Abstract
The bacterium Photorhabdus establishes a highly specific association with Heterorhabditis, its nematode host. Photorhabdus strains associated with Heterorhabditis bacteriophora or Heterorhabditis megidis were compared using a Photorhabdus DNA microarray. We describe 31 regions belonging to the Photorhabdus flexible gene pool. Distribution analysis of regions among the Photorhabdus genus identified loci possibly involved in nematode specificity.
Photorhabdus is an entomopathogenic gram-negative bacterium belonging to the Enterobacteriaceae. Both Photorhabdus luminescens and Photorhabdus temperata species are symbionts of entomopathogenic Heterorhabditis nematodes (7). Bacterial and nematode taxonomic data reveal a highly specific association between bacterial strains and nematode species. A recently described species, Photorhabdus asymbiotica, was never found to be associated with nematodes but was isolated from human infections (3, 12, 15, 22). Although a few studies have identified several Photorhabdus genes that are required for normal growth and development of the nematode (5, 6, 9, 19, 29), we have little molecular and functional data about the first step of nematode colonization and nematode specificity.
The genome sequence of P. luminescens subsp. luminescens strain TT01 revealed a high number of genes encoding proteins potentially involved in host-bacterium interaction (10). This genome also showed an impressive number of mobile or repeated genetic elements (phage remnants, IS, transposons, ERIC elements, and overrepresented families of paralogs). Furthermore, 32 genomic islands (GI) were predicted on the basis of in silico features.
The goal of this project was to identify bacterial genomic regions that are possibly involved in nematode specificity. The genomes of two strains harbored by two nematode species, P. luminescens subsp. laumondii TT01, associated with Heterorhabditis bacteriophora, and P. temperata subsp. temperata XlNach, associated with H. megidis, were compared using a Photorhabdus TT01 DNA microarray. Since the TT01 and XlNach strains belonged to different species, genomic differences could depend on the taxonomic difference. In order to avoid this bias, the microarray comparison was also performed between TT01 and the P. temperata C1 strain, which was isolated from an H. bacteriophora nematode (20, 23). The genomic regions present in both TT01 and C1 but that were missing in XlNach were considered potentially specific to strains associated with H. bacteriophora. Photorhabdus strains were stored at −80°C and grown in Luria-Bertani broth or on 1.5% nutrient agar (Difco) at 28°C. Genomic DNA (gDNA) was extracted according to the method of Brenner et al. (8) and stored at 4°C.
The Photorhabdus DNA microarray used in this study is representative of 4,144 genes out of the 4,909 predicted genes of the P. luminescens strain TT01 chromosomal sequence (accession number NC_005126). Paralogous genes (mainly IS and putative phages) were excluded. Primers were designed by use of a modified version of Primer 3 software (CAAT-Box [14]) to amplify specific fragments (300 to 600 bp). Probes were amplified with Titanium DNA polymerase (Clontech) from 30 ng of P. luminescens TT01 gDNA and purified on Multiscreen PCR filter plates (Millipore), and the probes' concentrations were adjusted to 30 ng/μl in 50% dimethyl sulfoxide. Quality and quantity of the final matrix were checked by gel electrophoresis of the amplified probes and sequencing of 96 randomly chosen amplified probes. Using the GenIII Amersham spotter, two replicates of each probe were spotted at different locations on glass slides (Microarray Type7 Star; Amersham). In each spotting replicate, 4,144 spots were gene probes, 100 were controls, and the remaining 364 were empty. The controls were composed of DNA from salmon sperm or Xenorhabdus and Photorhabdus genome or of housekeeping genes from Arabidopsis, rat, or Photorhabdus.
For the hybridization experiment, gDNA (1 μg) was labeled with Cy3 or Cy5 according to the Bioprime kit protocol (Invitrogen) except that the 10× deoxynucleoside triphosphate mixture was replaced by dATP, dGTP, dTTP (final, 0.12 mM; Promega), dCTP (0.06 mM; Promega), and Cy3- or Cy5-dCTP (0.02 mM; Amersham). Labeled gDNA was purified through QiaQuick minicolumns (QIAGEN) according to the nucleotide removal protocol except that the wash with PE buffer was performed three times. Cy5- and Cy3-labeled genomic DNAs were mixed, vacuum dried, and resuspended with 240 μl of hybridization buffer (30% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS]. Microarray slides were prehybridized for 1 h at 42°C in the saturation buffer (5× SSC, 0.1% bovine serum albumin, 0.1% SDS), rinsed under a continuous flow of deionized water, and, after a rapid isopropanol bath, blow dried using compressed gas. Hybridization was performed at 42°C for 12 h in an Amersham automated slide processor chamber. The microarrays were washed for 10 min in water, for 4 min in 1× SSC, 0.2% SDS, and four additional minutes in 0.1 × SSC, 0.2% SDS. Microarray slides were briefly washed in isopropanol, air dried, and scanned for fluorescence intensity by using an Amersham scanner.
One microarray comparison (TT01 versus XlNach or TT01 versus C1) included four slides with two dye-flipped replicates. Each slide contained two spotting replicates that were scanned in two sets of measurements. Therefore, we treated the eight sets of measurements as separate slides. The signal intensity of each spot in the microarray was quantified by using Arrayvision software (Amersham). Subsequent analysis was conducted by using Microsoft Excel software. Global normalization (using the global median) was applied on the data without background correction (11, 21). Since the different controls used on each slide demonstrate good quality for spotting and hybridization, we did not use statistical analysis. For each open reading frame tested, the median from the eight normalized values was calculated and used for determining Xlnach/TT01 or CI/TT01 ratios.
Ratios from 7.61 to 0.29 were obtained. In order to determine the ratio threshold that indicates the TT01 gene was missing in XlNach or C1, we selected regions of the XlNach or C1 strain, amplified them with the Herculase enhanced DNA polymerase (Stratagene), and sequenced them at MilleGen (Toulouse, France). Then, 24 genes with ratios ranging between 0.4 and 1.5 were randomly selected. When ratios were equal to or smaller than 0.6, genes had less than 20% identity with the probe spotted on the microarray. When ratios were equal to or higher than 0.98, genes had more than 70% identity with the probe spotted on the microarray. For ratios between 0.7 and 0.97, the identity percentage was variable. Therefore, we fixed the ratio threshold for missing genes to 0.6. According to this criterion, 449 (10.5%) and 357 (8.5%) of the TT01 genes present on the DNA microarray were missing in XlNach and C1, respectively.
In the XlNach strain, no large regions, such as canonic genomic islands, were absent relative to the reference strain TT01. Then, we searched for regions containing at least three contiguous genes missing from the XlNach genome that represent at least 50% of the TT01 genomic region. Thirty-one regions missing from XlNach were identified (Table 1). Genes contained in these regions mainly belong to the following putative functional classes: phage remnants, pilus biosynthesis, antibiotic biosynthesis, insecticidal toxins, iron uptake, and amino acid metabolism. Among the 31 regions missing from XlNach, only 8 were present in C1 (bold type in Table 1). Therefore, these eight regions may potentially be involved in H. bacteriophora interaction. Note that our DNA microarray analysis led to the identification of part of the flexible gene pool of Photorhabdus strains (17). Indeed, 29 of the regions missing from XlNach fit with in silico predicted mobile regions. Thirteen regions were or belonged to previously described GIs (10). Furthermore, using the Microbial Genome Annotation System (http://www.genoscope.cns.fr/agc/mage/wwwpkgdb/), 16 regions matched with enterobacterial variable regions (EVRs). The EVRs were gene blocks that were inserted at the location of synteny rupture in the enterobacterial core genome. Their sizes (3 to 62 kb) and their rich content in mobile elements evoked the Yersinia “difference regions,” which belong to the intra- and interspecific Yersinia flexible gene pool (18, 25).
TABLE 1.
Regions missing from XlNach described by whole-genome comparison using DNA microarraya
| Locus | Gene region | Size of region in TT01 (kb) | Products of interest (similarity or function) | Matching GIb | Matching EVR,c other features |
|---|---|---|---|---|---|
| 1 | plu0125-plu0132 | 10 | Unknown, Sai integrase | Part of GI plu0125-plu0169 | |
| 2 | plu0136-plu0156 | 18 | Unknown, transcriptional regulator, CoA metabolism, helicase | Part of GI plu0125-plu0169 | |
| 3 | plu0263-plu0269 | 8 | Plus cluster VI (Fim-like, type 1 pili) | Part of EVR plu0260-plu0271 (11.7 kb, recombinase) | |
| 4 | plu0280-plu0282 | 3 | Phage remnant | Part of EVR plu0275-plu0285 (10.4 kbases, truncated, transposase, DNA ligase, phage protein) | |
| 5 | plu0406-plu0418 | 12 | Phage remnant and plus cluster V (mrf-like, pili mannose resistant) | Part of GI plu0404-plu0419 | |
| 6 | plu0567-plu0577 | 13 | Sugar transport and metabolism, amino acid synthesis | Part of EVR plu0570-plu0574 (5.3 kb, ERIC sequences at 5′ extremity, IS) | |
| 7 | plu0597-plu0600 | 5 | Unknown, DNA methyltransferase | EVR plu0597-plu600 (4.7 kb, proximity of a truncated phage gene and a truncated transposase) | |
| 8 | plu0752-plu0764 | 17 | Peptide synthesis and transport, CoA metabolism | Part of GI plu0751-plu0798 | |
| 9 | plu0895-plu0899 | 16 | Cro/CI transcriptional regulator, antibiotic synthesis | Part of GI plu0884-plu0901 | |
| 10 | plu0960-plu0965 | 27 | Insecticidal toxins (loci tcd and tcc) | Part of GI plu 0958-plu1166 | |
| 11 | plu1002-plu1005 | 4 | Deshydratase, dioxygenase, cyanate, and benzoate transport | Part of GI plu 0958-plu1166 | |
| 12 | plu1207-plu1213 | 14 | Antibiotic synthesis | Part of GI plu1203-plu1238 | |
| 13 | plu1436-plu1443 | 11 | Antibiotic synthesis | Part of EVR plu1434-plu1448 (15.4 kb) | |
| 14 | plu2727-plu2729 | 3 | Enterobactin synthetase (entABE) | EVR plu2727-plu2729 (3.1 kb, low GC %, flanked by repeats) | |
| 15 | plu2792-plu2799 | 10 | Antibiotic synthesis | Part of EVR plu2787-plu2800 (18.3 kb, repeat-containing proteins) | |
| 16 | plu3135-plu3139 | 7 | Citrate synthase, efflux transporter, and unknown | Part of GI plu3111-plu3140 | |
| 17 | plu3144-plu3146 | 5 | lsr (luxS synthesis regulated) operon, AI-2 import | Proximity of transposases, ERIC sequence, plu3111-plu3140 | |
| 18 | plu3398-plu3405 | 6 | Phage remnant, unknown proteins | Part of GI plu3379-plu3538 | |
| 19 | plu3537-plu3539 | 5 | Aminotransferase, propionate metabolism | Overlaps the right border of GI plu3379-plu3538 | |
| 20 | plu3724-plu3726 | 4 | Aminobenzoyl-glutamate uptake and utilization | Flanks the GI plu3685-plu3723 | |
| 21 | plu4077-plu4081 | 5 | Truncated aldolase, deshydrogenase, transferase, unknown proteins | Part of EVR plu4075-plu4084 (12.3 kb, transposases) | |
| 22 | plu4143-plu4160 | 19 | ABC tranporter, amino acid metabolism, unknown | Part of GI plu4141-plu4246 | |
| 23 | plu4205-plu4219 | 16 | Transposase, unknown proteins | Part of GI plu4141-plu4246 | |
| 24 | plu4266-plu4269 | 5 | Amino acid metabolism, ABC transporter | Part of EVR plu4254-plu4310 (61.3 kb, transposase, Rhs family protein, low GC %) | |
| 25 | plu4324-plu4328 | 7 | Unknown proteins | Part of EVR plu4318-plu4331 (16.8 kb, phage proteins, truncated integrase) | |
| 26 | plu4336-plu4348 | 14 | Carotenoid biosynthesis, unknown proteins | Part of EVR plu4334-plu4348 (16.4 kb, transposase NTPase, C-terminal region of group II intron-associated maturase) | |
| 27 | plu4589-plu4591 | 3 | Unknown, transcription regulator LysR | Part of EVR plu4587-plu4594 (5.7 kb, tRNA-Gly site insertion at 3′ extremity) | |
| 28 | plu4621-plu4630 | 15 | Ferric enterobactin biosynthesis and uptake | EVR plu4621-plu4630 (14.7 kb, ATP-dependent DNA helicase RecQ at the 5′ border) | |
| 29 | plu4810-plu4823 | 15 | Lipopolysaccharide biosynthesis | Part of EVR plu4796-plu4833 (38.6 kb, transposase, low GC% by place) | |
| 30 | plu4873-plu4889 | 16 | Formate metabolism, O-methyltransferase, reverse transcriptase, macrolide-efflux protein, sugar kinase | Overlaps a part of EVR plu4872-plu4884 (11.1 kb, transposase, phage proteins, low GC% by place) | |
| 31 | plu4892-plu4895 | 6 | O-methyltransferase, transposase | Part of EVR plu4890-plu4895 (9.0 kb, transposase, low GC%) |
Bold type shows loci that are present in P. temperata C1 and that are consequently possibly involved in specific interaction with H. bacteriophora.
Genomic islands described in reference 10.
EVR shows synteny rupture relative to the Enterobacteriaceae core genome (http://www.genoscope.cns.fr/agc/mage/wwwpkgdb/).
Since nematodes are not able to grow on lawns of clinical strains (12), we assume that regions involved in nematode colonization are absent from clinical strain genomes. P. asymbiotica US3105-77, a clinical strain, is being sequenced (http://www.sanger.ac.uk/Projects/P_asymbiotica/). Using the BLASTP algorithm available from this site, we examined the presence of the eight regions possibly involved in H. bacteriophora interaction. Loci 3, 4, and 12 and the putative phage module of locus 5 were present in the clinical strain. Therefore, these loci are likely not involved in bacterial interaction with H. bacteriophora.
To further test the correlation between the TT01- and C1-specific regions and the interaction with H. bacteriophora, we studied the distribution of the putative pilus module of locus 5 (matching with a previously described GI), loci 15 and 26 (matching with EVRs), and locus 17 (not matching with any GI or EVR) in 14 Photorhabdus strains representative of the genus (Table 2) by PCR amplification. Primers were designed in flanking borders of the loci (primer sequences can be sent to readers upon request). gDNAs were amplified with the Herculase enhanced DNA polymerase (Stratagene). PCR products were analyzed by electrophoresis in 0.5% agarose gels (Table 3). When amplification succeeded, the PCR fragments were of various sizes. The amplification sizes of loci 5, 15, and 26 were not clearly correlated with the nematode host species. By contrast, locus 6 had a homogeneous size (5.2 kb) in bacteria carried by H. bacteriophora, Heterorhabditis indica, or a Heterorhabditis sp., whereas the fragment sizes were smaller than 1.1 kb in strains harbored by H. megidis or clinical strains. It is noteworthy that, except for locus 6, P. temperata gDNA was never amplified, suggesting a divergent core genome in this species.
TABLE 2.
Photorhabdus strains used in this study
| Strain | Nematode host | Geographical origin | Reference or source |
|---|---|---|---|
| Photorhabdus luminescens subsp. luminescens | |||
| HbT | Heterorhabditis bacteriophora Brecon | South Australia | 24 |
| FRG26 | Heterorhabditis sp. | Guadeloupe | H. Mauléon |
| Photorhabdus luminescens subsp. akhurstii | |||
| FRG04T | Heterorhabditis indica | Guadeloupe | 13 |
| JM12 | Heterorhabditis indica | Jamaica | 13 |
| Photorhabdus luminescens subsp. laumondii | |||
| TT01T | Heterorhabditis bacteriophora | Trinidad and Tobago | 13 |
| HP88 | Heterorhabditis bacteriophora HP88 | USAa (Utah) | 4 |
| Photorhabdus temperata | |||
| C1 = NC19 | Heterorhabditis bacteriophora NC1 | USA (North Carolina) | 1 |
| K122 | Heterorhabditis megidis | Ireland | 16 |
| Photorhabdus temperata subsp. temperata | |||
| XINachT | Heterorhabditis megidis | Russia | 2 |
| HL81 | Heterorhabditis megidis | The Netherlands | 26 |
| Photorhabdus asymbiotica subsp. asymbiotica | |||
| US3265-86T | Clinical specimen | USA | 12 |
| US3105-77 | Clinical specimen | USA | 12 |
| Photorhabdus asymbiotica subsp. australis | |||
| AU9802892T | Clinical specimen | Australia | 22 |
| AU9802397 | Clinical specimen | Australia | 22 |
USA, United States of America.
TABLE 3.
PCR assays for loci 5, 15, 17, and 26 in 14 Photorhabdus strains
| Host and bacterial strain | Amplicon size (kb)
|
|||
|---|---|---|---|---|
| Locus 5 pilus module | Locus 15 | Locus 17 | Locus 26 | |
| Host: Heterorhabditis bacteriophora | ||||
| TT01T | 8.5 | 14 | 5.2 | 18 |
| HbT | 0.5 | 7.4 | 5.2 | NA |
| HP88 | NAa | 14 | 5.2 | 17 |
| C1 | NA | NA | 5.2 | NA |
| Host: Heterorhabditis indica | ||||
| FRG04T | 0.5 | 6.9 | 5.2 | 9.4 |
| JM12 | 0.5 | 6.9 | 5.2 | NA |
| Host: Heterorhabditis sp. | ||||
| FRG26 | 0.5 | 6.9 | 5.2 | 10 |
| Host: Heterorhabditis megidis | ||||
| K122 | NA | NA | 0.8 | NA |
| XINAchT | NA | NA | 0.8 | NA |
| HL81 | NA | NA | 0.8 | NA |
| Clinical strains | ||||
| US3265-86T | 0.6 | 3 | 1.1 | 1.8 |
| US3105-77 | 0.6 | 3 | 1.1 | 1.8 |
| AU9802892T | NA | 2.6 | 0.9 | 0.5 |
| AU9802397 | NA | 2.6 | 0.9 | 0.5 |
NA, not amplified.
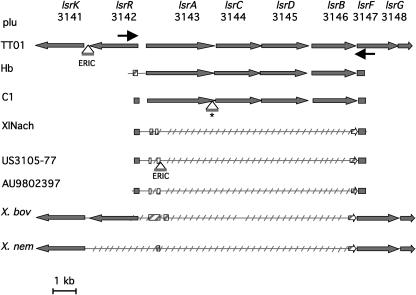
Locus 6 is similar to the Salmonella enterica serovar Typhimurium and Escherichia coli lsr region, which encodes an inner ABC transporter and a cytoplasmic phosphorylation-processing system of the autoinducer AI-2, involved in quorum sensing (27, 28, 30). In order to check the previous data, PCR products of locus 6 were purified by using the Montage PCR kit (Millipore) and sequenced with PCR primers at MilleGen (Toulouse, France). We also added for comparison the lsr-like locus of Xenorhabdus bovienii and Xenorhabdus nematophila ATCC 19061 (http://xenorhabdus.danforthcenter.org/), a genus closely related to Photorhabdus (7). Multiple alignments were performed with the ClustalW program (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html). The lsr locus was similar in TT01, Hb, and C1, three bacterial strain carried by H. bacteriophora (Fig. 1). In the US3105-77, AU9802397, and XlNach strains and in Xenorhabdus strains, the lsrA, lsrD, lsrC, and lsrB genes were missing. Furthermore, various lsrA, lsrB, and lsrR remnants were observed, showing that the lsr locus underwent independent deletions in these latter strains. Therefore, the lsr locus is an ancestral locus in the Photorhabdus and Xenorhabdus strains. The bacterial association with H. bacteriophora is possibly a selective pressure for the conservation of the lsr locus, whereas association with other nematode hosts leads to lsr locus loss by genomic decay.
FIG. 1.
Schematic representation of the deletions in the lsr region of several Photorhabdus and Xenorhabdus strains. Horizontal arrows represent primers designed for the long-range PCR analysis of the locus and for sequencing. Gray and hatched arrows or boxes symbolize, respectively, open reading frames and their remnants. Insertion of ERIC elements and a 27-nucleotide region (*) are represented.
Taken together, these data suggest that the lsr locus is possibly involved in the specific interaction with H. bacteriophora. In S. enterica serovar Typhimurium and E. coli, it was suggested that the Lsr transporter has a role in removing the AI-2 signal from the external environment in order to terminate cell-cell signaling (27, 28, 30). In nematode interaction, the termination of cell-cell signaling could be an important signal that allows a bacterial physiological shift, for example, in the insect cadaver, when bacteria recolonize the nematode intestinal tract.
In summary, this work shows that the DNA microarray is a powerful tool for selecting some genes or genomic regions potentially involved in bacterium-host interaction. Additionally, this study allowed the identification of a part of the flexible gene pool within the Photorhabdus genus.
Nucleotide sequences accession numbers.
Sequences of PCR products of Photorhabdus locus 6 have been deposited in EMBL under accession numbers AJ967010, AM039953, AJ966980, AJ966979, and AJ967009.
Acknowledgments
We thank Lionel Frangeul for the design of the DNA microarray primers, Cindy Aknin and Dany Séverac (Plateforme Transcriptome of the Génopole de Montpellier) for technical assistance in DNA microarray utilization, Claudine Médigue and David Vallenet (Atelier de Génomique Comparative, Genoscope, Evry, France) for giving access to the Microbial Genome Annotation System, the Xenorhabdus consortium and USDA-CREES 2004-35600-14181 for authorizing the use of unpublished Xenorhabdus sequence data. We also thank Patrick Tailliez for critical reading of the manuscript.
This work was supported by INRA (grant SPE 2004-1133-2) and by the Ministère de l'Industrie (AAV ASG no. 30; Contrat A01307).
REFERENCES
- 1.Akhurst, R. J. 1983. Taxonomic study of Xenorhabdus, a genus of bacteria symbiotically associated with insect pathogenic nematodes. Int. J. Syst. Bacteriol. 33:38-45. [Google Scholar]
- 2.Akhurst, R. J. 1987. Use of starch gel electrophoresis in the taxonomy of the genus Heterorhabditis (Nematoda: Heterorhabditidae). Nematologica 33:1-9. [Google Scholar]
- 3.Akhurst, R. J., N. E. Boemare, P. H. Janssen, M. M. Peel, D. A. Alfredson, and C. E. Beard. 2004. Taxonomy of Australian clinical isolates of the genus Photorhabdus and proposal of Photorhabdus asymbiotica subsp. asymbiotica subsp. nov. and P. asymbiotica subsp. australis subsp. nov. Int. J. Syst. Evol. Microbiol. 54:1301-1310. [DOI] [PubMed] [Google Scholar]
- 4.Akhurst, R. J., R. G. Mourant, L. Baud, and N. E. Boemare. 1996. Phenotypic and DNA relatedness between nematode symbionts and clinical strains of the genus Photorhabdus (Enterobacteriaceae). Int. J. Syst. Bacteriol. 46:1034-1041. [DOI] [PubMed] [Google Scholar]
- 5.Bennett, H. P., and D. J. Clarke. 2005. The pbgPE operon in Photorhabdus luminescens is required for pathogenicity and symbiosis. J. Bacteriol. 187:77-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bintrim, S. B., and J. C. Ensign. 1998. Insertional inactivation of genes encoding the crystalline inclusion proteins of Photorhabdus luminescens results in mutants with pleiotropic phenotypes. J. Bacteriol. 180:1261-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boemare, N. E. 2002. Biology, taxonomy and systematics of Photorhabdus and Xenorhabdus, p. 35-56. In R. Gaugler (ed.), Entomopathogenic nematology. CABI Publishing, Wallingford, United Kingdom.
- 8.Brenner, D. J., A. C. McWhorter, J. K. Knutson, and A. G. Steigerwalt. 1982. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 15:1133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciche, T. A., S. B. Bintrim, A. R. Horswill, and J. C. Ensign. 2001. A phosphopantetheinyl transferase homolog is essential for Photorhabdus luminescens to support growth and reproduction of the entomopathogenic nematode Heterorhabditis bacteriophora. J. Bacteriol. 183:3117-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J. F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Medigue, A. Lanois, K. Powell, P. Siguier, R. Vincent, V. Wingate, M. Zouine, P. Glaser, N. Boemare, A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 11.Fang, Y., A. Brass, D. C. Hoyle, A. Hayes, A. Bashein, S. G. Oliver, D. Waddington, and M. Rattray. 2003. A model-based analysis of microarray experimental error and normalisation. Nucleic Acids Res. 31:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farmer, J. J., III, J. H. Jorgensen, P. A. Grimont, R. J. Akhurst, G. O. Poinar, Jr., E. Ageron, G. V. Pierce, J. A. Smith, G. P. Carter, K. L. Wilson, et al. 1989. Xenorhabdus luminescens (DNA hybridization group 5) from human clinical specimens. J. Clin. Microbiol. 27:1594-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer-Le Saux, M., V. Viallard, B. Brunel, P. Normand, and N. E. Boemare. 1999. Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subsp. temperata subsp. nov. and P. asymbiotica sp. nov. Int. J. Syst. Bacteriol. 49:1645-1656. [DOI] [PubMed] [Google Scholar]
- 14.Frangeul, L., P. Glaser, C. Rusniok, C. Buchrieser, E. Duchaud, P. Dehoux, and F. Kunst. 2004. CAAT-Box, Contigs-Assembly and Annotation Tool-Box for genome sequencing projects. Bioinformatics 20:790-797. [DOI] [PubMed] [Google Scholar]
- 15.Gerrard, J., N. Waterfield, R. Vohra, and R. ffrench-Constant. 2004. Human infection with Photorhabdus asymbiotica: an emerging bacterial pathogen. Microbes Infect. 6:229-237. [DOI] [PubMed] [Google Scholar]
- 16.Griffin, C. T., J. F. Moore, and M. J. Downes. 1991. Occurrence of insect pathogenic nematodes (Steinernematidae, Heterorhabditidae) in the republic of Ireland. Nematologica 37:92-100. [Google Scholar]
- 17.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinchliffe, S. J., K. E. Isherwood, R. A. Stabler, M. B. Prentice, A. Rakin, R. A. Nichols, P. C. Oyston, J. Hinds, R. W. Titball, and B. W. Wren. 2003. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 13:2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joyce, S. A., and D. J. Clarke. 2003. A hexA homologue from Photorhabdus regulates pathogenicity, symbiosis and phenotypic variation. Mol. Microbiol. 47:1445-1457. [DOI] [PubMed] [Google Scholar]
- 20.Khan, A., and W. M. Brooks. 1977. A chromogenic bioluminescent bacterium associated with entomophilic nematode Chromonema heliothidis. J. Invertebr. Pathol. 29:253-261. [Google Scholar]
- 21.Park, T., S. G. Yi, S. H. Kang, S. Lee, Y. S. Lee, and R. Simon. 2003. Evaluation of normalization methods for microarray data. BMC Bioinformatics 4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peel, M. M., D. A. Alfredson, J. G. Gerrard, J. M. Davis, J. M. Robson, R. J. McDougall, B. L. Scullie, and R. J. Akhurst. 1999. Isolation, identification, and molecular characterization of strains of Photorhabdus luminescens from infected humans in Australia. J. Clin. Microbiol. 37:3647-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poinar, G. O., Jr. 1990. Biology and taxonomy of Steinernematidae and Heterorhabditidae, p. 23-61. In R. Gaugler and H. Kaya (ed.), Entomopathogenic nematodes in biological control. CRC Press, Boca Raton, Fla.
- 24.Poinar, G. O., G. M. Thomas, and R. Hess. 1977. Characteristics of the bacterium associated with Heterorhabitis bacteriophora (Heterorhabditidae: Rhabditida). Nematologiga 23:97-102. [Google Scholar]
- 25.Radnedge, L., P. G. Agron, P. L. Worsham, and G. L. Andersen. 2002. Genome plasticity in Yersinia pestis. Microbiology 148:1687-1698. [DOI] [PubMed] [Google Scholar]
- 26.Smits, P. H., and R.-U. Ehlers. 1991. Identification of Heterorhabitis spp. by morphometric characters and RFLP and of their symbiotic bacteria Xenorhabdus by species-specific DNA probes. IOBC/WPRS Bull. 14:195-201. [Google Scholar]
- 27.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 28.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 29.Watson, R. J., S. A. Joyce, G. V. Spencer, and D. J. Clarke. 2005. The exbD gene of Photorhabdus temperata is required for full virulence in insects and symbiosis with the nematode Heterorhabditis. Mol. Microbiol. 56:763-773. [DOI] [PubMed] [Google Scholar]
- 30.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]