Abstract
MreB, a major component of the recently discovered bacterial cytoskeleton, displays a structure homologous to its eukaryotic counterpart actin. Here, we study the assembly and mechanical properties of Thermotoga maritima MreB in the presence of different nucleotides in vitro. We found that GTP, not ADP or GDP, can mediate MreB assembly into filamentous structures as effectively as ATP. Upon MreB assembly, both GTP and ATP release the gamma phosphate at similar rates. Therefore, MreB is an equally effective ATPase and GTPase. Electron microscopy and quantitative rheology suggest that the morphologies and micromechanical properties of filamentous ATP-MreB and GTP-MreB are similar. In contrast, mammalian actin assembly is favored in the presence of ATP over GTP. These results indicate that, despite high structural homology of their monomers, T. maritima MreB and actin filaments display different assembly, morphology, micromechanics, and nucleotide-binding specificity. Furthermore, the biophysical properties of T. maritima MreB filaments, including high rigidity and propensity to form bundles, suggest a mechanism by which MreB helical structure may be involved in imposing a cylindrical architecture on rod-shaped bacterial cells.
Prokaryotic actin homologues MreB/ParM/Mbl are, along with tubulin homologue FtsZ and intermediate-filament homologue crescentin, the major components of what appears to be an extended filamentous cytoskeleton in bacteria (28). Recent studies have demonstrated the importance of these proteins in bacterial functions (1, 3, 5, 6, 23, 36). Fluorescence microscopy in vivo shows that MreB aggregates into a large filamentous spiral structure that lies underneath the cell membrane and spans the cell length (24). Several studies suggest an essential role for MreB in chromosome segregation (16, 25), polar localization of proteins (15, 36), maintenance of cell shape, and resistance to external mechanical stresses. When MreB is depleted, the bacterial cell wall displays gross morphological defects (47): vibrioid-shaped Caulobacter crescentus cells become lemon-shaped (12), and rod-shaped Bacillus subtilis (5) and Escherichia coli cells (47) become rounded. Peptidoglycan cell wall synthesis has been linked to the role of the MreB homolog, Mbl (5); however, the mechanism by which MreB may provide mechanical support directly to the cell or indirectly by affecting peptidoglycan wall integrity remains unclear.
In eukaryotic cells, cell stiffness is primarily provided by actin filaments, which organize into orthogonal arrays and ordered bundles that confer extraordinary elasticity to the cell (18). In physiological conditions, actin requires ATP or ADP to stabilize its folding and to polymerize (8). It has been reported that actin could polymerize in the presence of other nucleotides in vitro (31). Nevertheless, actin filament assembly and stability are highly favored in the presence of ATP and ADP (21, 31, 48). Yeast actin binds to and hydrolyzes GTP, but with much lower binding affinity and hydrolytic rate than ATP (21, 48), presumably because of a more open nucleotide cleft (2). Similarly, members of the actin superfamily, DnaK (27) and hexokinase (35), are known to bind and hydrolyze ATP more effectively than other nucleotides.
In contrast to actin, which forms double-stranded helical filaments, electron microscopy reveals that MreB forms nonhelical filaments in the presence of ATP (9, 46). Crystallography studies show that the MreB and actin share a similar three-dimensional crystal structure with a conserved nucleotide-binding domain. The residue differences in the nucleotide-binding domains between MreB and actin have been predicted to indicate dissimilar nucleotide-binding states (46). In the presence of GTP, MreB settles into the pellet during a high-speed centrifugation assay (46), which suggests that GTP mediates MreB aggregation. However, the assembly and nucleotide binding/hydrolysis properties and morphology of the structures formed by MreB in the presence of GTP are unclear. Moreover, since filamentous MreB plays a key structural role in the bacterial shape, whether MreB possesses sufficient mechanical properties to generate the forces needed to maintain the integrity of a rod-shaped cell remains a largely unanswered question. MreB has also been linked to chromosome segregation (6, 16, 25). Although the physical mechanism by which bacteria segregate their chromosome is still elusive, cells that are either depleted of MreB or express a mutant MreB protein, exhibit aberrant chromosome segregation (16, 25). There is no evidence that MreB binds directly to DNA; however, MreB could interact with chromosomes through auxiliary proteins. If MreB either segregates chromosomes in a manner similar to microtubules in eukaryotes or drives proteins to the poles of rod-shaped bacteria, then MreB filaments must possess, like microtubules, significant mechanical stiffness that will enable directed chromosomal pulling. The work presented here begins to address these issues by measuring the time-dependent micromechanics of MreB during and after assembly.
Using biochemical and biophysical methods, we study the assembly, morphology, and micromechanics of Thermotoga maritima MreB, as well as the kinetics of phosphate release during MreB assembly in the presence of different nucleotides. These studies suggest that, unlike for actin, ATP and GTP mediate the assembly of filamentous MreB equally effectively; the presence of either ADP or GDP completely precludes MreB assembly. T. maritima MreB catalyzes GTP hydrolysis and releases phosphate (Pi) at a rate similar to that for ATP. Quantitative rheometry reveals that both ATP and GTP mediate the extremely rapid formation of stiff gels. The micromechanical response of MreB filament arrays is assessed as a function of MreB concentration, as well as the frequency and amplitude of applied shear deformations. Our studies conclude that in the presence of both ATP and GTP, T. maritima MreB filaments assemble rapidly into bundled filament arrays, which possess sufficient toughness to contribute to bacterial cell mechanics.
MATERIALS AND METHODS
Purification of T. maritima MreB.
All reagents were purchased from Sigma (St. Louis, MO) unless otherwise stated. A pHis17 vector-derived construct containing the T. maritima MreB gene was kindly supplied by Fusinita Van den Ent (46). T. maritima MreB was purified according to the method of Esue et al. (9), as modified from Van den Ent et al. (46). The purified protein was dialyzed into polymerization buffer (100 mM Tris-HCl [pH 7.0], 4 mM MgCl2, 100 mM NaCl) and centrifuged (150,000 × g) for 1 h before use. MreB was assembled by adding 2 mM nucleotide and incubating the mixture at 65°C.
Actin purification and assembly.
Chicken skeletal muscle actin was prepared according to the method of Pardee et al. (39) with an added gel filtration step by using Sephacryl S-300 (50). Purified actin was continuously dialyzed at 4°C against buffer G (2 mM Tris-HCl [pH 8.0], 0.2 mM ATP, 0.5 mM dithiothreitol, 1 mM sodium azide, and 0.2 mM CaCl2). Mg2+-actin filaments were generated by adding 0.1 volume of 10× KMEI polymerizing salt (500 mM KCl, 10 mM MgCl2, 10 mM EGTA, 100 mM imidazole [pH 7.0]) to 0.9 volume of G-actin in buffer G.
EM.
MreB was incubated in polymerization buffer (100 mM Tris-HCl [pH 7.0], 4 mM MgCl2, 100 mM NaCl) with 2 mM ATP or GTP at 65°C. For each specimen, 10 μl of the mixture was placed on an electron microscopy (EM) grid coated with a collodion film. The grids were washed with 3 drops of polymerization buffer, stained with 2% uranyl acetate solution. The excess stain on the grid was drained off by positioning the grid at a 45° angle on a filter paper. When the grid was dried, EM was performed at the Integrated Imaging Center in Johns Hopkins University with a Philips 410 transmission electron microscope at magnifications between 65,000 and 105,000.
Time-resolved light scattering.
MreB assembly was monitored by multiple-angle light scattering (MALS; Wyatt Instruments, Santa Barbara, CA). We report the time-resolved light intensity measured at an angle of 90° from the axis of incident light. All experiments were conducted at 65°C, unless otherwise stated. The scattered light intensity was measured simultaneously by 18 photodiodes arranged circularly around the scattering volume. We also report the light intensity, I(q), as a function of the scattering wavevector amplitude, q = (4πn/λ0)sin(θ/2), where n is the refractive index, λ0 is the wavelength, and θ is the angles between the photodiodes and the incident light. The intensity of the q-dependent scattering-light profiles were fitted to a power law I(q) ∼ q−a. The exponent a reflects the structure of the “scatterers” in solution.
Phosphate release assay.
The production of inorganic phosphate by MreB solutions was measured by using the malachite green-sodium molybdate assay (9, 14, 41). Assembly of MreB was initiated by the addition of ATP or GTP at 65°C. The reaction was quenched at 1-minute time intervals with one volume of cold 0.6 M perchloric acid and stored on ice until all time points were collected. Two volumes of filtered malachite green solution (0.15 g of malachite green, 1 g of sodium molybdate, and 0.25 g of Triton X-100 in 500 ml of 0.7 M HCl) were added to the samples and incubated at room temperature for 30 min, and absorbance measured at A650 by using a precision microplate reader (Molecular Devices, Sunnyvale, CA). Curves were normalized with a phosphate standard and controls without protein.
Quantitative rheology.
The mechanical properties of MreB were measured by using a strain-controlled rheometer (TA Instruments, New Castle, DE) as described previously (10). Rheological concepts and definitions for the nonspecialist are described in reference 4). Assembly of MreB into filamentous structures was initiated by addition of 2 mM nucleotide; the solution was immediately placed between the temperature-controlled cone and plate of the rheometer. The plate is attached to a motor which applies an oscillatory shear deformation of controlled frequency and amplitude, while the cone which is attached to a torque transducer measures the stress induced by the shear deformation within the MreB networks. Applying an oscillatory shear deformation to the MreB specimen results in an induced stress with in-phase and out-of-phase components. The instrument divides the in-phase and out-of-phase stress component by the amplitude of oscillatory deformation to calculate the elastic (storage) modulus, G′, and the viscous (loss) modulus, G", respectively. The kinetics and extent of gelation of MreB solutions were monitored by measuring the elastic and viscous moduli, G′ and G", at 1 rad/s and 1-% strain every 30 s for 1 h. At steady state, the frequency-dependent elastic and viscous moduli, G′(ω) and G"(ω), were measured by applying 1-% amplitude oscillatory deformations of frequencies between 0.01 and 100 rad/s. The sample was then subjected to shear deformations of amplitude between 0.1 and 1,000% at a frequency of 1 rad/s. We also report the phase angle, δ, of MreB networks tested under various conditions.
RESULTS
GTP mediates MreB filament assembly.
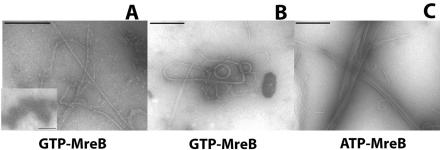
We used EM and negative staining to assess the morphology of the T. maritima MreB aggregates formed in the presence of different nucleotides, including ATP, ADP, GTP, GDP, cAMP, CTP, and TTP. We found that only ATP and GTP mediated the formation of filamentous structures (Fig. 1). ADP, GDP, cAMP, CTP, and TTP did not induce the formation of MreB filaments: MreB formed disordered structures similar to those formed in the absence of nucleotides (Fig. 1A, inset). GTP-MreB formed straight and curved filament bundles, whose morphologies were similar to those formed by ATP-MreB (Fig. 1). EM did not reveal significant morphological differences between ATP- and GTP-MreB filaments.
FIG. 1.
Electron micrographs (EM) of MreB structures. Electron micrographs of (A and B) GTP-MreB and (C) ATP-MreB filament structures are shown. The inset shows that MreB without nucleotides does not form filaments. The MreB concentration in panels A to C is 0.5 mg/ml. Scale bars, 0.2 μm.
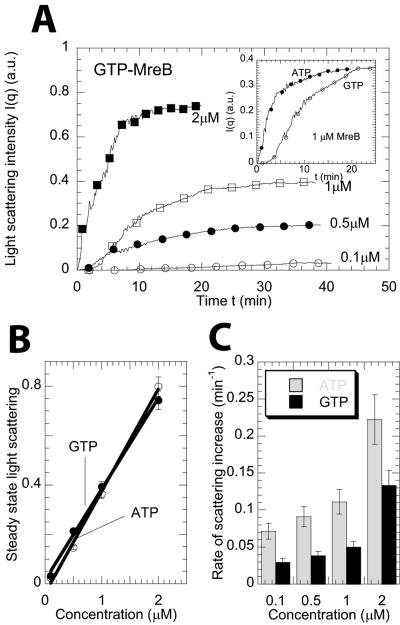
We used MALS to complement EM to probe the kinetics of assembly and morphological differences between GTP-MreB and ATP-MreB in solution. Upon addition of GTP, the scattered light intensity from MreB solutions increased with time (Fig. 2A). The steady-state scattered light intensity as a function of protein concentration and extrapolated to zero intensity determined an apparent critical concentration for GTP-MreB assembly (Fig. 2B). The extrapolated critical concentrations for the assembly of both ATP-MreB and GTP-MreB were low (<0.01 μM), much lower than for actin assembly (∼0.25 μM). The assembly kinetics of ATP-MreB and GTP-MreB were significantly different (Fig. 2A, inset). The rate of assembly of ATP-MreB was higher than that of GTP-MreB (Fig. 2C). Moreover, assembly rates, measured by the time it takes to reach 90% of its steady-state scattering intensity, increased readily with MreB concentration for both nucleotides. GTP-MreB assembly displayed, however, a short time lag that was independent of MreB concentration (Fig. 2A, inset).
FIG. 2.
Assembly of ATP-MreB and GTP-MreB. (A) Time-dependent light-scattering intensity for GTP-MreB assembly. Light scattering was measured at an angle of 90° from the direction of the incident light. Symbols correspond to 0.1 μM (○), 0.5 μM (•), 1 μM (□), and 2 μM (▪) GTP-MreB, respectively. The inset shows the intensity versus time for MreB in the presence of ATP (•) and GTP (○). (B) Steady-state light-scattering intensity as a function of total concentration of protein in solution for ATP-MreB and GTP-MreB. (C) Rates of increase of light scattering of MreB in the presence of ATP and GTP. The rate was calculated as the inverse of time it takes to reach 90% of its steady-state value.
MreB hydrolyzes GTP and ATP equally effectively.
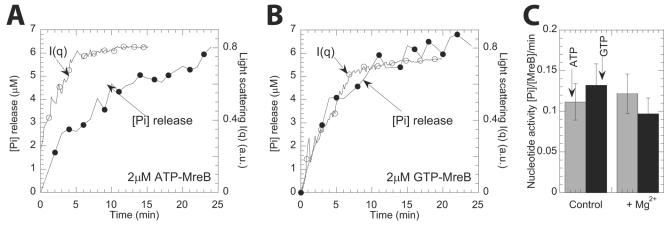
Phosphate release from polymerizing T. maritima MreB was monitored in the presence of either ATP or GTP (Fig. 3). Using a malachite green phosphate release assay (14), 100 μM nucleotide was used to induce MreB assembly; thereafter, the production of inorganic phosphate was measured at 1-min intervals for a wide range of MreB concentrations. This assay suggested that, during assembly, MreB hydrolyzed GTP and ATP equally effectively (Fig. 3A and B). However, the phosphate release profile of GTP-MreB showed no delay with assembly. This indicates that GTP was hydrolyzed promptly as MreB monomers were incorporated into the filaments. In contrast, a delay occurred between ATP-MreB assembly and phosphate release (9). The rate of phosphate release per mole of GTP-MreB and ATP-MreB was independent of MreB concentration (Fig. 3C). In polymerization buffer, the rate of nucleotide activity for GTP and ATP was 0.09 ± 0.02 and 0.12 ± 0.02 mol Pi/min/mol MreB, respectively (average ± the standard deviation; n = 4) (Fig. 3C).
FIG. 3.
Nucleotide hydrolysis monitored by phosphate release during MreB assembly. (A and B) Light-scattering intensity and phosphate release versus time for a 2 μM ATP-MreB solution (A) and a 2 μM GTP-MreB solution (B). (C) The specific nucleotide activity ([phosphate released]/min/[MreB]) is plotted as a function of Mg2+ ions in polymerization buffer. The rates of nucleotide activity for MreB in GTP and ATP were 0.09 ± 0.02 and 0.12 ± 0.02 mol Pi/min/mole MreB, respectively (average ± the standard deviation, n = 4).
Mg2+ ions may slow down the rate of MreB assembly (9). To exclude a potential role of Mg2+ in nucleotide hydrolysis, a control experiment was conducted in which MreB that was gel filtered with an EDTA-containing buffer (see Materials and Methods) and dialyzed into buffer without Mg2+ ions. This showed that Mg2+ ions did not affect the rate of nucleotide hydrolysis (Fig. 3C).
GTP-MreB filament rigidity in solution.
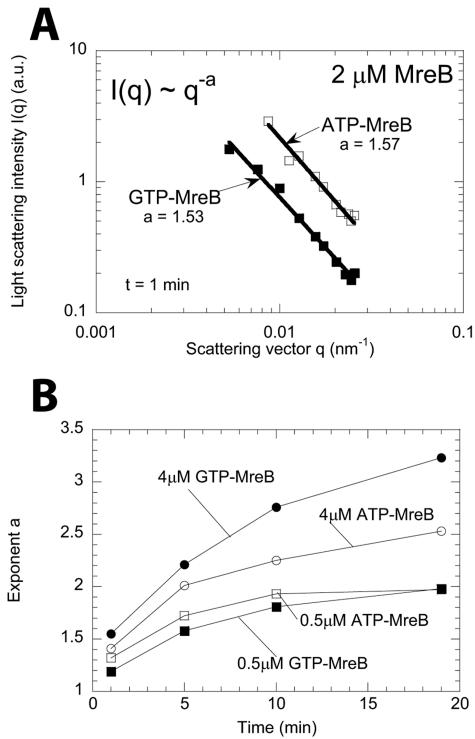
In EM experiments, MreB was placed on colloidion-coated grids, which may change the apparent morphology of the filaments (34). Therefore, time-dependent scattered light intensity profiles were collected to obtain real-time information about filament morphology in solution. We monitored the scattered light intensities, I(t; θ), simultaneously at 18 different angles. The rigidity of MreB filamentous structures in solution is defined by the time-dependent exponent, a(t), of the power-law fit of the light scattering profiles, I(t; q) ∼ q−a(t). The vector amplitude, q, is defined as q = (4πn/λ0)sin(θ/2) (see Materials and Methods). The predicted exponent a is 1 for straight rod polymers, 2 for highly flexible polymers, and 3 or 4 for sheets and smooth surfaces of three-dimensional objects (19). Actin filaments exhibit an exponent of ∼1.1 at similar concentrations (45). The exponent, a, for solutions containing MreB was ∼1.5 (Fig. 4A) one minute after the onset of assembly, which suggests that MreB had already formed semiflexible filaments. Then MreB filaments grew longer than their persistent length to become flexible filaments (a = 2) and later high-order structures (a = 3; sheets) (19), suggestive of two-dimensional sheets of filament bundles (Fig. 4B).
FIG. 4.
MreB filament structures in solution via multiple-angle light scattering. (A) Time-dependent exponent a was obtained from power-law fits of scattering spectra I(t; q) ∼ q−a(t). The exponent a for 2 μM MreB is shown 1 min after initiating assembly. (B) Time-dependent exponent a(t) for solutions of ATP-MreB and GTP-MreB. Symbols correspond to 0.5 μM GTP-MreB (▪), 0.5 μM ATP-MreB (□), 4 μM GTP-MreB (•), and 4 μM ATP-MreB (○).
Both ATP and GTP mediate the rapid formation of stiff MreB gels.
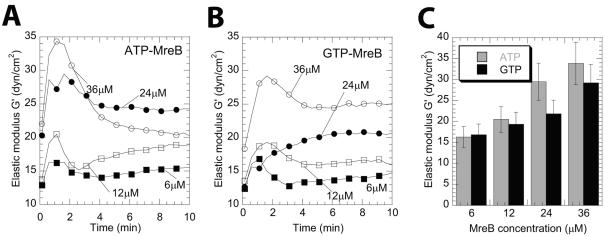
Quantitative rheology has been extensively used to probe the mechanical properties of filamentous proteins of the eukaryotic cytoskeleton (4, 22, 42, 49). However, the intrinsic mechanical properties of prokaryotic cytoskeleton proteins, and that of MreB in particular, are largely unknown. Here we used rheometry to determine whether filamentous MreB shared mechanical properties with those of its eukaryotic counterpart actin. See (4) for rheological concepts and definitions. Specimens were probed by using a cone-and-plate rheometer, which monitored the elastic modulus, G′ (which measures the propensity of the polymers to rebound after shear deformation), and the viscous modulus, G" (which measures how much the specimen flows), and phase angle, δ (which compares the elastic or viscous nature of a material) of filamentous MreB (Fig. 5). This phase angle, δ = tan−1(G"/G′), is directly obtained from values of G′ and G". The phase angle for an elastic solid like rubber is 0° (G′≫G"), which indicates that there is no delay between input deformation (strain) and output stress. For a viscous liquid such as water or glycerol, the phase angle is 90° (G"≫G′), which indicates that there is a maximum delay between input deformation (strain) and output stress. For a viscoelastic specimen, 0° < δ < 90°. The mechanical properties of MreB were assessed both during filament assembly and at steady state and compared to those of F-actin (Table 1). Suspensions of nonassembled MreB in storage buffer showed little viscosity and no elasticity (not shown). Upon assembly, the elasticity of MreB increased rapidly with time and reached a plateau value of 22 ± 3 dyn/cm2, for 24 μM GTP-MreB and 24 ± 4 dyn/cm2 for ATP-MreB (Fig. 5). The rate of gelation of MreB, measured as the inverse of the time required to reach 90% of the steady-state G′ value (see gelation profiles in Fig. 5A and B), was ∼10-fold higher that that of F-actin (Table 1) (43).
FIG. 5.
Gelation kinetics and viscoelastic properties of MreB filament networks. (A and B) Time-dependent increase in elasticity upon filament assembly of ATP-MreB (A) and GTP-MreB (B). The elastic modulus is measured every 30 s. Symbols in panels A and B correspond to 6 μM (▪), 12 μM (□), 24 μM (•), and 36 μM (○) MreB. The elastic modulus goes through a characteristic maximum at ∼2 min after initiating filament assembly. (C) The maximum elastic modulus is plotted as a function of MreB concentration in the presence of ATP (gray) and GTP (black).
TABLE 1.
Mechanical properties of MreB and F-actina
| Cytoskeleton protein | Mean ± SD
|
Exponent G′(c) ∼ c | pH | Source or reference | ||
|---|---|---|---|---|---|---|
| Elasticity (dyn/cm2) | Phase angle (°) | Resilience (%) | ||||
| F-actin | 10 ± 3 | 30 ± 5 | 10 ± 2 | 1.2 | 8.0 | 45 |
| GTP-MreB | 22 ± 3 | 17 ± 4 | 3 ± 1 | 0.29 | 7.0 | This work |
| ATP-MreB | 24 ± 4 | 15 ± 4 | 3 ± 1 | 0.44 | 7.0 | This work |
Elasticity, G′, and phase angle, δ, were measured by using a cone-and-plate strain-controlled rheometer, which applied oscillatory shear deformations of small amplitude (1%) and a frequency of 1 rad/s. Rheological parameters G′ and δ were measured at steady state, i.e., after these parameters had reached a steady state after onset of assembly. The phase angle measures the delay in the response of the stress induced in the filament networks by the rheometer. An elastic solid will show no delay (phase angle of 0°); a viscous liquid without elasticity like glycerol will show a maximum delay (phase angle of 90°). The mechanical resilience of cytoskeleton proteins is defined as the shear amplitude at which the elastic modulus started to decrease. The protein concentration for measurements of G′, δ, and mechanical resilience was 24 μM. For the range of concentrations for the concentration-dependent elasticity, G′(c), see the text and reference.
The gelation of both ATP-MreB and GTP-MreB went through a distinct maximum that occurred ∼2 min after onset of assembly (Fig. 5A and B). The elastic modulus of MreB gradually decreased after this maximum to a final steady-state value. A similar maximum occurs during the gelation of F-actin in the presence of cross-linking/bundling proteins but not F-actin alone (43). This phenomenon is presumably due to the reorganization of the filaments inside the forming networks before a steady state is reached (44).
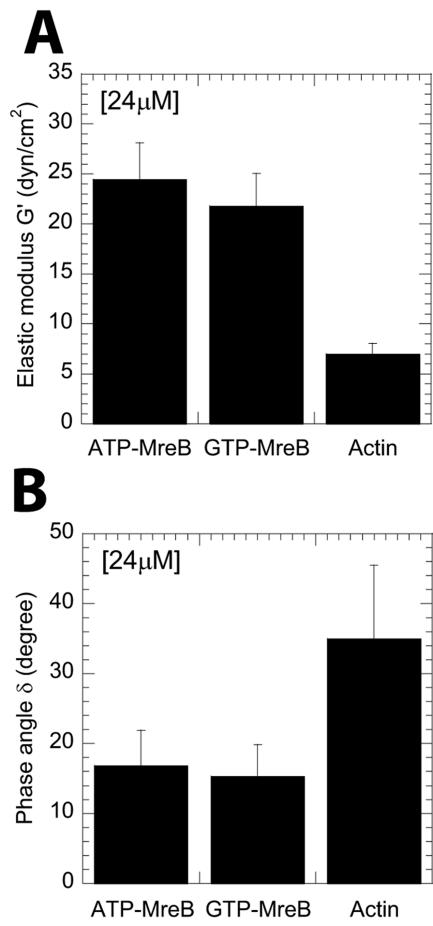
In polymerization buffer, MreB filaments formed rapidly solid-like structures (i.e., more elastic than viscous), as assessed by a rapidly decreasing phase angle, which reached a plateau value of 12° ± 3° within 10 min (Fig. 6A and B). In contrast, actin filaments featured a more liquid-like character, as assessed by a significantly higher phase angle (Fig. 6B). The steady-state elasticity of a 24 μM GTP-MreB (22 ± 3 dyn/cm2) and ATP-MreB (24 ± 4 dyn/cm2) was approximately 4- to 6-fold higher than that of the same concentration of F-actin, (7 ± 2 dyn/cm2) (Fig. 6A) (Table 1). Using a power law fit, G′ ∼ Cb, we observed that the elasticity of MreB depended weakly on its concentration ranging from 6 to 36 μM. The exponents, b, are 0.44 and 0.29 for ATP-MreB and GTP-MreB, respectively. F-actin has a much stronger concentration dependence, for which b = 1.2 (Table 1) (38).
FIG. 6.
Steady-state mechanical properties of MreB filament networks. (A and B) Comparison of phase angle (A) and elastic modulus (B) of filamentous MreB and F-actin. The protein concentration is 24 μM. A phase angle of 90° designates a perfectly viscous liquid such as glycerol, which displays no elasticity. A phase angle of 0° designates a perfectly elastic solid, which displays no or little viscosity.
Mechanical response of MreB filaments to shear stresses.
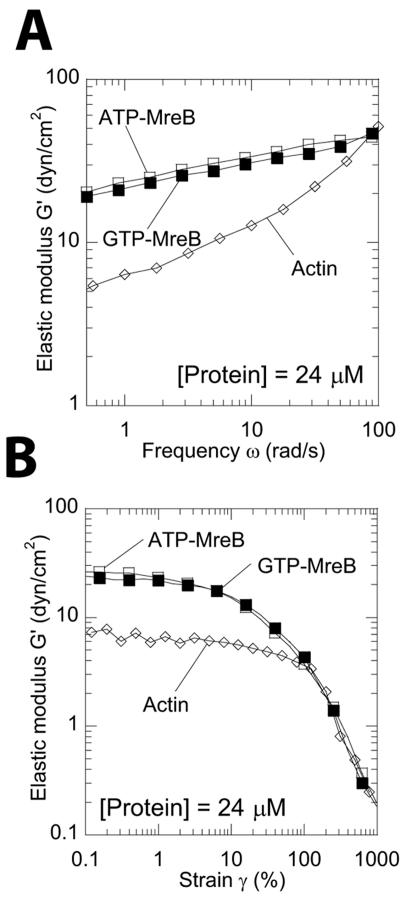
The propensity of MreB filaments to move in networks was assessed by subjecting MreB networks to a constant shear deformation of increasing frequency (see Materials and Methods). The elastic modulus of a covalently cross-linked or highly entangled network is independent of an increasing frequency, while that of an uncrosslinked (but untangled) filaments will depend more strongly on frequency (11). The elastic modulus of MreB networks, G′(ω), increased only weakly with frequency, from ∼20 to 45 dyn/cm2 at 24 μM for shear frequency between 0.5 and 100 rad/s (Fig. 7A). Over the same frequency range, the elastic modulus of a 24 μM F-actin network increased more than 10-fold, from ∼5 to 55 dyn/cm2 (Fig. 7A). This result suggests that MreB filaments are much less labile than actin filaments in networks, presumably due to nonsteric interfilament interactions.
FIG. 7.
Mechanical response of MreB filament networks to shear deformations. (A) Frequency-dependent elastic modulus of MreB and actin filament networks. MreB filaments are subject to shear deformations of fixed 1% amplitude and frequency between 0.5 and 100 rad/s. (B) Strain-dependent elastic modulus of MreB and actin filament networks. Filaments are subject to shear deformations at a fixed frequency (1 rad/s) and amplitudes between 0.1% and 1,000%. The protein concentration is 24 μM. Symbols correspond to GTP-MreB (▪), ATP-MreB (□), and F-actin (⋄).
Depending on the type of interfilament interactions, networks may stiffen or soften under mechanical stress, i.e., their elasticity may increase or decrease under increasing shear (4). We subjected MreB filaments to shear deformations of increasing amplitude (Fig. 7B) to assess the mechanical response. The modulus of MreB was independent of the applied deformation amplitude at low shear deformations, which defines the linear rheological regime for which applied deformation and induced stress are proportional, and declined steeply past a threshold value (Fig. 7B). MreB filaments showed no hint of strain-induced hardening whereby the modulus would first increase with deformation before softening under large deformations. MreB filaments softened at a strain amplitude of 2 to 3% (arrow in Fig. 7B).
DISCUSSION
GTP-mediated MreB filament assembly and GTPase activity of MreB.
Our results suggest that MreB assembles into filamentous structures in vitro in the presence of either ATP or GTP, but not other nucleotides, including ADP and GDP. EM indicates that the morphology of the filament structures formed by GTP-MreB is similar to that formed by ATP-MreB, including straight and curved filament bundles. Both ATP-MreB and GTP-MreB have an extremely low critical concentration for filamentous assembly. The equilibrium ATPase and GTPase activities ([Pi]/[MreB]/min) of MreB are also similar. However, the rate of ATP-MreB filament assembly is higher than that of GTP-MreB and Pi release from ATP-MreB is delayed with its filamentous assembly, while Pi release from GTP-MreB is almost simultaneous with its assembly. The presence of both straight and curved filaments may suggest that the state of hydrolysis of the nucleotide within the filament, whereby straight filaments begin to curve as ATP or GTP is hydrolyzed to ADP or GDP, respectively. This phenomenon has also been observed in filamentous proteins such as microtubules (37) and FtsZ (29, 30). The fact that in vitro MreB is not dynamically unstable (9) further suggests that ADP and GDP are able to maintain MreB in its filamentous form.
The cytoskeleton of differentiated eukaryotic cells is composed of three major classes of filamentous proteins: actin, which utilizes ATP to polymerize into filament to perform force-generation and structural functions; tubulin, which uses GTP to drive its assembly into microtubules to guide cell polarity, mitosis, and vesicle trafficking; and intermediate filaments (IFs), which do not require nucleotides for assembly to perform their mechanical function. Cytoskeleton proteins in bacteria do not seem to be as ubiquitous as their eukaryotic counterparts. Thus far, prokaryotic IFs have only been identified in C. crescentus, while MreB has been identified mostly in rod-shaped cells. Moreover, bacterial cytoskeleton proteins often have functions that are fundamentally different from those of their eukaryotic counterparts. Live-cell studies suggest that tubulin homolog FtsZ is involved in the Z-ring contractility (3), an activity analogous to the actomyosin contractile ring during eukaryotic cell division. Similarly, ParM, another prokaryotic homologue of actin, is involved in the segregation of bacterial plasmid DNA, an activity analogous to that of microtubules during mitosis (13). Cells expressing mutant forms of MreB or cells that are depleted of MreB exhibit aberrant chromosome segregation (16, 25). Thus, evolutionary ancestor proteins of eukaryotic cytoskeletal proteins do not necessarily possess the same function as those that would be predicted from structure or functions defined in eukaryotic cells. Here, we find that T. maritima MreB filaments have biochemical and biophysical properties much more similar to those of IFs than those of F-actin. High elasticity, low phase angle, weak concentration dependence of the elastic modulus, low critical concentration, and high propensity for bundling are properties also displayed by IFs and not F-actin (4).
It is unknown whether bacteria cells utilize ATP, GTP, or both to assemble MreB in vivo. Here we found that ATP-MreB and GTP-MreB display similar morphological and rheological properties. However, their different assembly kinetics and mode of phosphate release kinetics suggest the possibility that the cell may exploit both nucleotides for different MreB-based functions. Previous studies show that MreB is involved in chromosome segregation, polar movement of proteins, and the formation of the spiral that spans the length of rod-shaped bacterial cells. Like ParM's involvement in segregating plasmids (33), MreB may also possess the ability to pull chromosomes apart (16). Studies show that in B. subtilis, helical MreB structures exhibit a rapid poleward movement along its tracks (7), suggesting a possible mechanism for polar localization of certain proteins and also for chromosome segregation. Although there is no direct evidence of MreB interaction with DNA, it could perform this function with the aid of binding proteins that are not yet identified. The requirements for MreB to utilize ATP and GTP as forms of energy may be linked to MreB's diverse functions.
Micromechanics of ATP-MreB and GTP-MreB filaments.
MreB plays an essential role in the control of bacterial cell shape; however, it remains unknown whether MreB regulates bacterial morphology through a cascade of signal events, through its intrinsic mechanical properties, or both. Rapid perturbation of MreB structures using A22 results in rod-shaped bacteria gradually becoming rounded (16, 20, 36). The gradual change in cell shape may be due to the dose-dependent effect of A22 or possibly that the disassembly of MreB structures has a slow effect on the already stiff bacterial cell wall. Similarly, upon depletion of MreB from rod-shaped cells, the gradual morphological change observed can be attributed to a slow reduction in MreB protein levels over several cell division cycles. Rheometric results show that, at physiological concentrations, MreB filaments are able to form highly elastic gels within physiological time scales (e.g., the E. coli cell cycle lasts ∼30 min), much more rapidly than F-actin (∼2 to 3 min for MreB versus ∼30 min for F-actin). Thus, MreB has the necessary physical properties to contribute to bacterial shape (see more below).
The high elasticity of filamentous MreB and its fast gelation kinetics are partly due to the strong tendency of MreB filaments to form bundles in vitro (9, 46). Electron tomography suggests that similar bundles are formed in bacteria (26). MreB has an elastic modulus more than threefold higher than that of F-actin at the same concentration (24 μM or ∼1 mg/ml). Since the elasticity of a polymer network is predicted to depend weakly on the intrinsic rigidity of the filaments (i.e., their persistence length) (38), this high elastic modulus can be mostly accounted for by strong interfilaments interactions between MreB filaments. The elasticity and concentration of MreB are related by a power law: G′ ∼ C0.44 and G′ ∼ C0.29 for ATP-MreB and GTP-MreB, respectively, which is consistent to that predicted for cross-linked filaments. For comparison, uncrosslinked and entangled semiflexible polymers, such as F-actin, have a predicted power-law relation between network elasticity and protein concentration of G′ ∼ C1.4 (38); uncrosslinked and entangled flexible polymers, such as DNA, have a predicted power-law relation of G′ ∼ C2.25 (32). The lower concentration dependence index of GTP-MreB indicates that GTP-MreB filaments have a greater tendency to form cross-links than ATP-MreB filaments. This is also consistent with the exponent, a, describing light-scattering profiles, which suggests that although GTP mediates slower assembly, GTP-MreB structures also feature a higher structural rigidity than ATP-MreB structures in solution.
MreB filament lateral interactions are also supported by our MALS studies. The vector amplitude of light-scattering profiles from MreB filament solutions are similar to those displayed by F-actin in the presence of the filament-bundling protein fascin (45). Interfilament interactions are further supported by the low phase angle and the frequency-independent elasticity of MreB network. The phase angle of MreB network (∼15°) is much lower than that of F-actin (∼30°) but comparable to F-actin in the presence of cross-linking proteins, filamin, or α-actinin (43). A phase angle of 90° implies the rheological behavior of a liquid such as glycerol, whereas a phase angle close to 0° implies an elastic solid, such as a stiff rubber. The elasticity of MreB network is largely independent of the rate of shear. Polymer physics (11) predicts that sufficiently slow deformations applied to a network of uncrosslinked polymers leave time for polymers to rearrange and relax the stress (11). Rapidly applied deformations leave little time for these polymers to relax: they will elastically resist shear deformations. Consequently, the elasticity of uncrosslinked filaments displays a frequency-dependent spectrum, which is the case for F-actin. In contrast, the elasticity of cross-linked polymers (i.e., most gels) is independent of frequency (11). We speculate that our in vitro rheological and light-scattering assays could be used as a high-throughput functional assay to screen MreB mutants that could interfere with the mechanical function and shape-defining functions of MreB in vivo through modulation of the cross-linking/bundling activity of MreB and/or its assembly.
Potential impact of MreB filament mechanics on cell shape.
In eukaryotic cells, shape is primarily established and maintained by the actin cytoskeleton. Despite its cytoplasmic abundance (50 to 200 μM) (17, 40), a large pool of monomeric actin is unavailable for assembly due to the regulation of actin-sequestering proteins. Eukaryotic cells cope with this by regulating sequestering proteins and exploiting cross-linking/bundling proteins, such as α-actinin and filamin, to transform its actin cytoskeleton into stiff, yet dynamic arrays (43). When triggered by signaling events, an equilibrium amount of G-actin is released from the actin-sequestering proteins and undergoes polymerization followed by gelation, which generates the essential stiffness to mechanically support cell shape changes. In contrast to eukaryotic cells, it has long been believed that bacterial cells establish their shape mostly by the peptidoglycan-rich cell wall (24). However, recent work shows that filamentous MreB influences the shape of vibrioid-shaped C. crescentus and rod-shaped E. coli and B. subtilis (12, 24, 47). Peptidoglycan assembly in B. subtilis (5) has been connected to the MreB homolog, Mbl, while the localization of the PBP2-peptidoglycan biosynthesis complex in C. crescentus (1) has been connected to MreB function.
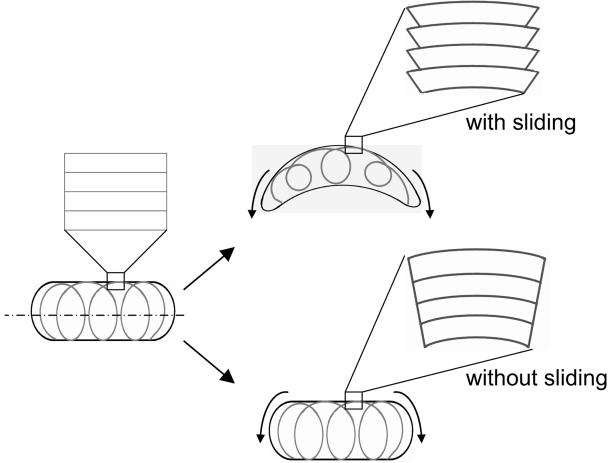
Whether MreB assembly affects bacterial shape directly, like its eukaryotic counterpart actin, or indirectly by affecting peptidoglycan-rich cell wall formation and integrity remains unclear. We do not test this hypothesis directly; however, our results show that MreB filaments possess the necessary biophysical properties (filament rigidity and tendency to bundle) to play an important role in bacterial cell mechanics. Cryo-electron tomography suggests that the helical structure in vivo is composed of bundled MreB filaments (26). A helical structure formed of filaments that do not interact strongly with each other (and therefore could slide) would offer little resistance to bending forces (Fig. 8). This is akin to a spring, which cannot be compressed laterally but bends readily. In this case, the MreB helical structure observed in vivo would not be able to play a major role in setting the shape of rod-shaped cells. Our light-scattering, EM, and rheological results indicate that MreB filaments have a high propensity to interact strongly with each other. These properties will endow the MreB helical structure with a high bending rigidity by preventing sliding between filaments within the bundle (Fig. 8). The MreB helical structure should be able to resist large bending and compression forces. Therefore, our results are consistent with the view that the MreB helical structure may play a direct role in segregating chromosomes, as well as in imposing a rigid cylindrical architecture to rod-shaped bacterial cells.
FIG. 8.
How high filament-filament interactions may provide MreB helical structures in vivo with bending rigidity. MreB filaments form a helical structure that spans the length of a rod-shaped bacterial cells. Our results in vitro suggest that filaments within the bundle do not slide under shear (bottom panel), which endows the MreB helical structure with a high resistance to bending. If sliding between filaments were allowed (top panel), the MreB helical structure would bend readily and it could not play a direct role in imposing a rigid cylindrical architecture to rod-shaped bacterial cells.
Acknowledgments
We thank Fusinita van den Ent for kindly providing the full-length cDNA of T. maritima MreB.
This study was funded by National Aeronautics and Space Administration grant NAG9-1563 and National Institutes of Health grant GM065835.
REFERENCES
- 1.Ausmees, N., J. R. Kuhn, and C. Jacobs-Wagner. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705-713. [DOI] [PubMed] [Google Scholar]
- 2.Belmont, L. D., A. Orlova, D. G. Drubin, and E. H. Egelman. 1999. A change in actin conformation associated with filament instability after Pi release. Proc. Natl. Acad. Sci. USA 96:29-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi, E. F., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 4.Coulombe, P. A., O. Bousquet, L. Ma, S. Yamada, and D. Wirtz. 2000. The “ins” and “outs” of intermediate filament organization. Trends Cell Biol. 10:420-428. [DOI] [PubMed] [Google Scholar]
- 5.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767-776. [DOI] [PubMed] [Google Scholar]
- 6.Defeu Soufo, H. J., and P. L. Graumann. 2005. Bacillus subtilis actin-like protein MreB influences the positioning of the replication machinery and requires membrane proteins MreC/D and other actin-like proteins for proper localization. BMC Cell Biol. 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Defeu Soufo, H. J., and P. L. Graumann. 2004. Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep. 5:789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De La Cruz, E. M., and T. D. Pollard. 1995. Nucleotide-free actin: stabilization by sucrose and nucleotide binding kinetics. Biochemistry 34:5452-5461. [DOI] [PubMed] [Google Scholar]
- 9.Esue, O., M. Cordero, D. Wirtz, and Y. Tseng. 2005. The assembly of MreB, a prokaryotic homolog of actin. J. Biol. Chem. 280:2628-2635. [DOI] [PubMed] [Google Scholar]
- 10.Esue, O., Y. Tseng, and D. Wirtz. 2005. The rapid onset of elasticity during the assembly of the bacterial cell-division protein FtsZ. Biochem. Biophys. Res. Commun. 333:508-516. [DOI] [PubMed] [Google Scholar]
- 11.Ferry, J. D. 1980. Viscoelastic properties of polymers. John Wiley & Sons, Inc., New York, N.Y.
- 12.Figge, R. M., A. V. Divakaruni, and J. W. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321-1332. [DOI] [PubMed] [Google Scholar]
- 13.Garner, E. C., C. S. Campbell, and R. D. Mullins. 2004. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306:1021-1025. [DOI] [PubMed] [Google Scholar]
- 14.Geladopoulos, T. P., T. G. Sotiroudis, and A. E. Evangelopoulos. 1991. A malachite green colorimetric assay for protein phosphatase activity. Anal. Biochem. 192:112-116. [DOI] [PubMed] [Google Scholar]
- 15.Gitai, Z., N. Dye, and L. Shapiro. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. USA 101:8643-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329-341. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, D. J., E. Eisenberg, and E. D. Korn. 1976. Characterization of cytoplasmic actin isolated from Acanthamoeba castellanii by a new method. J. Biol. Chem. 251:4778-4786. [PubMed] [Google Scholar]
- 18.Heidemann, S. R., and D. Wirtz. 2004. Towards a regional approach to cell mechanics. Trends Cell Biol. 14:160-166. [DOI] [PubMed] [Google Scholar]
- 19.Higgins, J. S., and H. C. Benoit. 1994. Polymers and Neutron Scattering. Oxford University Press, Oxford, United Kingdom.
- 20.Iwai, N., K. Nagai, and M. Wachi. 2002. Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci. Biotechnol. Biochem. 66:2658-2662. [DOI] [PubMed] [Google Scholar]
- 21.Iyengar, M. R., and H. H. Weber. 1964. The relative affinities of nucleotides to G-actin and their effects. Biochim. Biophys. Acta 86:543-553. [DOI] [PubMed] [Google Scholar]
- 22.Janmey, P. A., U. Euteneuer, P. Traub, and M. Schliwa. 1991. Viscoelastic properties of vimentin compared with other filamentous biopolymer networks. J. Cell Biol. 113:155-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen, R. B., and K. Gerdes. 1999. Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 colocalizes with its replicon during the cell cycle. EMBO J. 18:4076-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 25.Kruse, T., J. Moller-Jensen, A. Lobner-Olesen, and K. Gerdes. 2003. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 22:5283-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurner, J., A. S. Frangakis, and W. Baumeister. 2005. Cryo-electron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Science 307:436-438. [DOI] [PubMed] [Google Scholar]
- 27.Liberek, K., D. Skowyra, M. Zylicz, C. Johnson, and C. Georgopoulos. 1991. The Escherichia coli DnaK chaperone, the 70-kDa heat shock protein eukaryotic equivalent, changes conformation upon ATP hydrolysis, thus triggering its dissociation from a bound target protein. J. Biol. Chem. 266:14491-14496. [PubMed] [Google Scholar]
- 28.Lowe, J., F. van den Ent, and L. A. Amos. 2004. Molecules of the bacterial cytoskeleton. Annu. Rev. Biophys. Biomol. Struct. 33:177-198. [DOI] [PubMed] [Google Scholar]
- 29.Lu, C., and H. P. Erickson. 1999. The straight and curved conformation of FtsZ protofilaments-evidence for rapid exchange of GTP into the curved protofilament. Cell Struct. Funct. 24:285-290. [DOI] [PubMed] [Google Scholar]
- 30.Lu, C., M. Reedy, and H. P. Erickson. 2000. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol. 182:164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martonosi, A., and M. A. Gouvea. 1961. Studies on actin. VI. The interaction of nucleoside triphosphates with actin. J. Biol. Chem. 236:1345-1352. [PubMed] [Google Scholar]
- 32.Mason, T. G., A. Dhople, and D. Wirtz. 1998. Linear viscoelastic moduli of concentrated DNA solutions. Macromolecules 31:3600-3603. [Google Scholar]
- 33.Moller-Jensen, J., J. Borch, M. Dam, R. B. Jensen, P. Roepstorff, and K. Gerdes. 2003. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol. Cell 12:1477-1487. [DOI] [PubMed] [Google Scholar]
- 34.Mucke, N., L. Kreplak, R. Kirmse, T. Wedig, H. Herrmann, U. Aebi, and J. Langowski. 2004. Assessing the flexibility of intermediate filaments by atomic force microscopy. J. Mol. Biol. 335:1241-1250. [DOI] [PubMed] [Google Scholar]
- 35.Nagayama, F., H. Ohshima, H. Suzuki, and T. Ohshima. 1980. A hexokinase from fish liver with wide specificity for nucleotides as phosphoryl donor. Biochim. Biophys. Acta 615:85-93. [DOI] [PubMed] [Google Scholar]
- 36.Nilsen, T., A. W. Yan, G. Gale, and M. B. Goldberg. 2005. Presence of multiple sites containing polar material in spherical Escherichia coli cells that lack MreB. J. Bacteriol. 187:6187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nogales, E., H. W. Wang, and H. Niederstrasser. 2003. Tubulin rings: which way do they curve? Curr. Opin. Struct. Biol. 13:256-261. [DOI] [PubMed] [Google Scholar]
- 38.Palmer, A., J. Xu, S. C. Kuo, and D. Wirtz. 1999. Diffusing wave spectroscopy microrheology of actin filament networks. Biophys. J. 76:1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pardee, J. D., and J. A. Spudich. 1982. Purification of muscle actin. Methods Enzymol. 85(Pt. B):164-181. [DOI] [PubMed] [Google Scholar]
- 40.Pollard, T. D., L. Blanchoin, and R. D. Mullins. 2000. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545-576. [DOI] [PubMed] [Google Scholar]
- 41.Romberg, L., M. Simon, and H. P. Erickson. 2001. Polymerization of Ftsz, a bacterial homolog of tubulin. is assembly cooperative? J. Biol. Chem. 276:11743-11753. [DOI] [PubMed] [Google Scholar]
- 42.Sato, M., W. H. Schwartz, S. C. Selden, and T. D. Pollard. 1988. Mechanical properties of brain tubulin and microtubules. J. Cell Biol. 106:1205-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tseng, Y., K. M. An, O. Esue, and D. Wirtz. 2004. The bimodal role of filamin in controlling the architecture and mechanics of F-actin networks. J. Biol. Chem. 279:1819-1826. [DOI] [PubMed] [Google Scholar]
- 44.Tseng, Y., K. M. An, and D. Wirtz. 2002. Microheterogeneity controls the rate of gelation of actin filament networks. J. Biol. Chem. 277:18143-18150. [DOI] [PubMed] [Google Scholar]
- 45.Tseng, Y., E. Fedorov, J. M. McCaffery, S. C. Almo, and D. Wirtz. 2001. Micromechanics and ultrastructure of actin filament networks crosslinked by human fascin: a comparison with alpha-actinin. J. Mol. Biol. 310:351-366. [DOI] [PubMed] [Google Scholar]
- 46.Van den Ent, F., L. A. Amos, and J. Lowe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39-44. [DOI] [PubMed] [Google Scholar]
- 47.Wachi, M., and M. Matsuhashi. 1989. Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J. Bacteriol. 171:3123-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen, K. K., X. Yao, and P. A. Rubenstein. 2002. GTP-yeast actin. J. Biol. Chem. 277:41101-41109. [DOI] [PubMed] [Google Scholar]
- 49.Xu, J., W. H. Schwarz, J. A. Kas, T. P. Stossel, P. A. Janmey, and T. D. Pollard. 1998. Mechanical properties of actin filament networks depend on preparation, polymerization conditions, and storage of actin monomers. Biophys. J. 74:2731-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu, J., D. Wirtz, and T. D. Pollard. 1998. Dynamic cross-linking by alpha-actinin determines the mechanical properties of actin filament networks. J. Biol. Chem. 273:9570-9576. [DOI] [PubMed] [Google Scholar]