoctamer homology-binding factor/selenocysteine tRNA gene transcription activating factor, stimulates transcription of the human interferon regulatory factor-3 gene. J. Biol. Chem. 277:4853-4858. [DOI] [PubMed] [Google Scholar]
- 15.Maroney, P. A., G. J. Hannon, and T. W. Nielsen. 1990. Transcription and cap trimethylation of a nematode spliced leader RNA in a cell-free system. Proc. Natl. Acad. Sci. USA 87:709-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mital, R., R. Kobayashi, and N. Hernandez. 1996. RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol. Cell. Biol. 16:7031-7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mittal, V., M. A. Cleary, W. Herr, and N. Hernandez. 1996. The Oct-1 POU-specific domain can stimulate small nuclear RNA gene transcription by stabilizing the basal transcription complex SNAPc. Mol. Cell. Biol. 16:1955-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mittal, V., and N. Hernandez. 1997. Role for the amino-terminal region of human TBP in U6 snRNA transcription. Science 275:1136-1140. [DOI] [PubMed] [Google Scholar]
- 19.Mittal, V., B. Ma, and N. Hernandez. 1999. SNAPc: a core promoter factor with a built-in DNA-binding damper that is deactivated by the Oct-1 POU domain. Genes Dev. 13:1807-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy, S., J.-B. Yoon, T. Gerster, and R. G. Roeder. 1992. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol. Cell. Biol. 12:3247-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myslinski, E., A. Krol, and P. Carbon. 1998. ZNF76 and ZNF143 are two human homologs of the transcriptional activator Staf. J. Biol. Chem. 273:1998-2006. [DOI] [PubMed] [Google Scholar]
- 22.Rincon, J. C., S. K. Engler, B. W. Hargrove, and G. R. Kunkel. 1998. Molecular cloning of a cDNA encoding human SPH-binding factor, a conserved protein that binds to the enhancer-like region of the U6 small nuclear RNA gene promoter. Nucleic Acids Res. 26:4846-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts, S. G., B. Choy, S. S. Walker, Y. S. Lin, and M. R. Green. 1995. A role for activator-mediated TFIIB recruitment in diverse aspects of transcriptional regulation. Curr. Biol. 5:508-516. [DOI] [PubMed] [Google Scholar]
- 24.Sandaltzopoulos, R., and P. B. Becker. 1998. Heat shock factor increases the reinitiation rate from potentiated chromatin templates. Mol. Cell. Biol. 18:361-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schramm, L., P. S. Pendergrast, Y. Sun, and N. Hernandez. 2000. Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev. 14:2650-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuster, C., A. Krol, and P. Carbon. 1998. Two distinct domains in Staf to selectively activate small nuclear RNA-type and mRNA promoters. Mol. Cell. Biol. 18:2650-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka, M., J.-S. Lai, and W. Herr. 1992. Promoter-selective activation domains in Oct-1 and Oct-2 direct differential activation of an snRNA and mRNA promoter. Cell 68:755-767. [DOI] [PubMed] [Google Scholar]
- 29.Teichmann, M., Z. Wang, and R. G. Roeder. 2000. A stable complex of a novel transcription factor IIB-related factor, human TFIIIB50, and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc. Natl. Acad. Sci. USA 97:14200-14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, Z., and R. G. Roeder. 1995. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB-and high-mobility-group protein 2-related domains. Proc. Natl. Acad. Sci. USA 92:7026-7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willis, I. M. 2002. A universal nomenclature for subunits of the RNA polymerase III transcription initiation factor TFIIIB. Genes Dev. 16:1337-1338. [DOI] [PubMed] [Google Scholar]
- 32.Wong, M. W., R. W. Henry, B. Ma, R. Kobayashi, N. Klages, P. Matthias, M. Strubin, and N. Hernandez. 1998. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol. Cell. Biol. 18:368-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yudkovsky, N., J. A. Ranish, and S. Hahn. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225-229. [DOI] [PubMed] [Google Scholar]
- 34.Zawel, L., K. P. Kumar, and D. Reinberg. 1995. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 9:1479-1490. [DOI] [PubMed] [Google Scholar]
- 35.Zhao, X., P. S. Pendergrast, and N. Hernandez. 2001. A positioned nucleosome on the human U6 promoter allows recruitment of SNAPc by the Oct-1 POU domain. Mol. Cell 7:539-549. [DOI] [PubMed] [Google Scholar]