Abstract
CTnERL, a Bacteroides conjugative transposon, transferred DNA by an Hfr-type mechanism during conjugation when it was excision deficient due to an insertion in the integrase gene. Rescue of the conjugative transposon sequences required the recipient to be RecA proficient and to contain an integrated CTnERL. The transfer efficiency was only 10- to 30-fold lower than the normal element transfer efficiency, and the direction of transfer from the oriT gene showed that the integrase end was transferred first and that the transfer genes were transferred last.
Conjugative transposons (CTns) are integrated elements that normally initiate their transfer process by excising from the chromosome to form a double-stranded circular transfer intermediate. A single-stranded DNA (ssDNA) copy of the CTn is then transferred by conjugation into the recipient cell, where it becomes double-stranded and integrates into the chromosome (16, 17, 26). Excision and integration are independent of homologous recombination (6). Previously, Li et al. (9) demonstrated that the transfer origin (oriT) of the 52-kbp Bacteroides CTn, CTnERL, is located within a 1.4-kbp region that lies near the center of the element. The interior location of the oriT gene raised the question of whether this CTn might be able to transfer at least a portion of itself without excising from the chromosome, in a manner similar to F-plasmid Hfr transfer (12).
Construction of an excision-defective, transmissible form of CTnERL.
Excision of CTnERL requires the integrase gene intERL, as well as three other genes (orf2c, orf2d, and exc). The intDOT gene is located at one end of CTnERL (attR). To test the hypothesis that CTnERL is capable of Hfr-like transfer, we first needed to inactivate intERL in order to eliminate natural excision. This could not be done by inactivating any of the excision genes in the orf2c operon because recent work has shown that products of these genes are required for full expression of the genes involved in conjugal transfer (25). Since we did not know the direction of transfer, we also needed to mark the other end of the CTn in a way that did not interfere with the ability of the CTn to transfer. This was done by integrating a resistance gene into a gene, traP, which was shown previously to be interrupted without reducing the CTn's transfer frequency (3).
The mutant element is shown in Fig. 1. To construct this mutant element, a strain of Bacteroides thetaiotaomicron 5482 that contained a single copy of CTnERL in the chromosome, BT4104N1-3, was used as the starting point. The CTnERL element was used because it contains only one antibiotic resistance gene, tetQ. This feature allowed us to make best use of the limited number of resistance gene markers available for Bacteroides. To place a chloramphenicol resistance (Cmr) marker gene on the attL side of oriT, a suicide vector, pGW160.1 (Table 1; Fig. 1), was used to introduce a single-crossover disruption into the transfer gene, traP, generating BT4104ΩtraP. As expected, this construct was still able to transfer at the wild-type frequency of 10−6 transconjugants per recipient (3). Using an unrelated suicide vector, pGW161.1, which carried ermG, a single-crossover disruption was introduced into the integrase gene (attR side) of the oriT gene of the CTnERL element in BT4104ΩtraP, generating the double-insertion strain BT4104ΩtraPΩint. All gene disruptions were tested by PCR (Ωint) and/or Southern blot analysis (ΩtraP) to confirm that the single-crossover insertions resulted in the expected gene disruption or phenotype as described previously (2, 4, 5). No excision was detected by PCR amplification of the joined ends of the mutant element, demonstrating that the insertion in intERL had indeed eliminated normal excision.
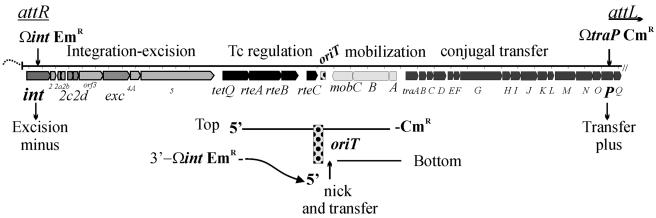
FIG. 1.
Functional map of CTnERL indicating locations of insertions to test for Hfr-type transfer and direction of transfer. The location of the ORFs determined from sequence data and experimental data in the region of interest are shown. The sites of the insertional mutations in the int gene (Ωint Emr), which is 800 bp from attR, and in traP (ΩtraP Cmr), which is 8 kb from attL, are indicated at the ends of the expanded region. There were only Emr transconjugants and no Cmr transconjugants observed, indicating that the int side of the element was transferred first. Assuming 5′-to-3′ transfer of the DNA, the nick occurs on the bottom strand of the element as drawn, with the resulting transfer of the integrase end of the element containing the Emr insertion.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant phenotypea | Source and/or description |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5αMCR | RecA | Gibco BRL |
| S17-1 | RecA Tpr Strr (ΩRP42-Tc::Mu-Kn::Tn7) | IncPα plasmid RP4 inserted into the S17-1 chromosome by bacteriophage Mu (22) |
| Bacteroides thetaiotaomicron | ||
| BT4001 | (Rifr) | Spontaneous rifampin mutant of B. thetaiotaomicron 5482A (21) |
| BT4100 | (Thy− Tpr) | A spontaneous Thy− Tpr mutant of B. thetaiotaomicron 5482A (1) |
| BT4104N1-3 | (Thy− Tpr Tcr) | BT4100 containing single copies of CTnERL and the mobilizable transposon NBU1 in its chromosome (21) |
| BT4104N1-3ΩintΩtraP | (Thy− Tpr Tcr Emr Cmr) | BT4104N1-3 in which the integrase gene (int) and transfer gene (traP) have been disrupted by homologous recombination with suicide vectors pGW160.1 and pGW161.1 (this study) |
| BT4004-28 | (Rifr Tcr) | BT4001 containing a single copy of CTnERL in the same integration site as BT4104N1-3 (this study) |
| BT4004-28RecA | (Rifr Tcr Cmr-RecA MMSs) | BT4004-28 in which the recA gene has been disrupted with pRAI49; this strain is RecA and MMSs (this study) |
| Plasmids | ||
| pNLY2 | Cmr (Cmr) | Bacteroides suicide vector that contains a pACYC184 E. coli replicon that is not homologous with pUC19-based suicide plasmids like pGERM (13) |
| pGERM | Apr (Emr) | Bacteroides suicide vector that contains a pUC19 E. coli replicon that is not homologous with pACYC184-based suicide plasmids like pNLY2 suicide plasmids (20) |
| pGW160.1 | Cmr (Cmr) | 449-bp PCR fragment amplified from an internal region of the CTnDOT transfer gene, traP, in pNLY2 (this study) |
| pGW161.1 | Apr (Emr) | 440-bp PCR fragment amplified from an internal region of the CTnDOT integrase gene, int, cloned into pGERM (this study) |
| pRAI49 | Cmr (Cmr) | pNLY2 containing a 433-bp RecA fragment; used to construct RecA insertional mutants of B. thetaiotaomicron 5482 strains (6) |
Phenotypes in parentheses are expressed in Bacteroides strain 5482A, and phenotypes not in parentheses are expressed in E. coli. Thy−, thymidine auxotroph.
Testing for transfer of a portion of the mutant CTnERL element.
The methods employed for the growth of Bacteroides and Escherichia coli strains, for DNA manipulation, and for measuring conjugal-transfer frequencies have been described elsewhere (7, 18, 19). We anticipated that the frequency of Hfr-type transfer might be very low because this was not a normal type of transfer for the element. Since the transfer frequency of CTnERL is normally only 10−5 to 10−6 transconjugants per recipient, we combined recipients from four filters to obtain a suspension that contained about (2 to 3) × 108 recipients per ml. A total of at least 2 ml of this mixture (in 0.2- or 0.5-ml portions, depending on the selective antibiotic) was plated, enabling us to detect frequencies as low as 10−9 transconjugants per recipient. In the case of erythromycin (Em) selection, we found that 0.5 ml of the resuspended cells could be applied to a single plate without having background growth. In the case of the Cm selection, however, only 0.2 ml or less could be applied on a single plate, so a larger number of plates was needed to reach the desired concentration of recipients. The antibiotic concentrations used were as follows: ampicillin (Ap), 100 μg/ml; cefoxitin, 20 μg/ml; Cm, 20 μg/ml; Em, 10 μg/ml; gentamicin, 200 μg/ml; rifampin (Rif), 10 μg/ml; tetracycline (Tc), 3 μg/ml; thymidine (Thy), 100 μg/ml; and trimethoprim (Tp), 100 μg/ml.
If transfer of one portion of CTnERL occurred to wild-type B. thetaiotaomicron BT4001 with no CTn, this transfer would not be detected since the joined ends of the CTn that come together in the circular form are essential for normal integration. Thus, the possibility of homologous recombination with a resident CTn was needed to allow for rescue of incoming CTn DNA. Accordingly, the recipient strain contained an integrated copy of CTnERL to rescue the transferred portion of CTnERL by homologous recombination (Fig. 2). To increase the frequency of homologous recombination, especially in the case of the insertion in intERL, where the insertion was close to one end, we wanted to have as large a region of sequence identity between the recipient DNA and the incoming mutant CTnERL DNA as possible. Since the intERL gene is only 800 bp from the attR end of the element, it might require a longer region of homology flanking the insertion in intERL to obtain the maximal integration frequencies when the mutant element integrates into the wild-type CTnERL. In this case, the integration event would have to occur between the intERL gene and the end of CTnERL in order to rescue the Emr insertion. Accordingly, we used a recipient that had a wild-type CTnERL integrated in the same site as the mutant CTnERL in the donor. To obtain such a recipient strain, BT4001 (Rifr) was the recipient in a mating with BT4104 (Thy− Tcr), which contained wild-type CTnERL. Matings were done as previously described (19). The transconjugants were isolated as Rifr Tcr colonies, and Southern blotting of DNA from different isolates was performed, using a hybridization probe that contained sequences of the joined ends of the excised form of CTnERL. Several restriction enzymes, including HincII and SspI, were used to identify transconjugants with CTnERL in the same site as in the mutant CTnERL in the donor. Of the 50 transconjugants screened by Southern blot analysis, only one transconjugant that had CTnERL inserted in or near the same site as the mutant element in the donor was found as determined by Southern blot analysis. This strain was designated BT4004-28.
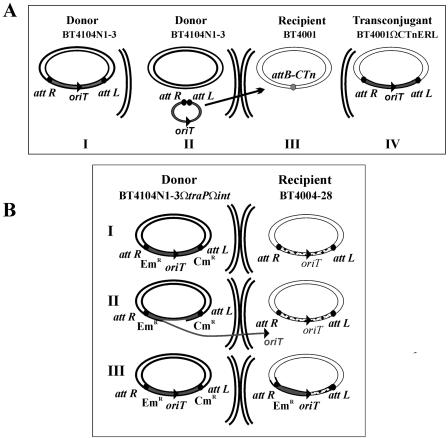
FIG. 2.
Model for transfer of DNA mediated by wild-type or excision-defective CTnERL. The stages of the transfer of wild-type CTnERL are shown in panel A. The element is normally found integrated in the donor (BT4104N1-3) chromosome (diagram I). The integrase end is indicated as attR, and the other end, near the transfer region, is called attL for consistency, irrespective of the orientation shown. Following induction by growth in the presence of tetracycline, the element excises and circularizes (diagram II) and is then transferred by conjugation to a recipient indicated by the arrow. One of the sites for integration (attB) in the recipient, BT4001, is shown in diagram III, and CTnERL will integrate by a RecA-independent mechanism (diagram IV). If the CTn is defective in its ability to excise normally, the process shown in panel B can occur. In this study, an insertion into int (Emr) was made at the attR end of the element and a second mutation in a nonessential gene, traP (Cmr), was made at the attL end of CTnERL in BT4104N1-3ΩtraPΩint (diagram I). When the donor strain is grown in the presence of tetracycline, the element cannot excise normally, but the transfer functions are still induced. Transfer presumably begins from the oriT region, located within a 1.4-kbp region near the center of the integrated element, to a recipient cell (diagram II). The CTn DNA once transferred can then find its homolog (a copy of the CTn) in the recipient, BT4004, and recombine into the regions of DNA sequence identity by RecA-dependent homologous recombination (III). If there was no copy of CTnERL in the recipient, there were no transconjugants. This study also showed that the int or attR end containing the Emr insertion was transferred first and that no transfer of the attL gene (Cmr) was detected. Since conjugal transfer occurs by a rolling circle mechanism (12), the donor would still be intact (diagram III).
BT4104ΩtraPΩint was utilized as the donor in conjugation experiments to test the transfer of the excision-deficient element. The donor strain was Thy−, trimethoprim resistant, and Rif sensitive (Rifs) as well as Cmr Emr. In the initial experiments, the strain with CTnERL in the same site (BT4004-28) was used as the recipient. In later experiments, isogenic strains with CTnERL in different sites in the recipient than in the donor were used as recipients. Transconjugants were then selected on medium containing Rif-Cm or Rif-Em. Transconjugants were subsequently patched onto plates containing Rif-Cm and plates containing Rif-Em to identify any transconjugants that contained both Emr and Cmr markers. This should be extremely rare since it would either be due to the donor sustaining two spontaneous mutations making it both Thy+ and Rifr (a frequency of about 10−12) or be due to cotransfer of the Emr and Cmr markers, indicating that the entire element, along with the entire 6.2-Mb B. thetaiotaomicron chromosome, was transferred.
Transfer of CTnERL requires induction of transfer genes by growth of the cells in tetracycline (15). In the presence of Tc, the Bacteroides strain that contained a wild-type copy of CTnERL (BT4104) transferred the element at a frequency of 10−5 to 10−6 transconjugants per recipient. When the Bacteroides strain containing the excision-deficient CTnERLΩtraPΩint element (BT4104ΩtraPΩint) was used as a donor in mating experiments with BT4004-28, transconjugants were obtained at a frequency of 10−7 transconjugants per recipient when the mating mixture was plated on medium containing Em. In contrast, when transconjugants were plated on medium containing chloramphenicol, the observed frequency of transfer was <10−9 (Table 2). All Emr transconjugants were sensitive to chloramphenicol, which is consistent with transfer of sequences only from the intERL side of the oriT gene.
TABLE 2.
Transfer frequency of wild-type and excision-deficient CTnERL elements
| Donor strain (Tpr Thy−)a | Recipient strain (Rifr)b | Frequency of transferc
|
Selection | |
|---|---|---|---|---|
| Without Tc | With Tc | |||
| Wild-type CTnERL (Tcr) strain | BT4001 | <10−9 | (1-3.5) × 10−6 | Rif Tc |
| Mutant CTnERL (Tcr) derivatives | ||||
| ΩtraP (Cmr) | BT4001 | <10−9 | (1-3) × 10−6 | Rif Cm |
| Ωint ΩtraP (Emr Cmr) | BT4001 | <10−9 | <10−9 | Rif Cm or Rif Em |
| Ωint ΩtraP (Emr Cmr) | BT4004-28 (Tcr) | <10−9 | (0.4-4.8) × 10−7 | Rif Em |
| Ωint ΩtraP (Emr Cmr) | BT4004-28 (Tcr) | <10−9 | <10−9 | Rif Cm |
The wild-type donor strain was BT4104N1-3, which carries a single chromosomal copy of CTnERL. ΩtraP indicates an insertion in traP which makes the strain Cmr, and Ωint indicates an insertion in int which makes the strain Emr.
BT4001 carries no CTnERL sequences. BT4004-28 carries a single chromosomal copy of CTnERL, integrated into the same site as the single copy of CTnERL in BT4104N1-3.
Frequencies of conjugal transfer are expressed as numbers of transconjugants per recipient cells at the end of the mating observed when donors were induced with tetracycline or not induced. The ranges of the frequencies are for three to five separate mating experiments, each done in duplicate. Matings were performed between Thy− Tpr donors and Rifr recipients. See Table 1.
As expected, transfer of the excision-deficient BT4104ΩtraPΩint element did not occur in the absence of Tc (<10−9 transconjugants per recipient). Also, no transfer of either resistance gene marker was seen if the recipient lacked a copy of wild-type CTnERL (Table 2). Such transfers were expected only if the mutant CTnERL was still able to excise normally from the chromosome and form the circular intermediate which transfers and integrates (Fig. 2).
Construction of a derivative of BT4004-28 that was deficient in homologous recombination.
Normal excision-dependent transfer of CTnERL is independent of homologous recombination (6). If, however, only a portion of CTnERL was transferred in the absence of excision, homologous recombination should be required to rescue the transferred DNA. The transferred CTnDOT DNA would lack one of the element's ends found in the excised circular form and would also lack a functional intERL gene (Fig. 2). To verify the requirement of RecA function for transfer of a portion of the mutant CTnERL, a RecA-deficient derivative of BT4004-28 was used as a recipient. A recA mutation in the BT4004-28 recipient was constructed by making an insertional mutation in the recA gene as described by Cooper et al. (6). Since the Cmr cassette from the attL end of CTnERL in traP did not transfer from the donor BT4104ΩtraPΩint (see above), the Cmr marker in the insertional vector pRAI49, which contained an internal region of recA, could be used. The site of the recA insertion was verified by Southern blot analysis, and the strain was shown to be sensitive to 0.01% methyl methanesulfonate (MMS). The strain was grown in supplemented brain heart infusion and handled carefully because of its decreased aerotolerance (6). The transfer of the Emr marker to this recipient was <10−9 transconjugants per recipient (Table 3). In the case of Hfr transfer by the F plasmid, the presynaptic RecB to -D system, which facilitates the RecA function by providing a ssDNA substrate for recombination, is also important. The genes encoding homologs of these proteins appear not to be present in the B. thetaiotaomicron genome, although there are putative homologs for the RecFOR presynaptic system (14). Conjugation provides at least a transient ssDNA substrate for RecA filaments to form, as is evident from the successful integration of phage CTX, which integrates as ssDNA when it is transferred into a recipient by conjugation (23). We do not know if any other Bacteroides recombination system contributes to the RecA-dependent recombination observed in these experiments.
TABLE 3.
Transfer of defective and wild-type CTn elements to BT4004 recipients
| Donor strain (Tpr Thy−) | Recipient strain (Rifr) | Frequency of transfer (with Tc)a |
|---|---|---|
| BT4104N1-3 CTnERLΩintΩtraP (Tcr Emr Cmr) | BT4004-28 (Tcr) | 4 × 10−8 |
| BT4004-28ΩrecA (Tcr Cmr) | <10−9 | |
| BT4004-6 (Tcr) | 5 × 10−8 | |
| BT4107 (CTnDOT) (Tcr Emr)b | BT4004-28 (Tcr) | 5 × 10−7 |
| BT4004-28ΩrecA (Tcr Cmr) | 5 × 10−7 | |
| BT4006-6 (Tcr) | 6 × 10−7 |
There was no detectible transfer seen without tetracycline induction. The frequency is the number of Rifr Emr transconjugants per recipient (Rifr) at the end of the mating. The frequency is the average of results from two separate experiments for both donors and all three recipients, done in duplicate for each experiment.
CTnDOT is nearly identical to CTnERL except that it has a 13-kbp region containing ermF between the oriT and int genes, immediately adjacent to ORF2 in Fig. 1 (24). It is used here as a wild-type CTn control containing an Emr marker to show that a copy of CTnERL in the recipient did not exclude the incoming CTn.
In the experiments just described, the recipient contained a copy of CTnERL that was integrated into the chromosome in the same site as the copy of mutant CTnERL provided in the donor, BT4104. This was done under the assumption that including identical regions of chromosomal DNA outside the end of CTnERL might be needed to maximize the frequency of acquisition by homologous recombination of the transferred portion of the mutated CTn. To test this assumption, we determined the frequency of transfer of the excision-deficient mutant of CTnERL to four recipients that had CTnERL located in four different chromosomal sites identified by Southern blotting in the screen for BT4004-28 described above.
The results for one of these recipients are shown in Table 3. The three other recipients with CTnERL integrated into other chromosomal sites gave the same results (data not shown). These results indicate that the location of the CTn in the recipient had little impact on the frequency of transfer and recombination of the CTnERL sequences, and that 800 bp flanking a 5-kbp insertion was sufficient to allow efficient homologous recombination to occur. As a control to monitor the effect, if any, of the recA mutation or the presence of a closely related CTn in the recipient on the integration of the incoming CTn, we used a donor carrying a closely related wild-type CTn, CTnDOT, which carries an Emr resistance gene. CTnERL is virtually identical at the DNA sequence level (>90%) to CTnERL except that CTnDOT has a 13-kbp insertion that contains the ermF gene (24). The results shown in Table 3 demonstrated that the transfer of CTnDOT was not impeded by the presence of the CTnERL in the recipient and that transfer and integration of CTnDOT was not affected by the absence of RecA in the recipient.
Conclusions.
CTnERL and related CTns have a complex regulatory network that coordinates the expression of the excision and transfer genes (5, 10, 17, 25). The coordination of excision and transfer should be important for the survival of the CTn in natural hosts, because if the entire CTn is not transferred, the transferred CTn could not be circularized in the recipient and would thus be incapable of normal integration, which requires the joined ends of the circular form. Yet, transfer of only a portion of the CTn could still promote the survival of some of genes of the CTn, if the transferred DNA was rescued by homologous recombination with an intact copy or even remnants of a closely related CTn in the recipient.
The results of the experiments described in this report demonstrate for the first time that transfer of a portion of the CTn is indeed possible and that the transferred DNA can be rescued if there is a closely related CTn present in the recipient chromosome. Our results also show that the incoming region of the transferred element is not excluded by the original CTn and is able to introduce a new gene into the CTn in the recipient. CTnERL/CTnDOT elements have a broad host range for transferring DNA, e.g., to E. coli recipients. A recombinational event might take place in a nonpermissive host under natural conditions even if the integrase gene is not expressed in the host. The CTn would then be trapped in the recipient after the recombinational event. Surprisingly, about 80 to 90% of the CTnERL-related elements examined so far transfer intact under laboratory conditions. There are, however, several examples of cryptic, fragmentary CTn-type elements with various amounts of sequence identity to the CTnERL/CTnDOT family in the chromosomes of sequenced Bacteroides strains (8, 28). Whether any of these CTns are still active for excision and transfer remains to be seen. Presumably, any CTn or part of a CTn that still possessed a functional mob-oriT region and transfer gene region could be capable of DNA transfer to a recipient, whether by normal excision-type transfer or by Hfr-type transfer. Our findings suggest the possibility that fragmentary CTns found in natural isolates might have resulted, in some cases, from the Hfr-type transfer of a portion of an active CTn and that some of these cryptic elements may be capable of mediating Hfr-type transfer.
The frequency with which the excision-independent transfer of CTnERL occurred was only 10- to 30-fold lower than that of normal transfer of the CTnERL to a recipient (Table 2). We had expected the frequency of the double crossovers needed to rescue the CTn DNA to be much lower than normal integration frequencies due to the need for homologous recombination. A possible explanation is suggested by results of studies of the F-mediated Hfr processes in E. coli that indicated that the recombination frequencies are especially high near the origin of transfer (11), and this could explain why we were successful in detecting the transfer of the Emr insertion. This may also explain why additional sequence identity beyond the end of the element did not show any effect on the survival of the 5-kb insertion. However, it may have been fortunate that we could even detect the recombination at all, since the F-mediated Hfr data suggest that markers beyond 45 kb may actually have a higher rate of recovery than the placement of the marker that we were using, which was only 20 kb from the origin of transfer (12).
An important result of the experiments described here is that they have allowed us for the first time to establish the direction of transfer of CTnERL. Our results suggest that the portion of CTnERL that contains the regulatory region and the integrase gene is transferred first and that the mobilization and transfer genes are the last to transfer (Fig. 1). In the normal type of transfer involving the circular form of the CTn, this would mean that the transfer starts with oriT; continues through the regulatory region, the excision operon, the intERL gene, and the joined ends; and only then proceeds to other genes (Fig. 1 and 2). That is, the genes essential for transfer of the CTn are the last to enter the recipient cell, whereas the portion of the CTn that contains essential integration functions are transferred first. The transfer genes are the last to be transferred in the case of F plasmid (12), but this is only partially true for IncP plasmids (27). However, the conjugal plasmids do not need to integrate for survival. The intERL gene is expressed constitutively and may allow the element to integrate in the recipient chromosome immediately after transfer is completed before the genes not required for the integration step are expressed. We do not know if any of the other early-entry genes on the CTn contribute to their survival in the recipient.
Acknowledgments
This work was supported by grant number AI/GM 22383 from the U.S. National Institutes of Health.
REFERENCES
- 1.Bedzyk, L. A., N. B. Shoemaker, K. E. Young, and A. A. Salyers. 1992. Insertion and excision of Bacteroides conjugative chromosomal elements. J. Bacteriol. 174:166-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonheyo, G., D. Graham, N. B. Shoemaker, and A. A. Salyers. 2001. Transfer region of a Bacteroides conjugative transposon CTnDOT. Plasmid 45:41-51. [DOI] [PubMed] [Google Scholar]
- 3.Bonheyo, G., B. Hund, N. B. Shoemaker, and A. A. Salyers. 2001. Transfer region of a Bacteroides conjugative transposon contains regulatory as well as structural genes. Plasmid 46:202-209. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, Q., B. J. Paszkiet, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2000. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J. Bacteriol. 182:4035-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, Q., Y. Sutanto, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2001. Identification of genes required for the excision of CTnDOT, a Bacteroides conjugative transposon. Mol. Microbiol. 41:625-632. [DOI] [PubMed] [Google Scholar]
- 6.Cooper, A. J., A. P. Kalinowski, N. B. Shoemaker, and A. A. Salyers. 1997. Construction and characterization of a Bacteroides thetaiotaomicron recA mutant: transfer of Bacteroides integrated conjugative elements is RecA independent. J. Bacteriol. 179:6221-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holdeman, L. V., and W. E. C. Moore. 1975. Anaerobe laboratory manual, 4th ed. Virginia Polytechnical Institute and State University, Blacksburg, Va.
- 8.Kuwahara, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh, N. Okada, S. Kuhara, M. Hattori, T. Hayashi, and Y. Ohnishi. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA 101:14919-14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, L. Y., N. B. Shoemaker, and A. A. Salyers. 1995. Location and characteristics of the transfer region of a Bacteroides conjugative transposon and regulation of transfer genes. J. Bacteriol. 177:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moon, K., N. B. Shoemaker, J. F. Gardener, and A. A. Salyers. 2005. Regulation of excision genes of the Bacteroides conjugative transposon, CTnDOT. J. Bacteriol. 186:5732-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pittard, J., and E. M. Walker. 1967. Conjugation in Escherichia coli: recombination events in terminal regions of transferred deoxyribonucleic acid. J. Bacteriol. 94:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porter, R. D. 2002. Conjugation, p. 463-506. In U. N. Streips and R. E. Yasbin (ed.), Modern molecular genetics, 2nd ed. Wiley-Liss, Inc., New York, N.Y.
- 13.Reeves, A. R., G. R. Wang, and A. A. Salyers. 1997. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J. Bacteriol. 179:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rocha, E. P., E. Cornet, and B. Michel. 2005. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLOS Genet. 1:247-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salyers, A. A., N. B. Shoemaker, and A. M. Stevens. 1995. Tetracycline regulation of conjugal transfer genes, p. 393-400. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 16.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salyers, A. A., G. Whittle, and N. B. Shoemaker. 2004. Conjugative and mobilizable transposons, p. 125-143. In R. V. Miller and M. J. Day (ed.), Microbial evolution: gene establishment, survival, and exchange. ASM Press, Washington, D.C.
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 19.Shoemaker, N. B., C. Getty, E. P. Guthrie, and A. A. Salyers. 1986. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 166:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoemaker, N. B., G. R. Wang, and A. A. Salyers. 2000. Multiple gene products and sequences required for excision of the mobilizable integrated Bacteroides element NBU1. J. Bacteriol. 182:928-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoemaker, N. B., G. R. Wang, A. M. Stevens, and A. A. Salyers. 1993. Excision, transfer, and integration of NBU1, a mobilizable site-selective insertion element. J. Bacteriol. 175:6578-6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 23.Val, M.-E., M. Bouvier, J. Campos, D. Sherratt, F. Cornet, D. Mazel, and F.-X. Barre. 2005. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell 19:559-566. [DOI] [PubMed] [Google Scholar]
- 24.Whittle, G., B. D. Hund, N. B. Shoemaker, and A. A. Salyers. 2001. Characterization of the 13 kb ermF region of Bacteroides conjugative transposon CTnDOT. Appl. Environ. Microbiol. 67:3488-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whittle, G., N. B. Shoemaker, and A. A. Salyers. 2002. Characterization of genes involved in modulation of conjugal transfer of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 184:3839-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittle, G., N. B. Shoemaker, and A. A. Salyers. 2002. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 59:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkins, B., and E. Lanka. 1993. DNA processing and replication during plasmid transfer between Gram-negative bacteria, p. 105-136. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Press, New York, N.Y.
- 28.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]