Abstract
Phase variation between smooth and rugose colony variants of Vibrio cholerae is predicted to be important for the pathogen's survival in its natural aquatic ecosystems. The rugose variant forms corrugated colonies, exhibits increased levels of resistance to osmotic, acid, and oxidative stresses, and has an enhanced capacity to form biofilms. Many of these phenotypes are mediated in part by increased production of an exopolysaccharide termed VPS. In this study, we compared total protein profiles of the smooth and rugose variants using two-dimensional gel electrophoresis and identified one protein that is present at a higher level in the rugose variant. A mutation in the gene encoding this protein, which does not have any known homologs in the protein databases, causes cells to form biofilms that are more fragile and sensitive to sodium dodecyl sulfate than wild-type biofilms. The results indicate that the gene, termed rbmA (rugosity and biofilm structure modulator A), is required for rugose colony formation and biofilm structure integrity in V. cholerae. Transcription of rbmA is positively regulated by the response regulator VpsR but not VpsT.
The etiologic agent of the diarrheal disease cholera (28), Vibrio cholerae, is a facultative human pathogen. It is naturally found in environmental aquatic habitats both as a free-living organism in the water column and in a biofilm state attached to different surfaces, including mucilaginous surfaces of phytoplankton, chitinous surfaces of zooplankton, and abiotic surfaces (16). V. cholerae causes periodic, seasonal outbreaks of disease in regions where it is an established member of the aquatic ecosystem (10, 16, 32). The occurrence of these outbreaks is likely to be linked to survival strategies used by V. cholerae in various aquatic habitats. In other words, enhanced survival of the organism in the environment increases the numbers of V. cholerae cells, thereby increasing the likelihood of disease. It has been proposed that the organism uses biofilm formation on surfaces and phase variation as survival strategies (57, 58, 62).
Biofilms are surface-attached microbial communities composed of microorganisms and the extrapolymeric substances they produce (12, 40, 43). Biofilm formation begins with the transport and attachment of the bacterium to surfaces. After the initial attachment, colonization of a surface is mediated by the movement and growth of attached bacteria. Surface colonization then leads to the formation of microcolonies, which are often surrounded by extrapolymeric substances. Further growth of bacteria and continued production of exopolysaccharides lead to the development of mature biofilm structures characterized by pillars and channels. It has been shown that development of these structures depends on biomass growth rate, twitching motility, signaling molecules, and exopolysaccharide production (12, 40, 43).
The sequence of events leading to the initiation of V. cholerae biofilm on abiotic surfaces under laboratory conditions and the genes required for these steps have been reported (57). The mannose-sensitive hemagglutinin type IV pilus and flagellum facilitate attachment to abiotic surfaces. The flagellum is also required for the spreading of bacteria along the surface. Under laboratory conditions, mannose-sensitive hemagglutinin is also required for efficient colonization of biotic surfaces, such as the chitinous exoskeleton of zooplankton (9) and cellulose fibers (56). Chitin-regulated pilus and toxin-coregulated pilus also facilitate attachment to chitinous surfaces (34, 44). Vibrio polysaccharide, VPS, is required for the development of mature biofilms (57, 62), which are characterized by macrocolonies separated by water channels. In addition, MbaA, a protein belonging to the family of GGDEF- and EAL-domain proteins that modulate levels of the second messenger c-di-GMP in cells, is also critical for maintenance of biofilm architecture in V. cholerae (5).
In response to environmental stresses, V. cholerae undergoes phase variation, which results in two morphologically distinct colonial variants, called smooth and rugose (36, 55, 58, 62), that differ greatly at the phenotypic level. The presence of phenotypic variants has also been reported for Vibrio vulnificus (19) and Vibrio parahaemolyticus (33), indicating that this process is common among clinically important Vibrio species. The rugose variant of V. cholerae forms corrugated colonies and well-developed biofilms and shows a higher level of resistance to osmotic and oxidative stresses than the smooth variant. Increased production of VPS in the rugose variant is largely responsible for these phenotypes (36, 45, 55, 62). In V. cholerae, VPS production is mediated by proteins encoded by genes located in the vpsI (vpsA-K, VC0917-27) and vpsII (vpsL-Q, VC0934-9) regions. Mutations in any of the vps genes cause smooth colonial morphology and reduced capacity to form biofilms (62). Two known positive regulators of vps gene expression, VpsR and VpsT, exhibit homology to response regulators of the two-component regulatory system. Disruption of either gene in the rugose genetic background prevents expression of vpsA and vpsL, blocks production of VPS, yields a smooth colonial morphology, and abolishes typical three-dimensional biofilm structure formation (8, 60).
The molecular consequences of smooth-to-rugose phase variations were determined by whole-genome expression profiling of the two variants during the exponential growth phase in nutrient broth (61). The study showed that 124 genes are differentially regulated between the phase variants. Among these, the expression of 77 genes was induced and that of 47 was repressed in the rugose compared to the smooth variant. To gain a better understanding of the consequences of phase variation, we identified changes in the proteome that accompany the smooth-to-rugose phase variation using two-dimensional gel electrophoresis on whole-cell extracts of V. cholerae cultures grown to exponential and stationary phase. We identified a protein (RbmA) that is produced at a higher level in the rugose variant. Using mutational analysis, we determined that RbmA is required for the full development of corrugated rugose colonial morphology and of “typical” pillar structures of biofilms. In addition, we established that RbmA production is regulated by VpsR.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used are listed in Table 1. All V. cholerae and Escherichia coli strains were routinely grown aerobically, in Luria-Bertani (LB) medium, at 30°C and 37°C, respectively, unless otherwise specified. The agar medium contained 1.5% (wt/vol) granulated agar (Difco), except for the motility studies, where the agar concentration was 0.3% (wt/vol). Concentrations of antibiotics used are as follows: for ampicillin, 100 μg/ml; for rifampin, 100 μg/ml; and for gentamicin, 30 μg/ml. Pellicle formation experiments were carried out in glass culture tubes (18 by 150 mm) containing 5 ml of LB medium inoculated with 200-fold dilutions of overnight cultures. The tubes were incubated at 30°C under static conditions.
TABLE 1.
Plasmids and strains used
| Plasmid or strain | Relevant properties | Reference or source |
|---|---|---|
| Plasmids | ||
| pGP704-sacB28 | Used for sucrose-based selection, Apr | Gary Schoolnik |
| pGPVC0928 | pGP704-sacB28 containing the SOEing PCR product of rbmA | This study |
| pACYC177 | Low-copy-number cloning vector, Apr | New England Biolabs |
| prbmA | pACYC177 harboring wild-type copy of rbmA | This study |
| pBAD/Myc-His B | Arabinose-inducible expression vector with C-terminal Myc epitope and six-His tags | Invitrogen |
| prbmA-myc | pBAD/Myc-His B containing the coding region of rbmA | This study |
| pRS415 | lacZ-based cloning vector for transcriptional fusion studies, Apr | 50 |
| prbmA::lacZ | pRS415 containing rbmA upstream regulatory fragment fused to lacZ | This study |
| pCC11 | pRS415 containing vpsA upstream regulatory fragment fused to lacZ | 8 |
| pCC12 | pRS415 containing vpsL upstream regulatory fragment fused to lacZ | 8 |
| pGP704::Tn7-GFP | Mini-Tn7 vector harboring a constitutively expressed gfp cassette | Gary Schoolnik |
| pUX-BF13 | Helper plasmid containing transposase gene for mini-Tn7 | Gary Schoolnik |
| Strains | ||
| V. cholerae 92A1552 | ||
| FY_Vc_0001 | O1 E1 Tor, Inaba, smooth, Rifr | 60 |
| FY_Vc_0002 | O1 E1 Tor, Inaba, rugose, Rifr | 60 |
| FY_Vc_0004 | Rugose ΔlacZ | 8 |
| FY_Vc_0005 | RΔvpsT ΔlacZ | 8 |
| FY_Vc_0006 | RΔvpsR ΔlacZ | 8 |
| FY_Vc_0007 | RΔvpsR ΔvpsT ΔlacZ | 8 |
| FY_Vc_102 | SΔrbmA | This study |
| FY_Vc_105 | RΔrbmA | This study |
| FY_Vc_222 | Rugose-gfp | This study |
| FY_Vc_224 | RΔrbmA-gfp | This study |
| E. coli | ||
| DH5α | F′ endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA) U169 (φ80dlacΔM15) | Promega |
| CC118 (λ pir) | D(ara-leu) araD DlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 λ pir | 21 |
| S17-1 (λ pir) | Tpr SmrrecA thi pro rK− mK+RP4:2-Tc:MuKm Tn7 λ pir | 15 |
Recombinant DNA techniques.
DNA manipulations were carried out by standard molecular techniques (46). Restriction enzymes and DNA modification enzymes were obtained from New England Biolabs. PCRs were carried out with primers purchased from Operon Technologies and with a High-Fidelity PCR kit (Roche).
2D SDS-PAGE.
Rugose and smooth V. cholerae wild-type cells were harvested during exponential and stationary growth phases and resuspended in 125 to 250 μl of lysis buffer containing 0.3% sodium dodecyl sulfate (SDS), 10 mM Tris (pH 7.4), 100 μg/ml DNase I (DNase I type II from bovine pancreas), and 50 μg/ml RNase A (RNase A type IIIA from bovine pancreas) in 5 mM MgCl2, 10 mM Tris-Cl (pH 7.0), and 1% protease inhibitor stock [20 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF), 1 mg/ml leupeptin, 0.36 mg/ml E-64, 500 mM EDTA, and 5.6 mg/ml benzamidine]. The cells were then vortexed and incubated on ice for 5 min. An equal volume of SDS boiling buffer, composed of 5% SDS, 10% glycerol, and 60 mM Tris (pH 6.8), was added, and protein concentrations were determined using a bicinchoninic acid assay (Pierce). Before loading the samples for SDS-polyacrylamide gel electrophoresis (SDS-PAGE), β-mercaptoethanol was added to a final concentration of 5% (vol/vol). Labeling of exponentially grown cultures (1 ml) was carried out with 40 μCi of [35S]methionine (Du Pont NEN, Wilmington, Del.) for 3 min followed by a 1-min chase with 0.2 mM unlabeled methionine. Labeling time was increased to 15 min for stationary-phase cultures, followed by a 5-min chase with unlabeled methionine. The samples were washed twice with phosphate-buffered saline (PBS). The amount of [35S]methionine incorporated was determined by liquid scintillation counting of a small portion of the cell lysate precipitated with 10% trichloroacetic acid (TCA). Protein samples were loaded at 2,200,000 cpm, and two-dimensional (2D) gel electrophoresis was carried out with a Protean II (Bio-Rad) device as described by the manufacturer. Labeled proteins were visualized by autoradiography. The following 14C molecular weight markers (Amersham) were added to a well in the agarose that sealed the tube gel to the slab gel: myosin (220 kDa), phosphorylase B (97 kDa), bovine serum albumin (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (30 kDa), and lysozyme (14 kDa). These markers appeared as faint bands at the basic end of the X-ray films. For Coomassie blue-stained gels (Fig. 1), 250 μg of each protein sample was loaded. The following proteins (Sigma) were added as molecular weight standards to a well in the agarose that sealed the tube gel to the slab gel: myosin (220 kDa), phosphorylase A (94 kDa), catalase (60 kDa), actin (43 kDa), carbonic anhydrase (29 kDa), and lysozyme (14 kDa). These standards appear as horizontal lines on the Coomassie brilliant blue R-250-stained 10% acrylamide slab gels. 2D protein electrophoresis was conducted according to the method of O'Farrell (39) by Kendrick Labs, Inc. (Madison, WI). Briefly, isoelectric focusing (IEF) was carried out in glass tubes with an inner diameter of 2.0 mm using 2.0%, pH 4 to 8 ampholines (BDH from Hoefer Scientific Instruments) at 9,600 V · h. One microgram of an IEF internal standard, tropomyosin protein, with a molecular weight of 33 kDa and pI 5.2, was added to the samples. The positions of the standard are indicated by arrowheads on the stained 2D gels (Fig. 1). After equilibration for 10 min in buffer O (10% glycerol, 50 mM dithiothreitol, 2.3% SDS, and 0.0625 M Tris, pH 6.8), the tube gels were sealed to the top of the stacking gels, which were above the 10% acrylamide slab gels (0.75 mm thick). SDS slab gel electrophoresis was carried out for about 4 h at 12.5 mA/gel. Protein spot 1 was excised from the Coomassie blue-stained gels and subjected to enzymatic digestion and internal sequencing. Following 2D SDS-PAGE analysis on two different biological replicates, proteins differentially produced in rugose and smooth cells were identified visually.
FIG. 1.
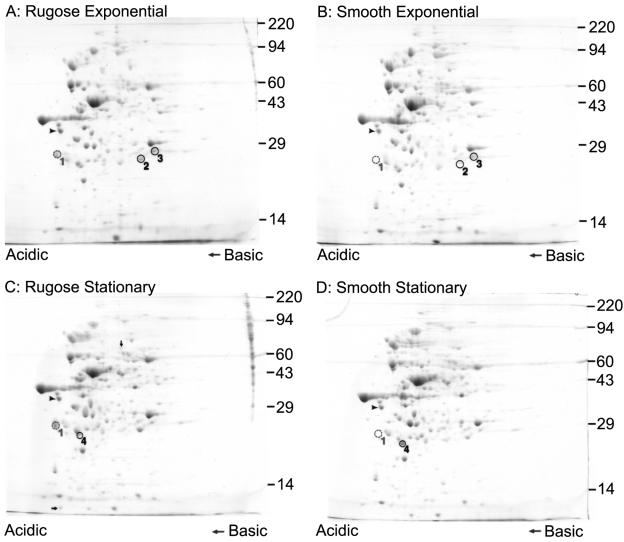
Protein profiles of smooth and rugose variants are different. Two-dimensional gel electrophoretic analyses of proteins in whole-cell lysates from rugose and smooth variants of V. cholerae during exponential and stationary growth phases are shown. The arrowheads indicate the IEF internal standard, tropomyosin protein (33 kDa), whereas protein spots that were differentially produced between the two variants are circled and numbered. Protein spot 1 corresponds to the protein RbmA. The arrows in panel C indicate protein spots that are present in larger amounts in rugose whole-cell lysates than in those from the smooth variant during stationary growth phase. Sizes (in thousands) of molecular weight markers are indicated on the right side of each panel.
Generation of in-frame rbmA deletion mutants in smooth and rugose variants of V. cholerae.
The 5′ (560 bp) and 3′ (417 bp) regions of the rbmA gene, which encodes VC0928, were amplified by PCR with primers VC0928_del_A (GATCTCTAGACTGTTTGTGGCGATCACATC) and VC0928_del_B (CAGAAACAATACAACTGGCGCTAAGGCTAA) and primers VC0928_del_C (CGCCAGTTGTATTGTTTCTGGTGAGCGAGT) and VC0928_del_D (GATCCCATGGTCGTTTCTTTCGAACATGTCA), respectively. The two PCR products generated (termed AB and CD) were then joined via SOEing (splicing by overlap extension) PCR (23, 24, 31) with primers VC0928_del_A and VC0928_del_D. Overlapping nucleotide sequences (20 bp) were incorporated into primers VC0928_del_B and VC0928_del_C to facilitate SOEing PCR. The amplified, 997-bp deletion fragment was cloned into pGP704-sacB28 via XbaI and NcoI restriction enzyme sites, which were incorporated into primers VC0928_del_A and VC0928_del_D, respectively. The accuracy of the deletion fragment was verified by DNA sequencing. The deletion recombinant plasmid (pGPVC0928) was used to transform the cloning strain E. coli CC118 (λ pir), purified, and then used to transform the conjugative strain E. coli S17-1 (λ pir). S17-1 (λ pir) was then conjugated with V. cholerae smooth and rugose variants separately. Transconjugants were selected on LB agar medium containing 100 μg/ml of ampicillin and rifampin. Sucrose-based selection, similar to that described by Fullner and Mekalanos (18), was used to select V. cholerae deletion clones that had undergone homologous recombination between the wild-type chromosomal copy of rbmA and the 997-bp deletion fragment. Briefly, 16 independent ampicillin- and rifampin-resistant transconjugants were randomly selected, streaked on LB agar plates containing 100 μg/ml of ampicillin and rifampin, and incubated at 37°C. Single colonies were then streaked on LB agar plates containing 100 μg/ml of ampicillin and 6% (wt/vol) sucrose without NaCl or 100 μg/ml of ampicillin alone to test for sucrose sensitivity and ampicillin resistance. Culture plates were incubated at 37°C, except for those containing sucrose, which were incubated at room temperature. Six ampicillin-resistant and sucrose-sensitive clones were selected, inoculated into 5 ml of LB medium, and incubated at 37°C with shaking at 200 rpm for 6 h. A loopful (1 μl) of the liquid culture grown for 6 h was then streaked on LB agar plates containing 6% (wt/vol) sucrose without NaCl and incubated at room temperature for 2 days. Deletion mutants growing on the sucrose-based medium were selected and streaked for single colonies on LB agar medium. Presence of the deletion was verified by PCR. Deletion mutants of rbmA generated in smooth and rugose backgrounds were designated as SΔrbmA and RΔrbmA, respectively.
Generation of complementation strains in RΔrbmA.
The complementation plasmid prbmA was constructed by cloning an amplified 1,113-bp fragment of rbmA (including 174 bp upstream to 123 bp downstream of the predicted rbmA open reading frame [ORF]), using primers VC0928_com F (GATCAAGCTTTTTGGTTTATTTTTGCTTTCTGG) and VC0928_com R (GATCCTCGAGTGAGGAAAGCGGTAAACTCC), into pACYC177 via HindIII and XhoI sites, which were incorporated into the primers. Following electroporation of prbmA into electrocompetent RΔrbmA cells, complemented clones were selected on LB agar medium containing ampicillin (100 μg/ml).
Generation of RΔrbmA expressing Myc-tagged RbmA.
The 813-bp coding region of rbmA (including the start codon but excluding the stop codon) was amplified with primers VC0928myc F (GATCCTCGAGGTTGTCTAACTTTAAAGGATCTATCA) and VC0928myc R (GATCTCTAGATTTTTTTTTACCACTGTCATTGACTG) and directionally cloned into plasmid pBAD/Myc-His B (Invitrogen) via XhoI and XbaI sites in frame with the downstream Myc epitope and six histidine codons. The resulting C-terminal Myc-tagged expression vector prbmA-myc was electroporated into electrocompetent RΔrbmA cells, and recombinant clones were selected on LB agar medium containing ampicillin (100 μg/ml).
Generation of rbmA-lacZ fusion strains in the V. cholerae rugose variant.
A 292-bp DNA fragment immediately upstream of the predicted rbmA ORF was amplified with primers P0928 F1 (GATCGAATTCGTTACAAGAACCCGGAAGAATG) and P0928 R1 (GATCGGATCCCCATTTGTTTTTACAACTGGCGCTAA). The amplified product was cloned into the promoterless lacZ vector pRS415 (50) via EcoRI and BamHI sites, generating the rbmA-lacZ fusion construct prbmA::lacZ, which was used to transform E. coli DH5α. In constructing V. cholerae rbmA-lacZ fusion strains, prbmA::lacZ was electroporated into electrocompetent rugose V. cholerae strains harboring a lacZ deletion (RugoseΔlacZ); vpsT and lacZ deletions (RΔvpsT ΔlacZ); vpsR and lacZ deletions (RΔvpsR ΔlacZ); and vpsR, vpsT, and lacZ deletions (RΔvpsR ΔvpsT ΔlacZ). Recombinant clones were selected on LB agar medium containing 100 μg/ml ampicillin.
β-Galactosidase assays.
β-Galactosidase activity was determined using a protocol similar to that described by Miller (35). Briefly, overnight cultures of V. cholerae were diluted 200-fold and inoculated into fresh LB medium. Cells in the exponential growth phase were harvested when the optical density at 600 nm (OD600) reached 0.4. Different dilutions of 100 μl of cell culture were pipetted into wells of a MultiScreen-HA 96-well microtiter plate fitted onto a MultiScreen filtration system (Millipore). Culture medium was removed by applying a vacuum to the filtration system. The cells were then washed with 200 μl of buffer Z (16.1 g/liter Na2HPO4 · 7H2O, 5.5 g/liter NaH2PO4 · H2O, 0.75 g/liter KCl, 0.246 g/liter MgSO4 · 7H2O) at pH 7.0. Cell lysis was carried out by adding 100 μl of buffer Z containing 0.69% (vol/vol) β-mercaptoethanol, 0.02% (wt/vol) cetyltrimethylammonium bromide, and 0.01% (wt/vol) deoxycholic acid sodium salt, followed by incubation at room temperature for 5 min. ONPG (o-nitrophenyl-β-d-galactopyranoside) solution (20 μl of 4 mg/ml) was added to the lysed cell suspensions, and these were further incubated at 30°C until sufficient color development was observed. The duration of color development was noted for each culture. The reaction was stopped by adding 50 μl of 1 M Na2CO3. The intensities of color were measured at ODs of 420 nm and 550 nm after transferring the samples to a new flat-bottom 96-well microtiter plate. Miller units for β-galactosidase activity were calculated using the following formula: Miller units = 1,000 × (OD420 − 1.75 × OD550)/(t × v × OD600), where t is the reaction time in minutes and v is the assay volume in milliliters. The data were presented as average values of at least three technical replicates from one representative experiment, which was repeated three times with different biological replicates showing similar data.
Quantitative biofilm assays.
Biofilm formation assays were carried out in polyvinyl chloride microtiter plates following a method previously described (60). Briefly, 100 μl of 200-fold-diluted overnight cultures were pipetted into the wells of 96-well microtiter plates. The plates were incubated at 30°C for 8 h. Liquid medium in each well was discarded, and the plates were washed once gently with water. Crystal violet staining and ethanol solubilization were carried out, as previously described, to quantify biofilm formation (41). The data are presented as average values of at least 15 technical replicates from one representative experiment, which was repeated three times with different biological replicates showing similar data.
Generation of GFP-tagged strains of V. cholerae wild-type variants and rbmA deletion mutants.
Green fluorescent protein (GFP)-tagged wild-type rugose and rbmA deletion mutants were generated by triparental conjugation with donor E. coli S17-1 (λ pir) cells carrying pGP704::Tn7-GFP (a mini-Tn7 transposon containing the gfpmut3 gene under the control of a constitutive ribosomal promoter) and helper E. coli S17-1 (λ pir) cells harboring pUX-BF13 containing the Tn7 transposase gene. The mini-Tn7 transposon targets the chromosomal region between loci VC0487 and VC0488. Transconjugants were selected on thiosulfate-citrate-bile salts-sucrose (Difco) agar medium containing 30 μg/ml gentamicin at 30°C. GFP-tagged strains were verified by PCR analysis and fluorescence microscopy.
Flow cells and confocal scanning laser microscopy (CSLM).
Overnight cultures, grown in LB liquid medium at 30°C with shaking at 200 rpm, were diluted to an OD600 of 0.02, and 200-μl aliquots were inoculated by injection into flow cell chambers. Before inoculation, the chambers were sterilized and equilibrated with 1 liter of 0.5% (vol/vol) hypochlorite, followed by 1 liter of sterile MilliQ water and 200 ml of 2% LB (0.2 g/liter tryptone, 0.1 g/liter yeast extract, 9 g/liter NaCl) at a flow rate of 0.26 ml/min. After inoculation, the chambers were allowed to stand inverted, with no flow, for 1 h. Flow was resumed at a rate of 0.26 ml/min with chambers standing upright for 30 min. Flow cell experiments were carried out at room temperature. Confocal images of biofilms that formed in flow cell chambers were captured with an LSM 5 PASCAL system (Zeiss) at 488-nm excitation and 543-nm emission wavelengths. Three-dimensional images of the biofilms were reconstructed using Imaris software (Bitplane) and quantified using COMSTAT (22). For the biofilm structure integrity studies, 0.2% (wt/vol) SDS was used.
Cellular fractionation.
Overnight cultures were diluted 100-fold and inoculated into fresh LB medium containing ampicillin. Cells in the exponential growth phase (OD600 = 0.3) were then induced with 0.1% (wt/vol) arabinose for 1 h. Cultures (30 ml) were harvested by centrifugation (10,000 × g), and cellular fractionation was carried out similarly to a procedure previously described (7). Briefly, the cell pellet was resuspended in PBS containing polymyxin B sulfate (10,000 U) and incubated on ice for 10 min. The soluble periplasmic fraction was collected after centrifugation (10,000 × g) at 4°C for 10 min. The spheroplast pellet was resuspended in 10 mM Tris-Cl (pH 8.0), and cells were lysed by sonication (10-s pulses repeated five times). Unlysed cells were removed by repeated centrifugations (10,000 × g) at 4°C for 15 min each. The soluble cytoplasmic fraction was separated from total membranes by centrifugation of the lysate (100,000 × g) at 4°C for 1 h. The inner and outer membranes were differentially solubilized by resuspending the total membrane pellet in 10 mM Tris-Cl containing 100 mM NaCl and 2.5% (wt/vol) Sarkosyl and incubated at room temperature with shaking for 30 min. The soluble inner membrane fraction was separated from the outer membrane pellet by centrifugation at 200,000 × g at 4°C for 1 h. The outer membrane was resuspended in 200 μl of 10 mM Tris-Cl (pH 8.0).
Proteins in the culture supernatant were precipitated using TCA (4). Briefly, 2 ml of 0.15% (wt/vol) deoxycholic acid sodium salt was added to 20 ml of culture supernatant, vortexed, and incubated at room temperature for 10 min. Then, 2 ml TCA was added, and the samples were vortexed and incubated on ice for 30 min. The protein precipitate was pelleted by centrifugation at 10,000 × g for 10 min at 4°C. The pellet was washed twice with ice-cold acetone and resuspended in 200 μl of 10 mM Tris-Cl (pH 8.0). Protein concentrations were estimated using a bicinchoninic acid protein assay kit (Pierce) with bovine serum albumin as the standard. Protein samples were loaded at 5 and 1 μg for immunoblot analysis of RbmA and OmpU, respectively.
Immunoblot analysis.
Proteins from various cellular and culture supernatant fractions were separated on a 10% SDS-polyacrylamide gel and electroblotted onto a nitrocellulose membrane with a Mini Trans-Blot Cell (Bio-Rad) apparatus in transfer buffer containing 25 mM Tris (pH 8.3), 192 mM glycine, 20% (vol/vol) methanol, and 0.1% (wt/vol) SDS. The blot was blocked with 2% (wt/vol) skim milk and washed with PBS buffer containing 250 mM NaCl and 0.1% (vol/vol) Tween 20. Rabbit polyclonal antiserum against V. cholerae OmpU (provided by K. Klose) was used at a dilution of 1:100,000, while mouse monoclonal antibody against the Myc epitope (Santa Cruz Biotechnology) was used at a dilution of 1:1,000. Horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse secondary antibodies (Santa Cruz Biotechnology) were used at a dilution of 1:2,500. Immunoblots were developed with a SuperSignal West Pico chemiluminescent kit (Pierce). Immunoblot analyses were conducted using two biological replicates and two technical replicates.
RESULTS
Protein profiles of the rugose and smooth variants are different.
To identify changes in total protein production patterns resulting from phase variation, smooth and rugose cultures were labeled with [35S]methionine during both exponential and stationary phases of growth, and labeled proteins were then identified by 2D gel electrophoresis. Results of such analyses (data not shown) show differences in total protein profiles between the two variants, whereby phase variation negatively and positively regulates production of multiple proteins.
To determine the identities of the proteins that are differentially produced, crude protein extracts from whole-cell lysates were separated by 2D gel electrophoresis, and the gels were stained with Coomassie blue. Analysis of the proteins produced by smooth and rugose variants during exponential and stationary growth phases revealed that at least four protein spots (indicated by circles and numbered in Fig. 1) were differentially produced. Protein spot 1 was produced at higher levels in cell lysates from the rugose variant than in those from the smooth one (Fig. 1). Similarly, protein spots 2 and 3 were present in larger amounts in crude protein extract from the rugose variant grown to exponential phase than in that from the smooth variant (Fig. 1A and B). Protein spot 4 was present in smaller amounts in lysates from the rugose variant grown to stationary phase than in lysates from the smooth variant (Fig. 1C and 1D). In addition, two other protein spots (indicated by arrows in Fig. 1C) appear to be present in slightly larger amounts in the protein sample from the rugose variant grown to stationary phase than the smooth one.
We focused our studies on protein spot 1 and determined the sequence of 12 amino acids corresponding to the internal part of the protein, which aligned to the amino acid sequence of the hypothetical protein annotated as VC0928 in the V. cholerae El Tor N16961 genome (Fig. 2A). We named the gene locus VC0928 rbmA (for rugosity and biofilm structure modulator A) because of evidence that it is involved in rugose colony and biofilm structure formation (discussed below). There are two possible start codons encoding methionine (M1 and M9) in rbmA (Fig. 2A). BLAST searches of the nonredundant protein database (NCBI) using the predicted polypeptide sequences beginning with either methionine did not reveal any significant homologs, suggesting that this protein may be present only in V. cholerae. The closest hit had an E value of 0.19 for a Mesorhizobium loti sequence of unknown function. More-sensitive searches using the Sequence Alignment and Modeling software system (SAM-T2K and SAM-T04 methods) (29, 30) were also unsuccessful in finding any similar sequences. Using the TMHMM (transmembrane helices prediction), SignalP (signal peptide cleavage sites prediction), and LipoP (lipoproteins and signal peptides prediction) programs, a strong prediction was given for a signal peptide with cleavage between A30 and E31 (probability 0.922 and 0.998 for the two SignalP methods) (27, 38, 51). We next attempted to understand the function of this protein using structure prediction programs; their outputs are presented in the Discussion.
FIG. 2.
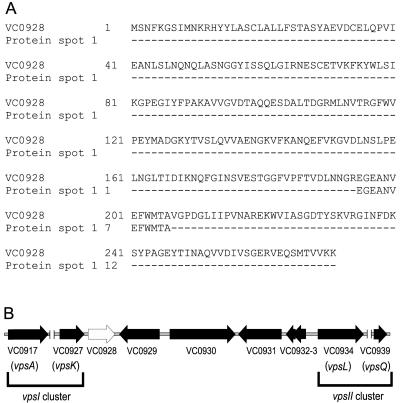
Amino acid sequence of protein spot 1 corresponds to that of VC0928. (A) Alignment of the 12-amino-acid sequence from protein spot 1 with a peptide sequence encoded by rbmA (locus VC0928), as obtained from TIGR. (B) The genetic organization of rbmA (open arrow) on the V. Cholerae chromosome is depicted with locus annotations under each ORF. The vpsI and vpsII gene clusters, as well as the first and last genes of each cluster (vpsA and vpsK; vpsL and vpsQ) are marked and labeled. Unlinked chromosomal DNA regions within the vps clusters are also indicated (∥). (Illustration is not to scale.)
rbmA is located in the vps region between the vpsI and vpsII clusters (Fig. 2B), which contain the genes required for VPS production (62). Putative regulatory elements (−10 and −35 regions and terminator sequences) have been predicted up- and downstream of the rbmA ORF (data not shown), suggesting that it is expressed as a monocistronic transcript and not part of an operon structure. Immediately upstream of rbmA is the last gene of the vpsI cluster (vpsK, VC0927), predicted to encode a UDP-N-acetyl-d-mannosamine transferase. Directly downstream of rbmA is the locus VC0929, predicted to code for a hypothetical protein 408 amino acids in length, and then several ORFs predicted to code for a hemolysin-related protein (VC0930), three hypothetical proteins (VC0931-3), and the first gene of the vpsII cluster (vpsL, VC0934), which encodes a capsular polysaccharide biosynthesis glycosyltransferase. Inactivation of locus VC0930 using transposon TnphoA insertion resulted in a switch from rugose to smooth colonial phenotype (2), suggesting that the genes located between the two vps clusters play a role in maintaining rugose colony formation in V. cholerae.
rbmA deletion affects rugose colonial morphology.
The close proximity of rbmA to the vps clusters prompted us to ask whether the gene is involved in cellular processes that are affected by the expression of vps genes (62). To identify the function of rbmA, deletion mutants were generated in both the rugose (RΔrbmA) and smooth (SΔrbmA) variants. Deletion of rbmA in the rugose variant decreased the corrugation of rugose colonies but did not lead to completely smooth colony formation (Fig. 3A1 and 3A3). Complementation of RΔrbmA using the plasmid prbmA, which harbors the wild-type copy of rbmA (including 174 bp upstream and 123 bp downstream from the predicted ORF) on a low-copy-number vector, restored the normal rugose colonial morphology (Fig. 3A6). In contrast, RΔrbmA transformed with plasmid vector alone retained its abnormal colonial morphology (Fig. 3A5). No obvious differences in colonial morphology were observed in the rbmA deletion mutant generated in the smooth variant compared to the wild type (Fig. 3A2 and A4). Together, these results show that RbmA is required for rugose colony formation in V. cholerae.
FIG. 3.
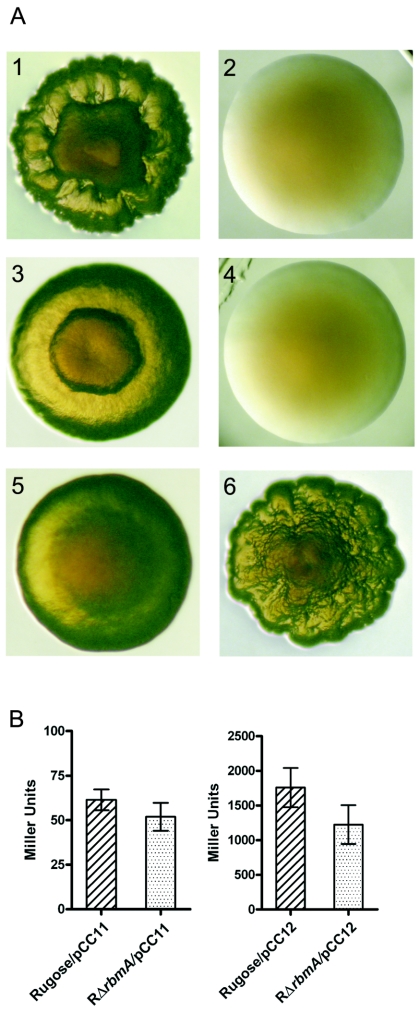
RbmA is critical to rugose colony formation, and the expression of vpsA and vpsL is not altered in RΔrbmA. Panel A shows the colony morphologies of the rugose variant (A1), smooth variant (A2), RΔrbmA (A3), SΔrbmA (A4), RΔrbmA/pACYC177 (A5), and RΔrbmA/prbmA (A6). (B) Expression of the vpsA-lacZ and vpsL-lacZ fusion genes (carried in the pCC11 and pCC12 plasmids, respectively) in the wild-type rugose and RΔrbmA variants grown in LB at 30°C to mid-exponential growth phase (OD600 = 0.4). Error bars represent standard deviations.
We also determined the abundance of transcripts corresponding to the vpsA and vpsL genes in RΔrbmA to investigate whether expressions of these two genes, known to be important to rugose colonial morphology, were altered. To this end, vpsA- and vpsL-lacZ fusion plasmids (pCC11 and pCC12, respectively) (8) were introduced into the wild-type rugose variant and RΔrbmA, and β-galactosidase activities were determined. As shown in Fig. 3B, there are no significant differences in transcriptional abundance of vpsA and vpsL between the wild type and RΔrbmA. Both wild-type and RΔrbmA rugose variants containing vector alone showed negligible background β-galactosidase activities (data not shown). These results demonstrate that vps gene expression is not altered in RΔrbmA, and therefore the less-corrugated colonial morphology of RΔrbmA is due to the loss of RbmA function.
rbmA is critical for biofilm structure and fitness.
The biofilm-forming capacity of the rugose variant is greatly enhanced compared to that of the smooth variant. Since RΔrbmA exhibited an altered colonial morphology, we asked whether the biofilm-forming property of RΔrbmA was also altered relative to the wild type. Thus, we first compared the biofilm-forming properties of the two strains by crystal violet staining of biofilms formed on microtiter plates after 8 h of incubation under static conditions at 30°C. The results, shown in Fig. 4A, revealed that the biofilm-forming capacity of RΔrbmA was not significantly different from that of the wild-type rugose variant. This finding suggests that rbmA has a different role than the genes that code for VPS biosynthesis, since vps mutants have reduced biofilm-forming capacities.
FIG. 4.
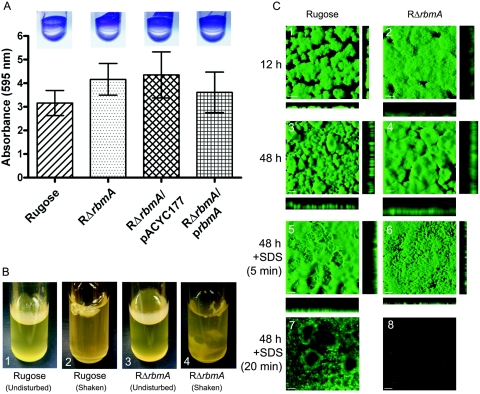
RbmA is required for biofilm structure. (A) Quantitative comparison of biofilm formation by the wild-type rugose variant, RΔrbmA, RΔrbmA/pACYC177, and RΔrbmA/prbmA after an 8-h incubation in LB medium at 30°C under static conditions. Error bars represent standard deviations. Crystal violet-stained biofilms formed in the wells of polyvinyl chloride microtiter plates, which were used in the quantitative biofilm assays, are shown in the inserts. (B) Pellicle formation by the wild-type rugose variant (B1 and B2) and RΔrbmA (B3 and B4) after 2 days of incubation in LB at 30°C. B2 and B4 show pellicles disturbed by shaking. (C) Confocal scanning laser microscopic images of horizontal (xy) and vertical (xz) projections of biofilm structures formed by the rugose variant and RΔrbmA in a flow cell system. Biofilm structures that developed after 12 and 24 h of incubation are shown in panels C1 to C4. Biofilm structure integrity studies using SDS are shown in C5 to C8, and the duration of SDS treatment is indicated on the left. In C1 and C2, bars represent 30 μm, whereas in C3 to C8, they represent 100 μm.
The rugose variant also forms a pellicle, which is a biofilm formed at air-liquid interface. When cultures are grown statically, RΔrbmA forms a pellicle that appears slightly less developed (that is, less wrinkled) than that of the rugose variant. Upon shaking, the pellicle formed by RΔrbmA broke into pieces, suggesting that it might have been more fragile (Fig. 4B). In contrast, the pellicle formed by the wild-type rugose variant detached as an aggregate. Differences in the stabilities of pellicles prompted us to find structural differences between the biofilms formed by RΔrbmA and the wild-type rugose variant.
To understand the dynamics of biofilm development in a flowing system, we first analyzed biofilm formation by CSLM as a function of time using a once-through flow cell reactor and GFP-tagged strains. After 12 h, the biofilm formed by RΔrbmA appears less developed (that is, less differentiated) than that of the wild-type rugose variant (Fig. 4C). COMSTAT analysis of the biofilms revealed that the RΔrbmA biofilm is thicker in height (average thickness of 61 μm compared to 14 μm for the wild type), less organized, and more disperse (that is, loosely packed) (Fig. 4C1 and C2). These differences were more evident after 48 h, when the average thickness of the rugose biofilm was 200 μm compared to 312 μm for RΔrbmA (Fig. 4C3 and C4). The increased thickness of the biofilm structure of RΔrbmA is not likely to be due to an increase in VPS production, since vpsA and vpsL gene expression and total biofilm mass (as determined by crystal violet quantification) are similar between RΔrbmA and the wild type. Treatment of biofilms with 0.2% (wt/vol) SDS for 5 min destroyed the biofilm formed by RΔrbmA (Fig. 4C6), and after 20 min of SDS treatment the biofilm had almost completely detached (Fig. 4C8). On the other hand, the biofilm formed by the rugose variant remained relatively intact after the same treatment (Fig. 4C5 and C7). The differences observed in pellicle and biofilm formation could not be attributed to an altered growth rate of the mutant, since there was no difference in the growth rates of RΔrbmA and the wild type. Together, these results show that RbmA is required for the formation of structurally mature biofilms and for biofilm fitness in V. cholerae.
RbmA is a secreted protein.
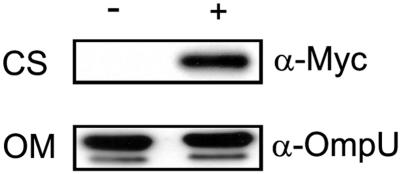
To investigate the cellular localization of RbmA, a chimeric protein was generated by translational fusion of the C terminus of RbmA to a Myc epitope and six histidine residues (in pBAD/Myc-His B). Transcription of the fusion protein was under the control of an arabinose-inducible promoter. RΔrbmA grown to exponential phase harboring the translational fusion plasmid prbmA-myc was induced with 0.1% (wt/vol) arabinose, harvested, and fractionated into cellular components. Anti-Myc antibody (α-Myc) detected an immunoreactive polypeptide of approximately 31 kDa (Fig. 5) in the TCA-precipitated culture supernatant (CS) fraction. In contrast, such a band was not detected in the uninduced CS fraction (Fig. 5) or in both induced and uninduced periplasmic, inner membrane, and outer membrane (OM) fractions (data not shown). A faint band of 31 kDa was also detected in the induced (but not in the uninduced) cytoplasmic fraction. As a control, immunoblot analysis of cellular fractions and TCA-precipitated CS fractions was also carried out with anti-OmpU antisera (α-OmpU). A strong signal corresponding to an immunoreactive polypeptide of the expected size for OmpU in V. cholerae was detected in the OM fractions from both induced and uninduced cultures (Fig. 5). Together, these results indicate that RbmA is predominantly present in the culture supernatant fraction of V. cholerae and thus likely to be a secreted protein.
FIG. 5.
RbmA is a secreted protein. Immunoblot analysis of proteins in the CS and OM fractions from arabinose-induced (+) and uninduced (−) cultures of RΔrbmA harboring plasmid prbmA-myc. Mouse monoclonal antibody against the Myc epitope (α-Myc) and rabbit polyclonal antisera against V. cholerae OmpU (α-OmpU) were used to detect RbmA-Myc and OmpU, respectively.
Expression of rbmA is regulated by VpsR.
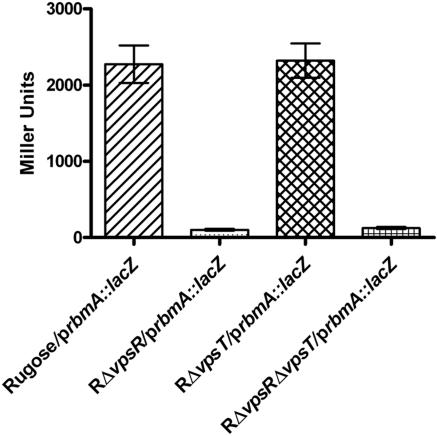
The response regulators VpsR and VpsT regulate the expression of vps genes and the formation of corrugated colonies. To study a possible effect of VpsR and VpsT on the expression of rbmA, an rbmA-lacZ transcriptional fusion (prbmA::lacZ) plasmid was constructed and electroporated into wild-type rugose, RΔvpsR, RΔvpsT, and RΔvpsR ΔvpsT strains (8). The chromosomal copy of lacZ in these strains has been inactivated by an in-frame deletion to prevent background β-galactosidase activity. High levels of β-galactosidase activity were detected in the wild-type rugose and RΔvpsT strains harboring prbmA::lacZ but not in RΔvpsR (β-galactosidase activity was 17-fold lower than in the wild type) or RΔvpsR ΔvpsT (14-fold lower than the wild type) (Fig. 6). All strains carrying the vector pRS415 showed negligible background β-galactosidase activity (data not shown). These results indicate that expression of rbmA is positively regulated by VpsR. Thus, rbmA is likely to be coordinately expressed with the vps genes.
FIG. 6.
VpsR is required for rbmA transcription. Expression of an rbmA-lacZ fusion gene (prbmA::lacZ) in the wild type and in vpsR, vpsT, and vpsR vpsT mutants of the rugose variant grown in LB at 30°C to mid-exponential growth phase (OD600 = 0.4) is shown. Error bars represent standard deviations.
DISCUSSION
Phase variation between smooth and rugose variants and the expression of associated properties are believed to be linked to the survival of V. cholerae in many aquatic environments. To better understand changes to the proteome that accompany phase variation, proteomic analyses were carried out on total cell proteins extracted from smooth and rugose variants grown to exponential and stationary growth phases (Fig. 1 and data not shown). The results showed that total protein profiles of smooth and rugose variants are different during both growth phases and that phase variation negatively and positively regulates the production of multiple proteins.
We identified a protein called RbmA that was produced at higher levels in the rugose variant than in the smooth variant during both exponential and stationary growth phases. This finding is congruent with transcriptome analysis of the phase variants, which showed that transcription of rbmA (VC0928 locus) is higher in the rugose variant (61). RbmA does not have any homologs in protein sequence databases. Thus, to gain insight into its possible function, we attempted to determine the likely structure of RbmA using different structure prediction programs. Because preliminary computer predictions and experimental results suggested that RbmA is a secreted protein, structure prediction analysis was conducted based on the assumption that RbmA is secreted and used only the portion of the protein starting with E31. We used the SAM-T04 protocol for structure prediction, though both local structure prediction and fold recognition are of limited reliability for orphan ORF sequences, as both rely heavily on signals from the sampling of sequence space done by evolution. Five different protein folds were found to have E values of less than 5.0, whereby the top two appeared to be secreted proteins, though their E values (1.48 and 2.49) indicate that the predictions are speculative at best. The top hit was to 1qfhA, F-actin cross-linking gelation factor (ABP-120) repeats from slime mold (Dictyostelium discoideum), which the program SCOP version 1.67 classifies as an E set domain, an immunoglobulin-like beta sandwich possibly related to the immunoglobulin and fibronectin type III domain (FnIIID) superfamilies (3). This fold family, initially characterized in fibronectin, is common among modular proteins. FnIIID is present in extracellular proteins, intracellular proteins, and the extracellular domains of membrane-receptor proteins (6). FnIIID is also found in some carbohydrases (enzymes that hydrolyze polymers composed of a six-carbon sugar backbone) from different bacteria. Structure analyses of the chitin binding protein, CBP21, from Serratia marcescens and chitinase A1 from Bacillus circulans WL-12 have also revealed the presence of FnIIID in these proteins (26, 54). The second hit was to 1jlxA, an agglutinin from Love-lies-bleeding (Amaranthus caudatus), which SCOP classifies as a beta-trefoil (37). The beta-trefoil family proteins include fibroblast growth factors, interleukin-1, mannose receptor, agglutinin, and Clostridium neurotoxins (59). The mannose receptor and agglutinin is a particularly interesting fold, as these proteins are known to bind sugars. Structure prediction suggests that RbmA may fold into either the FnIIID or beta-trefoil family of tertiary structures. Both superfamilies include proteins capable of binding carbohydrates (6, 59).
Protein localization studies revealed that RbmA is found predominantly in culture supernatants. The size of the approximately 31-kDa immunoreactive polypeptide that was detected in the arabinose-induced CS fraction is consistent with that of the chimeric protein RbmA-Myc without the signal peptide (predicted to be 29 kDa). The main terminal branch pathway of the type II general secretion pathway is involved in the secretion of several V. cholerae proteins, including cholera toxin (1, 14, 48), heat-labile enterotoxin (42), endochitinase (ChiA) (11, 17), and protease(s) (42, 47). This pathway may also be involved in the secretion of hemolysin and polysaccharides in V. cholerae (1). The secretory system by which RbmA translocates is yet to be determined, but it is possible that the main terminal branch pathway plays a role in this process.
In this study, we showed that RbmA is required for the maintenance of rugose colonial morphology (Fig. 3) and the development of wild-type biofilm architecture (Fig. 4). Both processes require the production of VPS, which is composed of nearly equal amounts of glucose and galactose and smaller amounts of N-acetylglucosamine and mannose. Hence, both structure predictions and phenotypic analyses suggest that RbmA may be a sugar-binding protein. The exact mechanism by which this protein functions (in other words, whether it binds carbohydrates and, if it does, what types) has yet to be determined. It is possible that RbmA binds to carbohydrates in the VPS matrix, thus giving rise to the tightly packed and organized biofilm structure observed in the wild-type rugose variant. It is also possible that RbmA acts like an agglutinin/adhesin, binding cells together or anchoring the biofilm and/or cells to a substratum that contains carbohydrates. Recent studies reported that agglutinin and adhesin proteins are present in E. coli and in Pseudomonas aeruginosa (25, 49, 52). The multipurpose, nonorganelle adhesin TibA mediates cell autoaggregration, binding, and invasion of E. coli to human cells and enhances biofilm formation (49). Similarly, the two carbohydrate-binding proteins (lectins), LecA and LecB, from P. aeruginosa have been shown to bind specifically to galactose and fucose, respectively. Furthermore, LecB, which localized on the outer membrane of the bacterium, is required for biofilm formation (25, 52).
In addition to altering biofilm architecture, absence of RbmA reduced the biofilm's resistance properties. Biofilms formed by RΔrbmA were more sensitive to SDS than wild-type biofilms. Similar findings have also been reported for the biofilms formed by toxin-coregulated pilus mutants of V. cholerae cultured on chitinous surfaces (44). These observations suggest that changes to the extrapolymeric substance composition of biofilms, at either the exopolysaccharide or the protein level, can alter biofilm fitness.
In V. cholerae, several members of the two-component superfamily of proteins are involved in regulating genes required (either directly or indirectly) for biofilm formation. The positive regulators VpsR and VpsT, which exhibit a high degree of homology to response regulators of the two-component superfamily, are required for the transcription of vps genes and the formation of mature biofilms in V. cholerae (8, 60). Data in this study show that expression of rbmA, unlike that of vps genes vpsA and vpsL, is positively regulated by only VpsR and not VpsT (Fig. 6). The significance of this result has yet to be determined. A recent study reported that VieA, the response regulator of the VieS/A/B three-component signal transduction system, represses the transcription of rbmA (VC0928 locus) and the vps genes (53). Interestingly, repression of rbmA in the ΔvieA mutant is observed only in the classical biotype and not in the El Tor biotype of V. cholerae, presumably because a functionally redundant protein in the El Tor biotype compensates for the loss of VieA (53). HapR, a negative regulator that represses the expression of vps genes at high cell density (20, 61, 63), also negatively regulates rbmA transcription (61). Altogether, these findings show that genes encoding proteins required for VPS production and biofilm architecture (including rbmA) are coordinately regulated by VpsR and HapR.
Biofilm formation by aquatic microorganisms is well documented (12). It has been proposed that the attachment of bacteria to surfaces and subsequent biofilm growth exemplify a survival strategy. This is because surfaces adsorb and concentrate nutrients from the surrounding liquid and provide protection from grazing predators and toxic compounds (12, 13, 43). Although the steps involved in biofilm formation and some of the required structural and regulatory genes have been identified, relatively little is known about the components of V. cholerae biofilms that control structure, function, and fitness. In this study, we identified one such protein, called RbmA, which is required for rugose colony formation and biofilm fitness. Its production enables cells to form more structured, detergent-resistant biofilms.
Acknowledgments
This work was supported by grants from The Ellison Medical Foundation and NIH (AI055987) to F.H.Y. The bioinformatics work was partially supported by NIH GM068570-01 to K.K.
We thank members of the Yildiz Laboratory for their suggestions; Michael Miller for providing pGP704-sacB28, pGP704::Tn7-GFP, and pUX-BF13; Karl Klose for providing the anti-OmpU antisera; Brooke Jude and Ronald Taylor for providing the cellular fractionation procedure; and Kivanc Bilecen for flow cell studies and CSLM.
REFERENCES
- 1.Ali, A., J. A. Johnson, A. A. Franco, D. J. Metzger, T. D. Connell, J. G. Morris, Jr., and S. Sozhamannan. 2000. Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production in and motility of Vibrio cholerae. Infect. Immun. 68:1967-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, A., Z. H. Mahmud, J. G. Morris, Jr., S. Sozhamannan, and J. A. Johnson. 2000. Sequence analysis of TnphoA insertion sites in Vibrio cholerae mutants defective in rugose polysaccharide production. Infect. Immun. 68:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreeva, A., D. Howorth, S. E. Brenner, T. J. Hubbard, C. Chothia, and A. G. Murzin. 2004. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 32:D226-D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bollag, D. M., M. D. Rozycki, and S. J. Edelstein. 1996. Protein methods, 2nd ed., p. 84-85. Eiley-Lis, Inc., New York, N.Y.
- 5.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185:1384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bork, P., and R. F. Doolittle. 1992. Proposed acquisition of an animal protein domain by bacteria. Proc. Natl. Acad. Sci. USA 89:8990-8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bose, N., and R. K. Taylor. 2005. Identification of a TcpC-TcpQ outer membrane complex involved in the biogenesis of the toxin-coregulated pilus of Vibrio cholerae. J. Bacteriol. 187:2225-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186:1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiavelli, D. A., J. W. Marsh, and R. K. Taylor. 2001. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl. Environ. Microbiol. 67:3220-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colwell, R. R. 1996. Global climate and infectious disease: the cholera paradigm. Science 274:2025-2031. [DOI] [PubMed] [Google Scholar]
- 11.Connell, T. D., D. J. Metzger, J. Lynch, and J. P. Folster. 1998. Endochitinase is transported to the extracellular milieu by the eps-encoded general secretory pathway of Vibrio cholerae. J. Bacteriol. 180:5591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 13.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, B. M., E. H. Lawson, M. Sandkvist, A. Ali, S. Sozhamannan, and M. K. Waldor. 2000. Convergence of the secretory pathways for cholera toxin and the filamentous phage, CTXφ. Science 288:333-335. [DOI] [PubMed] [Google Scholar]
- 15.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 16.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folster, J. P., and T. D. Connell. 2002. The extracellular transport signal of the Vibrio cholerae endochitinase (ChiA) is a structural motif located between amino acids 75 and 555. J. Bacteriol. 184:2225-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fullner, K. J., and J. J. Mekalanos. 1999. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect. Immun. 67:1393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grau, B. L., M. C. Henk, and G. S. Pettis. 2005. High-frequency phase variation of Vibrio vulnificus 1003: isolation and characterization of a rugose phenotypic variant. J. Bacteriol. 187:2519-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 21.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 23.Horton, R. M. 1997. In vitro recombination and mutagenesis of DNA. SOEing together tailor-made genes. Methods Mol. Biol. 67:141-149. [DOI] [PubMed] [Google Scholar]
- 24.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 25.Imberty, A., M. Wimmerova, E. P. Mitchell, and N. Gilboa-Garber. 2004. Structures of the lectins from Pseudomonas aeruginosa: insight into the molecular basis for host glycan recognition. Microbes Infect. 6:221-228. [DOI] [PubMed] [Google Scholar]
- 26.Jee, J. G., T. Ikegami, M. Hashimoto, T. Kawabata, M. Ikeguchi, T. Watanabe, and M. Shirakawa. 2002. Solution structure of the fibronectin type III domain from Bacillus circulans WL-12 chitinase A1. J. Biol. Chem. 277:1388-1397. [DOI] [PubMed] [Google Scholar]
- 27.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karplus, K., R. Karchin, C. Barrett, S. Tu, M. Cline, M. Diekhans, L. Grate, J. Casper, and R. Hughey. 2001. What is the value added by human intervention in protein structure prediction? Proteins Struct. Funct. Genet. 45:86-91. [DOI] [PubMed] [Google Scholar]
- 30.Karplus, K., S. Katzman, G. Shackleford, M. Koeva, J. Draper, B. Barnes, M. Soriano, and R. Hughey. 2005. SAM-T04: what's new in protein-structure prediction for CASP6. Proteins Struct. Funct. Bioinformat., in press. [DOI] [PubMed]
- 31.Lefebvre, B., P. Formstecher, and P. Lefebvre. 1995. Improvement of the gene splicing overlap (SOE) method. BioTechniques 19:186-188. [PubMed] [Google Scholar]
- 32.Lipp, E. K., A. Huq, and R. R. Colwell. 2002. Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 15:757-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarter, L. 1999. The multiple identities of Vibrio parahaemolyticus. J. Mol. Microbiol. Biotechnol. 1:51-57. [PubMed] [Google Scholar]
- 34.Meibom, K. L., X. B. Li, A. T. Nielsen, C. Y. Wu, S. Roseman, and G. K. Schoolnik. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. USA 101:2524-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Assay of β-galactosidase, p. 352-355. In J. H. Miller (ed.), Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Morris, J. G., Jr., M. B. Sztein, E. W. Rice, J. P. Nataro, G. A. Losonsky, P. Panigrahi, C. O. Tacket, and J. A. Johnson. 1996. Vibrio cholerae O1 can assume a chlorine-resistant rugose survival form that is virulent for humans. J. Infect. Dis. 174:1364-1368. [DOI] [PubMed] [Google Scholar]
- 37.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536-540. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 39.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 40.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 41.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 42.Overbye, L. J., M. Sandkvist, and M. Bagdasarian. 1993. Genes required for extracellular secretion of enterotoxin are clustered in Vibrio cholerae. Gene 132:101-106. [DOI] [PubMed] [Google Scholar]
- 43.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 44.Reguera, G., and R. Kolter. 2005. Virulence and the environment: a novel role for Vibrio cholerae toxin-coregulated pili in biofilm formation on chitin. J. Bacteriol. 187:3551-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rice, E. W., C. H. Johnson, R. M. Clark, K. R. Fox, D. J. Reasoner, M. E. Dunnigan, P. Panigrahi, J. A. Johnson, and J. G. Morris, Jr. 1993. Vibrio cholerae O1 can assume a ‘rugose’ survival form that resist killing by chlorine, yet retains virulence. Int. J. Environ. Health Res. 3:89-98. [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 47.Sandkvist, M., L. O. Michel, L. P. Hough, V. M. Morales, M. Bagdasarian, M. Koomey, V. J. DiRita, and M. Bagdasarian. 1997. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 179:6994-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandkvist, M., V. Morales, and M. Bagdasarian. 1993. A protein required for secretion of cholera toxin through the outer membrane of Vibrio cholerae. Gene 123:81-86. [DOI] [PubMed] [Google Scholar]
- 49.Sherlock, O., R. M. Vejborg, and P. Klemm. 2005. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect. Immun. 73:1954-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 51.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 52.Tielker, D., S. Hacker, R. Loris, M. Strathmann, J. Wingender, S. Wilhelm, F. Rosenau, and K. E. Jaeger. 2005. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 151:1313-1323. [DOI] [PubMed] [Google Scholar]
- 53.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaaje-Kolstad, G., D. R. Houston, A. H. Riemen, V. G. Eijsink, and D. M. van Aalten. 2005. Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J. Biol. Chem. 280:11313-11319. [DOI] [PubMed] [Google Scholar]
- 55.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watnick, P. I., K. J. Fullner, and R. Kolter. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White, P. B. 1938. The rugose variant of Vibrios. J. Pathol. 46:1-6. [Google Scholar]
- 59.Xu, R., and Y. Xiao. 2005. A common sequence-associated physicochemical feature for proteins of beta-trefoil family. Comput. Biol. Chem. 29:79-82. [DOI] [PubMed] [Google Scholar]
- 60.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPSETr-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497-515. [DOI] [PubMed] [Google Scholar]
- 62.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647-656. [DOI] [PubMed] [Google Scholar]