Abstract
C signaling plays a key role in coordinating cell movement and differentiation during the multicellular developmental process of Myxococcus xanthus. C signaling regulates expression of genes induced after about 6 h into development, when cells are forming mounds. One gene whose expression depends absolutely on C signaling was identified by insertion of a transposable element at site Ω4406 which generated a transcriptional fusion between lacZ and an upstream promoter. We have investigated regulation of the Ω4406 promoter. A 5′ deletion revealed a negative regulatory element located between bp −533 and −100 relative to the transcriptional start site. In the absence of this element, the promoter was still developmentally regulated but about fourfold more active. Also, the truncated promoter region retained normal dependence on two developmental regulators, FruA and DevS, but lost its dependence on the C-signaling protein CsgA. We infer that C signaling partially overcomes the negative effect of the upstream element on activity of the Ω4406 promoter. Deletion of downstream DNA between bp 50 and 140 caused a threefold loss in expression, suggesting that a positive regulatory element lies in this region. Additional positive and negative regulatory elements are present in the region from bp −69 to −49, based on the effects of multiple-base-pair mutations. Within this region, a 5-bp element and a C-box-like sequence resemble sequences found in other developmentally regulated M. xanthus promoter regions, but the effects of single-base-pair changes in these sequences suggest that each functions uniquely. We conclude that regulation of the Ω4406 promoter involves multiple positive and negative regulatory elements located upstream and downstream of the region typically bound by RNA polymerase.
Myxococcus xanthus is a gram-negative, soil-dwelling bacterium that undergoes a multicellular developmental process involving signaling between cells and changes in gene expression (4). When these bacteria are starved at a high cell density on a solid surface, the rod-shaped cells move in a synchronized fashion, forming aggregation centers, where cells pile on top of one another. Within the resulting mounds, cells differentiate into dormant, spherical spores that are resistant to heat and desiccation. The spore-filled mounds are called fruiting bodies and are typically about 0.1 mm in height and width, containing on the order of 105 spores.
Five extracellular signals called A, B, C, D, and E have been inferred to control M. xanthus development, based on genetic and cell mixing experiments (3, 10), but only the A and C signals have been identified. A signaling is important at the start of development and involves the release of proteases, peptides, and amino acids that are believed to permit quorum sensing (25-27, 35). At a sufficient cell density, the population embarks on development. C signaling is required later, beginning at about 6 h into development, as judged from expression of developmentally regulated gene fusions to a reporter (23). C signaling has been hypothesized to occur through end-to-end contact between cells (36) and involve interaction of a proteolytic cleavage product of CsgA from the donor cell (31) with a receptor on the surface of the recipient cell (reviewed in reference 18). C signaling controls cell movements, with a low level sufficing for rippling (cells accumulate in parallel ridges that appear to travel as waves in time-lapse microscopy) and a higher level necessary for aggregation (reviewed in references 17 and 41). Alignment of cells in fruiting bodies may permit an even higher level of C signaling which triggers sporulation (21, 28).
In addition to coordinating cell movement and differentiation, C signaling also coordinates gene expression during M. xanthus development. This regulation involves both temporal and spatial control. Genes that depend on C signaling for expression are activated at different times after 6 h into development, and some show partial expression in a csgA mutant, whereas others show no expression (23, 29). Expression of all such genes examined thus far could be rescued by codeveloping the csgA mutant with wild-type cells (to supply C signal) (1, 6, 7, 23, 32) or by adding a 17-kDa fragment of CsgA (22). A higher level of the CsgA fragment was needed to rescue expression of a gene normally induced at the onset of sporulation than for a gene induced earlier, indicating different thresholds for response (21). Possibly related to this, C-signal-dependent genes have been shown to be expressed specifically in cells within fruiting bodies and not in the peripheral rods that remain outside of fruiting bodies (16, 37).
How are genes regulated by C signaling during M. xanthus development? One component of the C-signal transduction pathway is FruA, a protein similar to response regulators of two-component systems (34). This protein has a critical aspartate residue, suggesting it may be phosphorylated by an unidentified histidine protein kinase in response to C-signaling (5). Recently, the C-terminal domain of FruA has been shown to bind specifically to the promoter region of fdgA, a gene whose expression depends partially on C signaling (45). Hence, FruA (or its phosphorylated form) may directly activate fdgA transcription. Other potential targets of FruA, or regulators that function downstream of FruA in the C-signal transduction pathway, have been identified by random insertion of the transposon Tn5 lac into the M. xanthus chromosome (24). Insertions that generated a transcriptional fusion between a developmentally regulated M. xanthus promoter and the Escherichia coli lacZ gene have been tested for dependence of expression on csgA (23, 29). The promoter regions upstream of two insertions that exhibited partial dependence on csgA (Ω4400 and Ω4499) and two that exhibited absolute dependence on csgA (Ω4403 and Ω4406) have been identified (1, 6, 7, 32). Three of these promoter regions have been subjected to mutational analysis to identify important regulatory elements (46-48).
Here, we report mutational analysis of the fourth promoter region, which drives expression of the Ω4406 locus. Unlike the three other promoters, the Ω4406 promoter is regulated by an upstream DNA element that acts negatively. This element is necessary for expression from the promoter to be C-signal dependent. Our results provide the first example of an upstream negative element in an M. xanthus promoter region and the first example of C-signal dependence mediated by negative regulation. We also obtained evidence that a downstream regulatory element boosts Ω4406 expression and that the Ω4406 promoter region contains positive elements at bp −64 to −49 which resemble those in other developmentally regulated promoters but appear to function uniquely in each case.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids used in the present study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | φ80 lacZΔM15 ΔlacU169 recA1 endA1 hsdR17 supE44 thi-1 gyrA relA1 | 11 |
| M. xanthus | ||
| DK1622 | Wild type | 19 |
| LS203 | csgA653 | 40 |
| fruA::TcΩ5 | fruA::Tcr | 34 |
| DK11209 | ΔdevS | A. Garza and D. Kaiser |
| MPV1727-8 | attB::pREG1727 | 46 |
| MKV4 | attB::pKV4 (pREG1727 with 1.0-kb XhoI-BamHI fragment from pPV4406-1.0)a | This study |
| MKV6 | attB::pKV6 (pREG1727 with 150-bp XhoI-BamHI fragment from pKV3) | This study |
| MKV11 | attB::pKV11 (pREG1727 with 0.6-kb XhoI-BamHI fragment from pKV9) | This study |
| MKV20 | attB::pKV20 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV12) | This study |
| MPV251 | attB::pPV0251 (pREG1727 with 351-bp XhoI-BamHI fragment from pPV251) | This study |
| MPV182 | attB::pPV0182 (pREG1727 with 282-bp XhoI-BamHI fragment from pPV182) | This study |
| MKV31 | attB::pKV31 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV28) | This study |
| MKV32 | attB::pKV32 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV29) | This study |
| MKV33 | attB::pKV33 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV30) | This study |
| MKV37 | attB::pKV37 (pREG1727 with 240-bp XhoI-BamHI fragment from pKV34) | This study |
| MKV39 | attB::pKV39 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV36) | This study |
| MKV55 | attB::pKV55 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV46) | This study |
| MKV56 | attB::pKV56 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV47) | This study |
| MKV57 | attB::pKV57 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV48) | This study |
| MKV58 | attB::pKV58 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV49) | This study |
| MKV59 | attB::pKV59 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV50) | This study |
| MKV60 | attB::pKV60 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV51) | This study |
| MKV61 | attB::pKV61 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV52) | This study |
| MKV63 | attB::pKV63 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV53) | This study |
| MKV65 | attB::pKV65 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV62) | This study |
| MKV67 | attB::pKV67 (pREG1727 with 420-bp XhoI-BamHI fragment from pKV64) | This study |
| MKV4ΔCP | csgA653 attB::pKV4 | This study |
| MKV11ΔCP | csgA653 attB::pKV11 | This study |
| MKV4ΔF | fruA::TcrattB::pKV4 | This study |
| MKV11ΔF | fruA::TcrattB::pKV11 | This study |
| MKV4ΔdevS | ΔdevS attB::pKV4 | This study |
| MKV11ΔdevS | ΔdevS attB::pKV11 | This study |
| Plasmids | ||
| pCR2.1-TOPO | Apr Kmr; lacZα | Invitrogen |
| pREG1727 | Apr Kmr; P1-inc attP lacZ | 7 |
| pPV4406-1.0 | pCR2.1-TOPO with 1.0-kb Ω4406 DNA segment from bp −533 to 485 generated by PCR | 32 |
| pKV3 | pCR2.1-TOPO with Ω4406 DNA segment from bp −100 to 50 generated by PCR | This study |
| pKV9 | pCR2.1-TOPO with 0.6-kb Ω4406 DNA segment from bp −100 to 485 generated by PCR | This study |
| pKV12 | pCR2.1-TOPO with Ω4406 DNA segment from bp −100 to 320 generated by PCR | This study |
| pPV251 | pCR2.1-TOPO with Ω4406 DNA segment from bp −100 to 251 generated by PCR | This study |
| pPV182 | pCR2.1-TOPO with Ω4406 DNA segment from bp −100 to 182 generated by PCR | This study |
| pKV28 | pKV12 with CATCGTG to ACGATGT mutation from bp −55 to −49 | This study |
| pKV29 | pKV12 with GGACA to TTCAC mutation from bp −69 to −65 | This study |
| pKV30 | pKV12 with GAACC to TCCAA mutation from bp −64 to −60 | This study |
| pKV34 | pCR2.1-TOPO with Ω4406 DNA segment from bp −100 to 140 generated by PCR | This study |
| pKV36 | pKV12 with GGCGTTTCA to TTATGGGAC mutation from bp −90 to −82 | This study |
| pKV46 | pKV12 with C to A mutation at bp −55 | This study |
| pKV47 | pKV12 with A to C mutation at bp −54 | This study |
| pKV48 | pKV12 with T to G mutation at bp −53 | This study |
| pKV49 | pKV12 with C to A mutation at bp −52 | This study |
| pKV50 | pKV12 with G to T mutation at bp −51 | This study |
| pKV51 | pKV12 with T to G mutation at bp −50 | This study |
| pKV52 | pKV12 with G to T mutation at bp −49 | This study |
| pKV53 | pKV12 with GTC to TGA mutation from −59 to −57 bp | This study |
| pKV62 | pKV12 with T to G mutation at bp −56 | This study |
| pKV64 | pKV12 with C to A mutation at bp −61 | This study |
Where possible, the plasmid description is given in parentheses after the strain description.
Growth and development.
Escherichia coli DH5α strains containing plasmids were grown at 37°C in Luria-Bertani medium (38) containing 50 μg of ampicillin per ml. M. xanthus strains were grown at 32°C in CTT broth or agar (1.5%) plates (1% Casitone, 10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4, 8 mM MgSO4 [final pH = 7.6]) (13). When necessary, 40 μg of kanamycin (Km) per ml and 12.5 μg of oxytetracycline (Tc) per ml were used for selection. Fruiting body development was performed on TPM agar (1.5%) plates (10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4-K2HPO4, 8 mM MgSO4 [final pH = 7.6]) as described previously (24).
Construction of plasmids.
The 1.0-kb XhoI-BamHI fragment from pPV4406-1.0, which contains DNA from bp −533 to 485 relative to the Ω4406 transcriptional start site, was subcloned into XhoI-BamHI-digested pREG1727 to construct pKV4. This plasmid served as the template for PCR amplification of Ω4406 DNA segments using upstream primers containing a XhoI site and downstream primers containing a BamHI site. Each PCR product was cloned using pCR2.1-TOPO as described by the manufacturer. Each DNA insert was sequenced at the Michigan State University Genomics Technology Support Facility to ensure the correct sequence was obtained. Each pCR2.1-TOPO derivative was digested with XhoI and BamHI, and the DNA insert was purified by agarose gel extraction and subcloned into pREG1727 that had been digested with XhoI and BamHI.
A QuikChange site-directed mutagenesis kit (Stratagene) was used to create mutations in the Ω4406 promoter region that were A↔ C or G↔ T single-base-pair or multiple-base-pair transversions. The plasmid pKV12 containing Ω4406 DNA from bp −100 to 320 served as the DNA template for the mutagenesis with various combinations of primers. Candidate mutant plasmids were sequenced at the Michigan State University Genomics Technology Support Facility to identify plasmids with only the desired mutations. Each mutant plasmid was digested with XhoI and BamHI, and the mutant DNA insert was purified by agarose gel extraction and subcloned into pREG1727 that had been digested with XhoI and BamHI.
Construction of M. xanthus strains and determination of lacZ expression during development.
Strains containing pREG1727 derivatives integrated at the Mx8 phage attachment site (designated attB in Table 1) were constructed by electroporation of M. xanthus (20), and transformants were selected on CTT-Km plates. Based on previous experience in our laboratory (1, 6, 7, 12, 32), the majority of transformants have a single copy of the plasmid integrated at attB. To eliminate colonies with unusual developmental lacZ expression, we screened at least 10 transformants on TPM agar plates containing 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside per ml. Any colonies with unusual expression of lacZ were discarded and, of the remaining candidates, three independent isolates of each mutant construct were chosen for development. In all cases, the three transformants gave similar results when developmental β-galactosidase activity was measured as described previously (24).
RESULTS
Deletion analysis reveals an upstream negative regulatory element in the Ω4406 promoter region.
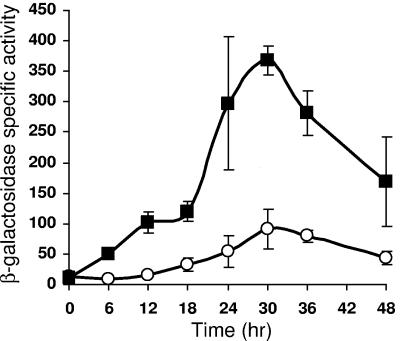
An ∼1.0-kb DNA segment spanning from bp −533 to 485 relative to the start site of Ω4406 transcription was shown previously to drive developmental expression of a lacZ reporter when the fusion was integrated at the Mx8 phage attachment site in the M. xanthus chromosome (32). The maximum level of expression (about 200 U) was similar to that observed for Tn5 lac Ω4406-Tcr (32). The fusion at the phage attachment site had been created in a plasmid (pPV12K) that encodes Tcr. For the present study, we fused the 1.0-kb segment to lacZ in a plasmid (pREG1727) that encodes Kmr. After integration at the Mx8 phage attachment site, this fusion exhibited developmental lacZ expression, reaching a maximum of 90 U at 30 h (Fig. 1 and Table 2). This pattern is similar to that reported previously for Tn5 lac Ω4406-Kmr (24, 32). Although we do not understand the reason for the higher expression of fusions associated with Tcr than Kmr, this effect was observed previously with a much larger (3.2-kb) segment spanning the Ω4406 promoter region (32).
FIG. 1.
An upstream DNA element negatively regulates Ω4406 expression. Developmental expression from the 1.0-kb (bp −533 to 485) (○) or 0.6-kb (bp −100 to 485) (▪) DNA segment containing the Ω4406 promoter fused to lacZ and integrated at the Mx8 phage attachment site in the chromosome of M. xanthus wild-type DK1622, was measured for three independently isolated transformants (see Materials and Methods). The graph shows the average β-galactosidase specific activity expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein, and error bars show 1 standard deviation of the data.
TABLE 2.
Summary of activities of mutant Ω4406 promoters
| Promoter assayed | Avg maximum β-galactosidase specific activity during developmenta ± SD | % Control activity measured in the same experimentb ± SD |
|---|---|---|
| Vector (no insert) | 16 ± 5 | |
| bp −533 to 485 | 90 ± 32 | 100 ± 36 |
| Deletions | ||
| bp −100 to 485 | 368 ± 24 | 476 ± 7 |
| bp −100 to 320 | 272 ± 35 | 346 ± 13 |
| bp −100 to 251 | 182 ± 12 | 205 ± 7 |
| bp −100 to 182 | 220 ± 50 | 244 ± 23 |
| bp −100 to 140 | 199 ± 30 | 247 ± 15 |
| bp −100 to 50 | 57 ± 6 | 55 ± 11 |
| Mutations in the C box-like sequence | ||
| CATCGTG −55 to −49 ACGATGT | 31 ± 10 | 11 ± 32 |
| C −55 A | 161 ± 3 | 59 ± 2 |
| A −54 C | 797 ± 199 | 293 ± 25 |
| T −53 G | 24 ± 9 | 9 ± 38 |
| C −52 A | 176 ± 34 | 65 ± 19 |
| G −51 T | 33 ± 4 | 12 ± 12 |
| T −50 G | 98 ± 22 | 36 ± 22 |
| G −49 T | 568 ± 93 | 209 ± 16 |
| Mutations farther upstream | ||
| T −56 G | 350 ± 6 | 129 ± 2 |
| GTC −59 to −57 TGA | 160 ± 25 | 59 ± 16 |
| GAACC −64 to −60 TCCAA | 67 ± 22 | 25 ± 32 |
| C −61 A | 95 ± 31 | 35 ± 33 |
| GGACA −69 to −65 TTCAC | 835 ± 175 | 307 ± 21 |
| GGCGTTTCA −90 to −82 TTATGGGAC | 421 ± 23 | 155 ± 5 |
The maximum β-galactosidase specific activity in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein (average ± 1 standard deviation) is listed for three independently isolated M. xanthus transformants (one determination each), except in the case of the vector control (one transformant, seven determinations) and the deletion with Ω4406 DNA from bp −100 to 320 (three transformants, two determinations each). Samples were assayed at 0, 6, 12, 18, 24, 30, 36, and 48 h during development.
Controls were included in each experiment. For the deletions, the controls were Ω4406 DNA from bp −533 to 485 and the vector with no insert. The maximum for each deletion is expressed as a percentage of the maximum observed for bp −533 to 485 in the same experiment, after subtraction from both values of the maximum observed for vector alone in that experiment. Likewise, the maximum for each mutation is expressed as a percentage of the maximum observed for bp −100 to 320 in the same experiment, after subtraction from both values of the maximum observed for vector alone in that experiment. The average percentage ±1 standard deviation is listed.
To localize the DNA elements important for Ω4406 transcription, we performed deletion analysis, beginning with a 5′ deletion. A DNA segment spanning from bp −100 to 485 was fused to lacZ in the Kmr-encoding plasmid (pREG1727) and integrated at the Mx8 phage attachment site. Developmental lacZ expression reached a higher maximum (368 U, Table 2) than for the 1.0-kb segment (Fig. 1). We conclude that DNA between bp −533 and −100 inhibits expression of a downstream lacZ reporter by at least fourfold.
The upstream negative element mediates C-signal dependence of the Ω4406 promoter.
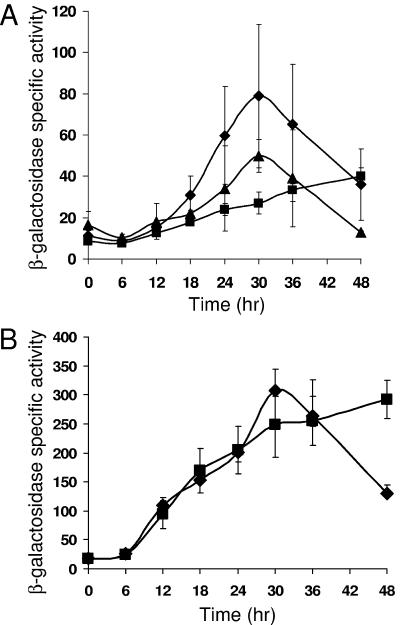
Previously, expression from the 1.0-kb segment spanning the Ω4406 promoter region was shown to be abolished in a csgA mutant incapable of C signaling (32). Developmental lacZ expression was restored upon codevelopment of the fusion-bearing csgA mutant with an equal number of wild-type cells, which supply C signal (32). These experiments used the fusion created in the Tcr-encoding plasmid (pPV12K). We repeated these experiments with the fusion of the 1.0-kb segment to lacZ in the Kmr-encoding plasmid (pREG1727). Expression was greatly reduced in the csgA mutant background (Fig. 2A). Upon codevelopment with wild type, induction of lacZ expression was partially restored, suggesting that transcription from the Ω4406 promoter within the 1.0-kb segment depends on extracellular C signaling.
FIG. 2.
C-signal dependence of Ω4406 expression requires the upstream negative element. (A) Developmental expression from the 1.0-kb DNA segment containing the Ω4406 promoter fused to lacZ and integrated at the Mx8 phage attachment site in the chromosome of M. xanthus wild-type DK1622 (⧫) or csgA mutant LS203 in the absence (▪) or presence (▴) of an equal number of wild-type DK1622 cells (lacking lacZ but capable of C signaling). (B) Developmental expression from the 0.6-kb (bp −100 to 485) segment in wild-type DK1622 (⧫) or csgA mutant LS203 (▪). The meaning of points and error bars is the same as described in the Fig. 1 legend.
We next tested the segment spanning from bp −100 to 485, fused to lacZ in the Kmr-encoding plasmid (pREG1727) and integrated at the Mx8 attachment site of the csgA mutant. Developmental expression in the csgA mutant was indistinguishable from that in wild type, except at 48 h (Fig. 2B). In the absence of the negative element located between bp −533 and −100, expression from the Ω4406 promoter appeared to be csgA independent and therefore independent of C signaling.
Expression from the Ω4406 promoter depends on fruA and devS even in the absence of the upstream negative element.
FruA mediates both motility and gene regulatory responses to C signaling (5, 34, 42). Since expression from the 1.0-kb segment containing the Ω4406 promoter was strongly dependent on C signaling, but expression from the ∼0.6-kb segment (i.e., bp −100 to 485) lacking the upstream negative element was C-signal independent (Fig. 2), we tested the effect of a fruA mutation on expression from these segments by integrating the fusion-containing, Kmr plasmids at the Mx8 attachment site of M. xanthus bearing the fruA::TcΩ5 mutation, which confers Tcr (34). Expression from both segments was greatly reduced in the fruA mutant (Fig. 3). These results demonstrate that FruA positively regulates Ω4406 expression and that it does not do so solely by counteracting the negative effect of the upstream element. Our results do not rule out the possibility that in response to C signaling, FruA both counteracts the negative upstream element and causes transcriptional activation (directly or indirectly) through a DNA element(s) located downstream of bp −100 in the Ω4406 promoter region.
FIG. 3.
Dependence of Ω4406 expression on fruA and devS. Developmental expression from the 1.0-kb (A) or 0.6-kb (B) DNA segment containing the Ω4406 promoter fused to lacZ and integrated at the Mx8 phage attachment site in the chromosome of M. xanthus wild-type DK1622 (⧫), fruA mutant fruA::TcΩ5 (▪), or devS mutant DK11209 (▴) was measured for three independently isolated transformants (see Materials and Methods). The meaning of points and error bars is the same as described in the Fig. 1 legend. The error bars are too small to be seen for the fruA mutant in both panels.
The dev locus functions after fruA on a pathway leading to sporulation (5). Previously, expression of Tn5 lac Ω7536-Kmr was shown to be abolished in csgA and devRS mutants (30). Like the devRS mutation, an in-frame deletion in devS causes a severe sporulation defect (9). This devS mutation reduced expression from both the 1.0- and 0.6-kb segments containing the Ω4406 promoter, by ∼2-fold (Fig. 3). We conclude that expression of Ω4406 depends less strongly on devS than on fruA, and neither of these effects relies on the upstream negative element.
A downstream element may boost expression from the Ω4406 promoter.
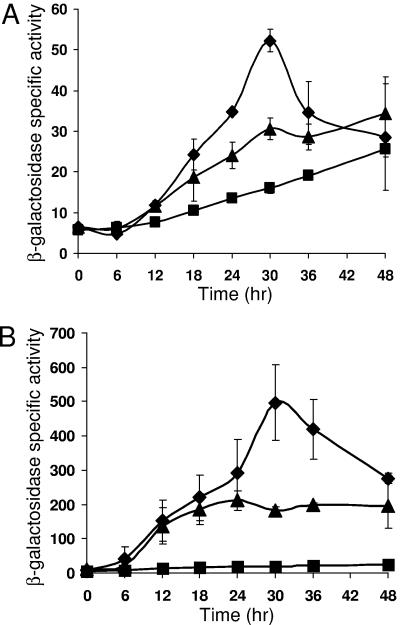
To further characterize the DNA elements important for Ω4406 expression, we performed 3′ deletion analysis. We chose to construct 3′ deletions with a common 5′ end at bp −100 (i.e., lacking the upstream negative element described above) since the 0.6-kb segment spanning from bp −100 to 485 drove a high level of developmental lacZ expression (Fig. 1). Fusions with 3′ ends at bp 320 (data not shown) or bp 251 (Fig. 4) exhibited similar timing of expression during development as the fusion at bp 485, but the maximum level of expression progressively decreased (Table 2). Fusions with 3′ ends at bp 182 or 140 showed similar expression as the fusion at bp 251 (Table 2). A fusion with a 3′ end at bp 50 exhibited much less developmental lacZ expression (Fig. 4), reaching less than 30% the maximum level observed for the fusion at bp 140 (Table 2). These results indicate that DNA between bp 50 and 140 boosts expression of a downstream reporter. The effect could be on transcription initiation from the Ω4406 promoter (e.g., due to a transcriptional activator protein that binds between bp 50 and 140 bp) or on a postinitiation event (e.g., transcription termination, mRNA stability, or translation). Another possibility, which we think is unlikely, is that the fusion at bp 50 results in reduced expression of the downstream lacZ reporter, due to a polar effect. Each fusion with a different 3′ end was designed to create an in-frame fusion between an open reading frame predicted to start at bp 37 and trpA, which precedes lacZ in pREG1727 (32). If translation instead begins between bp 50 and 140, so that only the fusion at bp 50 fails to create an in-frame fusion with trpA, it might cause a polar effect on expression of lacZ. However, it is unlikely that translation begins between bp 50 and 140, because the only potential translation start codon in this region, an ATG that begins at bp 97, is not preceded by a sequence likely to serve as a ribosomal binding site. On the other hand, the ATG that begins at bp 37 is preceded 5 bp upstream by the sequence AGGAG, which likely serves as a ribosomal binding site (32). Therefore, it is likely that DNA between bp 50 and 140 not only boosts expression of the downstream lacZ reporter in our fusions, but also boosts expression of the M. xanthus gene downstream of the Ω4406 promoter in its native context.
FIG. 4.
Effect of 3′ deletions on Ω4406 expression. Developmental expression from DNA segments spanning from bp −100 to 485 (▪) (i.e., the 0.6-kb segment; the data shown in Fig. 1 is also shown here), bp −100 to 251 (▴), or bp −100 to 50 (⧫) relative to the Ω4406 transcriptional start site, fused to lacZ and integrated at the Mx8 phage attachment site in the chromosome of M. xanthus wild-type DK1622, was measured for three independently isolated transformants (see Materials and Methods). The meaning of points and error bars is the same as described in the Fig. 1 legend. The error bars are too small to be seen for the bp −100 to 50 segment.
A C-box-like sequence centered at bp −52 is important for Ω4406 promoter activity.
Inspection of the promoter regions of several developmentally regulated M. xanthus genes revealed sequences that conform to the consensus CAYYCCY (Y means C or T), which was called the C box (6). Mutational analyses of C boxes centered near bp −50 have demonstrated the importance of this DNA element for promoter activity (43, 46-48). In the Ω4406 promoter region, the sequence CATCGTG centered at bp −52 matches the C box consensus at only the first four positions. To test whether this C-box-like sequence is important for Ω4406 promoter activity, we changed the sequence to ACGATGT in the context of DNA from bp −100 to 320. This mutant promoter region was fused to lacZ in a plasmid (pREG1727) and integrated at the Mx8 phage attachment site in the M. xanthus chromosome as described above. Developmental lacZ expression was nearly abolished (Table 2), demonstrating the importance of the C-box-like sequence for Ω4406 promoter activity.
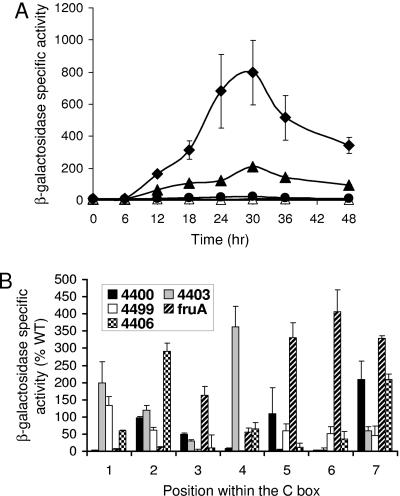
Despite their sequence similarity, C boxes appear to function differently in the promoter regions thus far tested, since single-base-pair transversion mutations in each C box produced a different pattern of effects on promoter activity (43, 46-48). We subjected the C-box-like sequence centered at bp −52 in the Ω4406 promoter region to this type of analysis. These mutations, as well as others described below, were analyzed in the context of DNA from bp −100 to 320. Certain single-base-pair changes in the C-box-like sequence had a dramatic effect on Ω4406 promoter activity. Developmental expression from the two mutant promoter regions with the highest and lowest activity is compared to that from the wild-type promoter region in Fig. 5A. The timing of expression from mutant promoter regions was similar to that from wild type (Fig. 5A and data not shown). The maximum level of expression during development for each mutant promoter region is compared to that for the wild-type promoter region (bp −100 to 320) in Table 2. The pattern of effects on promoter activity for mutations in the C-box-like sequence centered at −52 bp in the Ω4406 promoter region is compared to the effects of mutations in C boxes of other promoter regions in Fig. 5B. The pattern is unique for the C-box-like sequence in the Ω4406 promoter region, even considering just the first two positions, at which in each case the C at position 1 was changed to an A and, separately, the A at position 2 was changed to a C. Like the C boxes in other promoter regions, the C-box-like sequence in the Ω4406 promoter region appears to function in a unique fashion (e.g., due to binding of a different transcription factor in each case).
FIG. 5.
Single-base-pair changes in the C-box-like sequence centered at bp −52 have dramatic effects on Ω4406 expression, and the pattern of effects is different than for C boxes mutated previously. (A) Developmental expression from the wild-type DNA segment spanning from bp −100 to 320 (▴) or the same segment with a T-to-G change at bp −53 (•) or an A-to-C change at bp −54 (⧫), fused to lacZ and integrated at the Mx8 phage attachment site in the chromosome of M. xanthus wild-type DK1622, was measured for three independently isolated transformants (see Materials and Methods). The vector with no insert was included as a control (▵). The meaning of points and error bars is the same as described in the Fig. 1 legend. The error bars are too small to be seen except in the case of the base pair segment from −100 to 320 with the A-to-C change at −54 bp. (B) Comparison of the effects of single-base-pair transversion mutations in the C-box-like sequence centered at bp −52 in the Ω4406 promoter region and in four different C boxes. The x-axis represents the position in the C box (or C-box-like sequence) corresponding to the consensus sequence CAYYCCY with the A being position 2, etc. The bars represent the average maximum developmental lacZ activity expressed as a percentage of the wild-type promoter activity for the C boxes centered at bp −49 in the Ω4400 (47) and Ω4403 (46) promoter regions, at bp −33 in the Ω4499 promoter region (48), at bp −51 in the fruA promoter region (43), or for the C-box-like sequence centered at bp −52 in the Ω4406 promoter region (Table 2). Error bars show one standard deviation of the data.
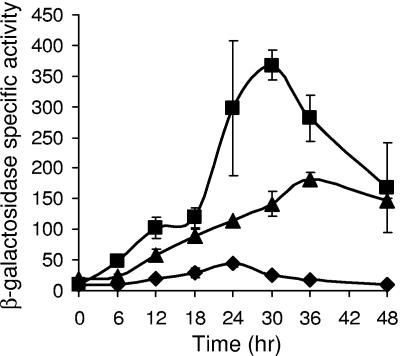
Mutations upstream of the C-box-like sequence reveal differences and similarities compared to other developmental promoter regions.
C boxes are preceded 5 to 8 bp upstream by sequences similar to the consensus GAACA in the promoter regions of several developmentally regulated M. xanthus genes (46). These sequences, called 5-bp elements, have been shown to be important for promoter activity (43, 46-48). The sequence of DNA between the 5-bp element and the C box also affects promoter activity, leading to speculation that the 5-bp element, the C box, and the sequence in between together constitute a recognition site for a transcription factor, which may be different for each promoter examined thus far (48). To test the importance of these sequences in the Ω4406 promoter region, we made four mutations and measured the activity of the mutant promoter regions as described above (Table 2). The timing of expression from mutant promoter regions was similar to that from wild type (data not shown). Changing T-to-G at position −56, immediately upstream of the C box-like sequence, caused a slight rise in the maximum level of developmental lacZ expression, to 130% of the maximum level observed for the wild-type promoter region (bp−100 to 320) in the same experiment (summarized in Fig. 6A). This is a unique result compared to three other C-signal-dependent promoter regions (Ω4400, Ω4403, and Ω4499), where transversion mutations at the position preceding a C box greatly reduced promoter activity (46-48). Changing GTC to TGA at base pair positions −59 to −57 slightly decreased promoter activity (Fig. 6A). In contrast, multiple-base-pair changes in sequences between 5-bp elements and C boxes abolished promoter activity (Ω4400 and Ω4499) or had no effect (Ω4403) (46-48). Changing the 5-bp element, GAACC, to TCCAA at base-pair positions −64 to −60 reduced activity ∼4-fold, and changing just the perfectly conserved C at position −61 bp to A reduced activity about threefold (Fig. 6A). As in other promoter regions (Ω4400, Ω4403, Ω4499, and fruA) (43, 46-48), the 5-bp element is very important for Ω4406 promoter activity. However, a threefold reduction in activity has not been observed previously for a C-to-A change at position 4 within a 5-bp element; the corresponding change in the Ω4403 (46), Ω4400 (47), and fruA (43) promoter regions resulted in 180, 1, and 270% of wild-type activities, respectively. We conclude that the Ω4406 promoter region has an architecture similar to that of other developmental promoter regions that have been examined in detail (Ω4400, Ω4403, Ω4499, and fruA) (43, 46-48), which includes a C-box-like sequence preceded 4 bp upstream by a 5-bp element. The different effects of many mutations in this region compared to the corresponding region of the other promoters is consistent with the idea (48) that each constitutes a recognition site for one or more transcription factors that differ between the promoters.
FIG. 6.
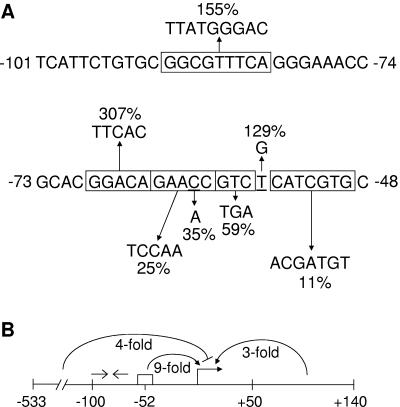
Summary of the Ω4406 promoter region. (A) Effects of mutations upstream of the Ω4406 promoter. The indicated changes were made in the context of DNA spanning from bp −100 to 320 bp relative to the Ω4406 transcriptional start site. Upward and downward arrows indicate that developmental lacZ expression was increased or decreased, respectively, and numbers indicate the maximum β-galactosidase specific activity observed for the mutant promoter region, expressed as a percentage of wild-type promoter activity measured in the same experiment (Table 2). (B) Summary of deletion analysis and the effect of a multiple-base-pair mutation in the C-box-like sequence. Deletions show that DNA between bp −533 and −100 exerts a fourfold negative effect on Ω4406 promoter activity and suggest that DNA between bp 50 and 140 exerts a threefold positive effect. A multiple-base-pair change in the C-box-like sequence centered at bp −52 (rectangle) indicates it exerts a ninefold positive effect. Open arrows indicate the position of an imperfect inverted repeat sequence that appears to be involved in negative regulation.
A second sequence similar to the 5-bp element consensus sequence was noted in the Ω4406 promoter region previously (32). This sequence, GGACA, is immediately upstream of the 5-bp element described above. Changing this sequence to TTCAC at base pair positions −69 to −65 increased promoter activity threefold (Table 2 and Fig. 6A). The timing of expression from the mutant promoter region was similar to that from wild type (data not shown). Clearly, the effect of mutating this sequence is different than for 5-bp elements located 5 to 8 bp upstream of a C box (43, 46-48) or for the 5-bp element located 4 bp upstream of the C-box-like sequence in the Ω4406 promoter region (Fig. 6A). It is worth noting that the GGACA sequence at base pair positions −69 to −65 is near the 3′ end of a 39-bp segment with an imperfect inverted repeat (−101 TCATTCTGTGCGGCGTTTCAGGGAAACCGCACGGACAGA−63) recognized previously (32). Also within this repeat is the sequence GGCGTTTCA, centered at bp −86 (32), which resembles an essential 10-bp element, GGCATGTTCA, centered at bp −74.5 in the Ω4403 promoter region (46). Changing this sequence to TTATGGGAC at base pair positions −90 to −82 increased promoter activity ∼1.6-fold (Table 2 and Fig. 6A) without altering the timing of expression (data not shown). Clearly, the sequence does not function like the 10-bp element in the Ω4403 promoter region. Rather, since both mutations in the large inverted repeat resulted in increased promoter activity, we speculate that it might be a site for binding of a dimeric repressor. This putative negative regulatory site would be in addition to the negative element located between bp −533 and −100 (Fig. 6B).
DISCUSSION
We have performed mutational analysis of the Ω4406 promoter region. Our most novel findings are that developmental activity of this promoter is governed by an upstream negative DNA element and that this element mediates the promoter's dependence on C signaling. We also obtained evidence that DNA downstream of the transcriptional start site boosts expression of the gene. Despite these unusual regulatory features, we found that the Ω4406 promoter region resembles that of other developmentally regulated M. xanthus genes in its general architecture. It has a C-box-like sequence and a 5-bp element, both of which are important for promoter activity. However, our detailed mutational analysis revealed unique features of the Ω4406 promoter region in comparison to other M. xanthus promoter regions studied thus far. We conclude that regulation of Ω4406 expression is complex and involves multiple positive and negative cis-acting DNA elements located upstream and downstream of the region typically bound by RNA polymerase (Fig. 6B).
Our results indicate that DNA between bp −533 and −100 negatively regulates Ω4406 promoter activity by about fourfold. Important goals of further research are to localize this cis-acting element and determine whether it is bound by a repressor protein. Upstream repressor binding sites typically function in cooperation with repressor bound farther downstream, forming a DNA loop (reviewed in reference 39). We speculate that the large inverted repeat between bp −101 and −63 might be such a downstream repressor binding site, since two different multiple-base-pair mutations in the repeat each caused Ω4406 promoter activity to increase. We searched for a sequence similar to the large inverted repeat, located between bp −533 and −100. Interestingly, the sequence GCGTTTGATTGCAACCGC at bp −317 to −300 matches the center of the large inverted repeat at 14 of 18 positions.
In the absence of the negative element located between bp −533 and −100, expression from the Ω4406 promoter was still developmentally regulated, but it did not depend on C signaling. The finding that loss of the negative element did not result in constitutive expression during growth indicates the existence of additional mechanisms of developmental control, and our analysis uncovered multiple positive regulatory DNA elements, which will be discussed below. The finding that loss of DNA upstream of bp −100 rendered expression from the Ω4406 promoter independent of C signaling is striking. It implies that the role of C signaling at this promoter is to overcome the negative effect of the upstream element. In fact, our results suggest the C signaling only partially overcomes this negative regulation, since developmental expression reached a fourfold-higher level in the absence of the upstream negative element. The only other M. xanthus promoter for which a regulatory DNA element mediating C-signal-dependent expression has been defined is the Ω4400 promoter. In this case, DNA from bp −86 to −81 exerts a two- to fourfold positive effect on expression and is at least partially responsible for C-signal dependence (47).
Because FruA mediates responses to C signaling during development (5, 34, 42), it was of interest to determine whether FruA relieves negative regulation of the Ω4406 promoter. We found that FruA is required positively for Ω4406 expression, even when the upstream negative element is absent (Fig. 3). This prevented us from determining whether FruA counteracts the negative effect of the upstream element. In the absence of the upstream element, Ω4406 regulation is like that of a few other M. xanthus genes, including tps and dofA, whose expression depends on FruA but not on C signaling (14, 15, 23, 34).
Expression of the dev locus depends on fruA (5), and expression of the Ω7536 locus depends on dev (30). This placed the Ω7536 locus downstream of dev on a pathway that leads to sporulation, since all of these genes are required for sporulation (30, 34, 44). Although the disruption caused by Tn5 lac Ω4406-Kmr did not cause a sporulation defect (24), we showed that Ω4406 expression depends partially on dev, being reduced about twofold in a devS mutant (Fig. 3). The effect was independent of the upstream negative element. It appears that dev function is not exclusively devoted to regulation of genes that are required for sporulation.
DNA between bp 50 and 140 boosted expression from a downstream lacZ reporter more than threefold when the upstream end of the Ω4406 promoter region was at bp −100. A similar observation was made previously using 3′ deletions and lacZ fusions to analyze the M. xanthus crtI promoter region (33). CrtI is involved in light-induced carotenoid biogenesis and crtI is induced in response to blue light (8). Martinez-Argudo et al. (33) found that a downstream regulatory element located between bp 30 and 130 is required for light-induced expression from the crtI promoter. This element has enhancer-like properties since it remained active when placed upstream of the crtI promoter. Although enhancers are common in promoter regions recognized by σ54 RNA polymerase, they are unusual in promoter regions recognized by holoenzymes bearing a σ in the σ70 family (reviewed in reference 2). The Ω4406 promoter does not resemble M. xanthus promoters believed to be transcribed by σ54 RNA polymerase (32, 48). It will be interesting to determine whether Ω4406 downstream DNA contains an enhancer-like regulatory element. We searched Ω4406 downstream DNA between bp 1 and 140 for inverted repeats but found nothing strongly suggestive of a binding site for a dimeric transcription factor or a transcription terminator that might be indicative of an antitermination regulatory mechanism.
The Ω4406 promoter region has blocks of sequence that are strikingly similar to sequences in the promoter regions of other developmentally regulated M. xanthus genes (32). These alignments helped us identify targets for mutagenesis in the Ω4406 promoter region, such as the C-box-like sequence centered at bp −52. Our mutational analysis showed that single-base-pair changes within this sequence can dramatically decrease or increase promoter activity. Likewise, single-base-pair changes in C boxes have decreased or increased activity of other promoters, but in each case the pattern of effects has been different (32, 43, 46-48). In keeping with this trend, the pattern of effects of single-base-pair changes in the C-box-like sequence centered at −52 bp in the Ω4406 promoter region is also unique (Fig. 5B). On the other hand, a multiple-base-pair change in the Ω4406 C-box-like sequence (Table 2 and Fig. 6) or in C boxes in other promoter regions (32, 43, 46-48) has in each case strongly decreased promoter activity. This suggests that these sequences are positive regulatory elements, and we have speculated that each is bound by a different transcriptional activator protein (43, 48), although these proteins may be evolutionarily related since they recognize similar sequences. According to this model, single-base-pair changes in C boxes (or the Ω4406 C-box-like sequence) that increase promoter activity do so by making the sequence a better binding site for the putative C-box-binding protein and therefore might be useful for biochemical approaches aimed at identifying these proteins.
The putative C-box-binding proteins may in fact recognize not only the C box but also the 5-bp element located 5 to 8 bp upstream of the C box (43, 48). The 5-bp element also appears to be a positive regulatory element, since multiple-base-pair changes have decreased promoter activity in every case examined thus far (43, 46-48). Four base pairs upstream of the C-box-like sequence in the Ω4406 promoter region is the sequence GAACC, which matches the 5-bp element consensus sequence, GAACA, in four of five positions. A multiple-base-pair change in this sequence reduced promoter activity fourfold. Hence, the Ω4406 promoter region has a similar architecture (i.e., a 5-bp element located 4 to 8 bp upstream of a C-box or C-box-like sequence) as the Ω4400, Ω4403, Ω4499, and fruA promoter regions (43, 46-48). DNA between the 5-bp element and the C box has also been shown to be important for activity of the Ω4400, Ω4403, and Ω4499 promoters (46-48); however, two mutations in the corresponding region had only modest effects on Ω4406 promoter activity. Also, changing C to A at position 4 of the 5-bp element had a different effect on Ω4406 promoter activity than on the activity of the Ω4400, Ω4403, or fruA promoters (43, 46, 47). These differences strengthen the impression that the 5-bp element, the C-box (or C-box-like) sequence, and the DNA in between function differently in each developmentally regulated promoter examined thus far.
Further differences between the Ω4406 promoter region and the promoter regions of other developmentally regulated M. xanthus genes that have been subjected to detailed mutational analysis are evident from the results of two mutations we made upstream of the 5-bp element. Most strikingly, changing GGACA at base pair positions −69 to −65 to TTCAC dramatically increased promoter activity. Despite its similarity to the 5-bp element consensus sequence, the GGACA sequence does not appear to function like 5-bp elements that are located 4 to 8 bp upstream of C-box or C-box-like sequences, since multiple-base-pair changes in these 5-bp elements greatly reduced promoter activity (43, 46-48). Neither does the GGCGTTTCAsequence centered at −86 bp in the Ω4406 promoter region appear to function like the 10-bp element, GGCATGTTCA, which is centered at bp −74.5 in the Ω4403 promoter region (46). Rather, our mutational analysis suggests that these sequences might be part of a repressor binding site, and we have speculated above that repressor bound to the large inverted repeat between bp −101 and −63 might interact with repressor bound farther upstream to form a repression loop.
In conclusion, our analysis of the Ω4406 promoter region revealed an upstream negative DNA element that mediates C-signal dependence, a likely downstream positive regulatory element, and upstream 5-bp and C-box-like elements. The latter act positively and may do so in conjunction, as observed for other developmentally regulated M. xanthus promoters, but somehow functioning uniquely in each promoter region, based on detailed mutational analyses. In the future, we hope to identify activator and repressor proteins that bind to the positive and negative regulatory DNA elements we have uncovered in the Ω4406 promoter region.
Acknowledgments
We thank L. Shimkets, S. Inouye, D. Kaiser, and A. Garza for providing bacterial strains, and we thank A. Garza for sharing information prior to publication.
This research was supported by NSF grant MCB-0416456 and by the Michigan Agricultural Experiment Station.
REFERENCES
- 1.Brandner, J. P., and L. Kroos. 1998. Identification of the Ω4400 regulatory region, a developmental promoter of Myxococcus xanthus. J. Bacteriol. 180:1995-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buck, M., M. T. Gallegos, D. J. Studholme, Y. Guo, and J. D. Gralla. 2000. The bacterial enhancer-dependent σ54 (σN) transcription factor. J. Bacteriol. 182:4129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Downard, J., S. V. Ramaswamy, and K. Kil. 1993. Identification of esg, a genetic locus involved in cell-cell signaling during Myxococcus xanthus development. J. Bacteriol. 175:7762-7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dworkin, M., and D. Kaiser. 1993. Myxobacteria II. American Society for Microbiology, Washington, D.C.
- 5.Ellehauge, E., M. Norregaard-Madsen, and L. Sogaard-Andersen. 1998. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in Myxococcus xanthus development. Mol. Microbiol. 30:807-817. [DOI] [PubMed] [Google Scholar]
- 6.Fisseha, M., D. Biran, and L. Kroos. 1999. Identification of the Ω4499 regulatory region controlling developmental expression of a Myxococcus xanthus cytochrome P-450 system. J. Bacteriol. 181:5467-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisseha, M., M. Gloudemans, R. Gill, and L. Kroos. 1996. Characterization of the regulatory region of a cell interaction-dependent gene in Myxococcus xanthus. J. Bacteriol. 178:2539-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontes, M., R. Ruiz-Vazquez, and F. J. Murillo. 1993. Growth phase dependence of the activation of a bacterial gene for carotenoid synthesis by blue light. EMBO J. 12:1265-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garza, A., and D. Kaiser. 2005. Personal communication.
- 10.Hagen, D. C., A. P. Bretscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 12.Hao, T., D. Biran, G. J. Velicer, and L. Kroos. 2002. Identification of the Ω4514 regulatory region, a developmental promoter of Myxococcus xanthus that is transcribed in vitro by the major vegetative RNA polymerase. J. Bacteriol. 184:3348-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of motility in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. USA 74:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horiuchi, T., T. Akiyama, S. Inouye, and T. Komano. 2002. Analysis of dofA, a fruA-dependent developmental gene, and its homologue, dofB, in Myxococcus xanthus. J. Bacteriol. 184:6803-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horiuchi, T., M. Taoka, T. Isobe, T. Komano, and S. Inouye. 2002. Role of fruA and csgA genes in gene expression during development of Myxococcus xanthus: analysis by two-dimensional gel electrophoresis. J. Biol. Chem. 277:26753-26760. [DOI] [PubMed] [Google Scholar]
- 16.Julien, B., A. D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser, D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nat. Rev. Microbiol. 1:45-54. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser, D. 2004. Signaling in myxobacteria. Annu. Rev. Microbiol. 58:75-98. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kashefi, K., and P. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthux frzF− defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S. K., and D. Kaiser. 1991. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J. Bacteriol. 173:1722-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, S. K., and D. Kaiser. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of Myxococcus xanthus. Cell 61:19-26. [DOI] [PubMed] [Google Scholar]
- 23.Kroos, L., and D. Kaiser. 1987. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1:840-854. [DOI] [PubMed] [Google Scholar]
- 24.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 25.Kuspa, A., L. Kroos, and D. Kaiser. 1986. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev. Biol. 117:267-276. [DOI] [PubMed] [Google Scholar]
- 26.Kuspa, A., L. Plamann, and D. Kaiser. 1992. Identification of heat-stable A-factor from Myxococcus xanthus. J. Bacteriol. 174:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuspa, A., L. Plamann, and D. Kaiser. 1992. A-signalling and the cell density requirement for Myxococcus xanthus development. J. Bacteriol. 174:7360-7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, S.-F., B. Lee, and L. J. Shimkets. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 29.Li, S.-F., and L. J. Shimkets. 1993. Effect of dsp mutations on the cell-to-cell transmission of CsgA in Myxococcus xanthus. J. Bacteriol. 175:3648-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Licking, E., L. Gorski, and D. Kaiser. 2000. A common step for changing cell shape in fruiting body and starvation-independent sporulation of Myxococcus xanthus. J. Bacteriol. 182:3553-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lobedanz, S., and L. Sogaard-Andersen. 2003. Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev. 17:2151-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loconto, J., P. Viswanathan, S. J. Nowak, M. Gloudemans, and L. Kroos. 2005. Identification of the Ω4406 regulatory region, a developmental promoter of Myxococcus xanthus, and a DNA segment responsible for chromosomal position-dependent inhibition of gene expression. J. Bacteriol. 187:4149-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Argudo, I., R. M. Ruiz-Vazquez, and F. J. Murillo. 1998. The structure of an ECF-sigma-dependent, light-inducible promoter from the bacterium Myxococcus xanthus. Mol. Microbiol. 30:883-893. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa, M., S. Fujitani, X. Mao, S. Inouye, and T. Komano. 1996. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol. Microbiol. 22:757-767. [DOI] [PubMed] [Google Scholar]
- 35.Plamann, L., A. Kuspa, and D. Kaiser. 1992. Proteins that rescue A-signal-defective mutants of Myxococcus xanthus. J. Bacteriol. 174:3311-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sager, B., and D. Kaiser. 1994. Intercellular C-signaling and the traveling waves of Myxococcus. Genes Dev. 8:2793-2804. [DOI] [PubMed] [Google Scholar]
- 37.Sager, B., and D. Kaiser. 1993. Spatial restriction of cellular differentiation. Genes Dev. 7:1645-1653. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Semsey, S., K. Virnik, and S. Adhya. 2005. A gamut of loops: meandering DNA. Trends Biochem. Sci. 30:334-341. [DOI] [PubMed] [Google Scholar]
- 40.Shimkets, L. J., and S. J. Asher. 1988. Use of recombination techniques to examine the structure of the csg locus of Myxococcus xanthus. Mol. Gen. Genet. 211:63-71. [DOI] [PubMed] [Google Scholar]
- 41.Sogaard-Andersen, L., M. Overgaard, S. Lobedanz, E. Ellehauge, L. Jelsbak, and A. A. Rasmussen. 2003. Coupling gene expression and multicellular morphogenesis during fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 48:1-8. [DOI] [PubMed] [Google Scholar]
- 42.Sogaard-Andersen, L., F. Slack, H. Kimsey, and D. Kaiser. 1996. Intercellular C-signaling in Myxococcus xanthus involves a branched signal transduction pathway. Genes Dev. 10:740-754. [DOI] [PubMed] [Google Scholar]
- 43.Srinivasan, D., and L. Kroos. 2004. Mutational analysis of the fruA promoter region demonstrates that C-box and 5-base-pair elements are important for expression of an essential developmental gene of Myxococcus xanthus. J. Bacteriol. 186:5961-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thony-Meyer, L., and D. Kaiser. 1993. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J. Bacteriol. 175:7450-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ueki, T., and S. Inouye. 2005. Identification of a gene involved in polysaccharide export as a transcription target of FruA, an essential factor for Myxococcus xanthus development. J. Biol. Chem. 280:32279-32284. [DOI] [PubMed] [Google Scholar]
- 46.Viswanathan, P., and L. Kroos. 2003. cis elements necessary for developmental expression of a Myxococcus xanthus gene that depends on C signaling. J. Bacteriol. 185:1405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoder, D., and L. Kroos. 2004. Mutational analysis of the Myxococcus xanthus Ω4400 promoter region provides insight into developmental gene regulation by C signaling. J. Bacteriol. 186:661-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoder, D., and L. Kroos. 2004. Mutational analysis of the Myxococcus xanthus Ω4499 promoter region reveals shared and unique properties in comparison with other C-signal-dependent promoters. J. Bacteriol. 186:3766-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]