Abstract
Yersinia pestis, the agent of plague, is usually transmitted by fleas. To produce a transmissible infection, Y. pestis colonizes the flea midgut and forms a biofilm in the proventricular valve, which blocks normal blood feeding. The enteropathogen Yersinia pseudotuberculosis, from which Y. pestis recently evolved, is not transmitted by fleas. However, both Y. pestis and Y. pseudotuberculosis form biofilms that adhere to the external mouthparts and block feeding of Caenorhabditis elegans nematodes, which has been proposed as a model of Y. pestis-flea interactions. We compared the ability of Y. pestis and Y. pseudotuberculosis to infect the rat flea Xenopsylla cheopis and to produce biofilms in the flea and in vitro. Five of 18 Y. pseudotuberculosis strains, encompassing seven serotypes, including all three serotype O3 strains tested, were unable to stably colonize the flea midgut. The other strains persisted in the flea midgut for 4 weeks but did not increase in numbers, and none of the 18 strains colonized the proventriculus or produced a biofilm in the flea. Y. pseudotuberculosis strains also varied greatly in their ability to produce biofilms in vitro, but there was no correlation between biofilm phenotype in vitro or on the surface of C. elegans and the ability to colonize or block fleas. Our results support a model in which a genetic change in the Y. pseudotuberculosis progenitor of Y. pestis extended its pre-existing ex vivo biofilm-forming ability to the flea gut environment, thus enabling proventricular blockage and efficient flea-borne transmission.
Flea-borne transmission of Yersinia pestis is a relatively recent adaptation that occurred within the past 20,000 years, the estimated time frame during which Y. pestis diverged from its Yersinia pseudotuberculosis ancestor (1). Y. pseudotuberculosis is an enteric pathogen transmitted primarily through contaminated food and water, whereas Y. pestis causes plague and is transmitted primarily by fleas. The two species are indistinguishable based on 16S rRNA sequences (40). More-recent comparative genomic studies show that there are relatively few genes unique to each species, and most of their orthologous genes are ≥97% identical at the nucleotide level (9, 20). This close phylogenetic relationship implies that it may be possible to trace the evolution of flea-borne transmission by determining the effect of Y. pestis- or Y. pseudotuberculosis-unique genes on phenotypes relevant to flea infection.
Flea-borne transmission depends on a characteristic development of Y. pestis infection in the vector with several key stages. When a flea takes a blood meal from a mammal with plague, the bacteria must survive and replicate within the inhospitable environment of the flea midgut and avoid being eliminated in the feces. Initial survival in the midgut depends on the Yersinia murine toxin (Ymt), a phospholipase D encoded on the Y. pestis-specific pFra plasmid, which protects the bacteria from lysis in the flea midgut by an as-yet-unidentified mechanism (23). Within 1 week, the bacteria begin to form dense aggregates in the midgut that are enclosed in a brown extracellular matrix (27). This bacterial biofilm can adhere to the cuticle covering the spines that line the interior of the proventriculus, a valve-like structure between the midgut and the esophagus. In 25 to 50% of infected Xenopsylla cheopis fleas, the adherent biofilm grows to fill the spaces between the proventricular spines and blocks the flea's digestive tract. Starvation induced by proventricular blockage promotes increased feeding attempts, during which bacteria from the periphery of the biofilm can be dislodged and become the inoculum for the next infection (27).
The ability of Y. pestis to block fleas correlates with the ability to form biofilm in glass flow cells at 21°C (27). Both abilities require the hemin storage (hms) gene products located in the hmsHFRS operon (22, 27). The hms genes are also required for the Y. pestis pigmentation (Pgm) phenotype, the ability when grown at temperatures <28°C to adsorb hemin or the structurally analogous dye Congo red and form densely pigmented colonies (30). Orthologues of the hms genes in Staphylococcus aureus, Staphylococcus epidermidis, and Escherichia coli synthesize an extracellular β-1,6-N-acetyl-d-glucosamine polymer that is required for biofilm formation (11, 18, 41, 42). An extracellular hms-dependent matrix is also produced by Y. pestis (27). The structure of the hms-dependent matrix has not been determined, although an enzyme that hydrolyzes polymeric β-1,6-N-acetyl-d-glucosamine prevents Y. pestis biofilm formation (26). Although Y. pseudotuberculosis also possesses the hms genes (9), indicating that they were acquired before the divergence of Y. pestis, most Y. pseudotuberculosis strains form nonpigmented colonies on Congo red agar (7).
Studying flea infection and blockage in the laboratory is labor intensive, not easily amenable to high-throughput investigations, and requires dedicated facilities for rearing and maintaining fleas. The nematode Caenorhabditis elegans has recently been proposed as an attractive surrogate for identifying both host and bacterial factors required for flea transmission of Y. pestis (10, 13, 25, 29, 39). This is based on the observation that a Y. pestis biofilm accumulates on the cuticle of the head and mouthparts of worms moving through a bacterial lawn (13). This phenotype, which prevents the worms from feeding, is also dependent on the hms genes (13). Although there is considerable variation, many Y. pseudotuberculosis strains are unable to colonize worms, but some bind avidly and form larger biofilms than Y. pestis strains under the same conditions (13, 29). Like hms-negative strains of Y. pestis, Y. pseudotuberculosis strains lacking hmsF (29) or hmsT (13) are unable to colonize the surfaces of worms, suggesting a molecular as well as functional similarity between Yersinia interactions with worms and fleas.
The ability of Y. pseudotuberculosis to infect fleas has not been well characterized. Previous experiments demonstrated that a serotype O1b strain of Y. pseudotuberculosis could survive over a 4-week period after being taken up in a blood meal (23). Blanc and Balthazard (5) also found, using a single Y. pseudotuberculosis strain, that fleas could maintain an infection up to 35 days after feeding on septicemic guinea pigs, but these fleas did not transmit the Y. pseudotuberculosis infection. Y. pseudotuberculosis strains have been assigned to 21 different O-antigen serotypes (37) and are more genetically variable than those of Y. pestis (20). We show here that most Y. pseudotuberculosis strains are able to persist in the flea digestive tract, but that none form biofilm in the flea or cause proventricular blockage, even those that are able to form cohesive biofilms in vitro or on C. elegans. These results suggest that a Y. pestis progenitor strain with an ability to survive in fleas adapted its pre-existing biofilm-forming ability specifically to the flea digestive tract, thus enabling flea-borne transmission of plague.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The Y. pseudotuberculosis strains used are listed in Table 1. Strains designated K were from the London School of Hygiene and Tropical Medicine and were previously described in reference 29; other strains were obtained from Elizabeth Carniel (Institute Pasteur; strains designated IP), J.-A. Bengoechea and Mikael Skurnik (University of Helsinki); strain PB1 [8] and its O-antigen-negative ΔddhD-wzz (derivative), and Creg Darby (University of Alabama—Birmingham; strain YPIII (16). All K strains lacked the pYV virulence plasmid except K199, which contains the plasmid, and K163, which contained a mixture of bacteria with and without pYV (29). All other Y. pseudotuberculosis strains contained pYV. As with Y. pestis, the presence or absence of the pYV virulence plasmid does not affect the flea infectivity of Y. pseudotuberculosis (21, 22; B. J. Hinnebusch, unpublished data). Y. pestis KIM6+ and KIM6, the pigmentation-negative (Pgm−) KIM6+ derivative that lacks the chromosomal locus containing hmsHFRS (35), and plasmid pHMS1.2, which contains the hmsHFRS genes in pBR322 (34), were provided by Robert Perry (University of Kentucky). Plasmid pCH16 contains the ymt gene from the Y. pestis plasmid pMT1 cloned into pACYC177 (24). Y. pseudotuberculosis strains PB1 and PB1 ΔddhD-wzz were transformed by electroporation with pGFP (Clontech; Mountain View, Calif.), which contains the gene for green fluorescent protein.
TABLE 1.
Percentage of X. cheopis fleas that remained infected and the CFU per infected flea at different time points after infection with different Y. pseudotuberculosis and one Y. pestis strain
| Species | Strain | Serotype | % Fleas infecteda
|
CFU/infected fleab
|
||||
|---|---|---|---|---|---|---|---|---|
| 1 day | 7 days | 28 days | 1 day | 7 days | 28 days | |||
| Y. pseudotuberculosis | IP32953 | O1b | 55 | 60 | 40 | 9.5 × 102 ± 9.6 × 102 | 1.4 × 103 ± 7.7 × 102 | 3.4 × 103 ± 3.2 × 103 |
| K163 | O1b | 45 | 25 | 15 | 3.4 × 103 ± 5.2 × 10 | 2.1 × 103 ± 2.1 × 103 | 6.5 × 101 ± 2.3 × 101 | |
| PB1 | O1b | 54 (13/24) | 57 (28/49) | 55 (30/55) | 2.2 × 104 ± 4.8 × 104 | 3.1 × 104 ± 3.4 × 104 | 2.9 × 104 ± 2.2 × 104 | |
| IP32951 | O2 | 55 | 55 | 42 (5/12) | 7.4 × 103 ± 5.6 × 103 | 2.8 × 103 ± 3.8 × 103 | 2.3 × 103 ± 2.1 × 103 | |
| K170 | O2c | 55 | 50 | 35 | 3.6 × 103 ± 3.5 × 103 | 1.3 × 103 ± 1.0 × 103 | 1.9 × 103 ± 3.6 × 103 | |
| IP32984 | O3 | 11 (2/18) | 0 | 0 | 1.5 × 102 ± 0 | 0 | 0 | |
| YPIII | O3 | 12 (7/57) | 5 (3/60) | 0 (0/60) | 1.2 × 103 ± 1.9 × 103 | 2.6 × 103 ± 2.2 × 103 | 0 | |
| YPIII(pCH16) | O3 | 59 (23/39) | 40 (24/60) | 50 (30/60) | 3.0 × 103 ± 5.4 × 103 | 2.7 × 104 ± 3.5 × 104 | 9.0 × 103 ± 8.3 × 103 | |
| K171 | O3 | 0 | 8 (3/39) | 0 (0/40) | 0 | 4.0 × 103 ± 4.8 × 103 | 0 | |
| K171(pCH16) | O3 | 43 (6/14) | 56 (24/43) | 54 (30/56) | 5.7 × 104 ± 8.8 × 104 | 1.5 × 104 ± 3.1 × 104 | 1.7 × 104 ± 2.1 × 104 | |
| IP31411 | O4 | 5 | 5 | 0 | 2.0 × 101c | 2.7 × 102c | 0 | |
| K174 | O4a | 35 | 30 | 44 (4/9) | 8.5 × 103 ± 1.5 × 104 | 5.6 × 102 ± 6.5 × 102 | 5.5 × 102 ± 6.6 × 102 | |
| K175 | O4b | 45 | 60 | 50 | 4.9 × 103 ± 1.3 × 104 | 1.3 × 103 ± 2.1 × 103 | 3.6 × 103 ± 4.8 × 103 | |
| IP32821 | O5 | 95 | 80 | 15 | 1.1 × 103 ± 1.3 × 103 | 2.0 × 103 ± 2.2 × 103 | 6.0 × 101 ± 6.0 × 101 | |
| K177 | O5a | 40 | 50 | 35 | 1.2 × 104 ± 1.0 × 104 | 1.2 × 104 ± 6.1 × 103 | 2.0 × 104 ± 8.8 × 103 | |
| IP31553 | O6 | 50 | 20 | 0 | 1.4 × 103 ± 1.7 × 103 | 3.0 × 102 ± 4.7 × 102 | 0 | |
| K186 | O9 | 25 | 45 | 10 | 4.3 × 103 ± 4.4 × 103 | 1.8 × 103 ± 2.6 × 103 | 5.4 × 103 ± 2.1 × 102 | |
| K199 | Unknown | 45 | 50 | 55 | 1.4 × 104 ± 1.2 × 104 | 3.5 × 103 ± 6.3 × 103 | 4.0 × 103 ± 5.1 × 103 | |
| PB1 ΔddhD-wzz | Rough | NDd | 26 (5/19) | 45 | NDd | 4.6 × 104 ± 2.2 × 104 | 1.6 × 104 ± 1.4 × 104 | |
| PB1 ΔddhD-wzz(pHMS1.2) | Rough | 70 | 60 | 55 | 2.6 × 103 ± 2.3 × 103 | 3.5 × 103 ± 3.9 × 103 | 2.3 × 103 ± 2.3 × 103 | |
| Y. pestis | KIM6+ | Rough | 81 | 89 | 54 | 4.9 × 104 ± 3.1 × 104 | 4.0 × 103 ± 1.1 × 105 | 4.0 × 105 ± 6.8 × 104 |
Results are for 20 fleas except where noted [e.g., (13/24)].
Results are expressed as means±standard deviations.
Only a single flea in the sample was infected.
ND, not determined.
Flea infections.
X. cheopis fleas were infected with either Y. pseudotuberculosis or Y. pestis by allowing them to feed on heparanized mouse blood containing 1 × 108 to 5 × 108 CFU/ml in an artificial feeding system (21, 22). Bacteria for flea infections were grown in 100 ml Brain Heart Infusion broth (Difco) at 37°C for 24 h, counted using a Petroff-Hauser chamber, and resuspended in phosphate buffered saline (PBS) before being added to the blood meal. Fleas (50 males and 50 females) that took an infected blood meal were collected, kept at 21°C and 75% relative humidity, and fed twice weekly on normal mice for 28 days. Fleas were monitored for proventricular blockage during the 28 days, as previously described (22). To determine infection rates, additional samples of 20 female fleas were collected immediately after infection, and at 7 and 28 days after infection, were individually triturated in glass-sand slurry in PBS, diluted, and plated on Yersinia-selective agar base (Difco). The plates were incubated at 37°C for Y. pseudotuberculosis or 28°C for Y. pestis for 48 h, and the CFU were counted to determine the bacterial load per infected flea. To assess biofilm formation in the flea, midguts were dissected 1 to 3 weeks after infection and examined by light microscopy for the presence of dark clumps of bacteria. Bacteria expressing green fluorescent protein were visualized using fluorescence microscopy. Alternatively, indirect fluorescent antibody assays were performed on dissected flea midgut contents as previously described (27), using rabbit anti-Y. pestis polyclonal antiserum as the primary antibody, followed by goat anti-rabbit immunoglobulin G labeled with fluorescein. The anti-Y. pestis antibody cross reacted with Y. pseudotuberculosis strains in vitro.
Flow cell biofilm assays.
Bacteria grown for 48 h at room temperature in N minimal media (15) containing 0.1% Casamino Acids, 38 mM glycerol, and 1 mM MgCl2 were counted and then diluted in fresh media to a concentration of 1 × 107 cells/ml. The bacterial suspension (0.4 ml) was injected into one channel of a flow cell (Stovall; Greensboro, N.C.) that was connected to a reservoir of sterile media via a peristaltic pump at the influent end and to a discard reservoir at the effluent end. After a 20-min period to allow bacteria to attach to the glass surface (designated t = 0), sterile medium was pumped through the flow cell at 0.3 ml/min. After 24 or 48 h, the media flow was stopped and 0.4 ml of 5 mM Syto 9 stain (Molecular Probes; Eugene, Oreg.) was injected into the flow cell. After a 20-min staining period, media flow was resumed for 5 min to remove unbound dye. Biofilm attached to the borosilicate glass surface of the flow cell was visualized with a Zeiss LSM 510 scanning confocal laser microscope. Stacks of z-section images, approximately 1-μm thick, acquired from each flow cell were used to create three-dimensional representations of the biofilms, using the Zeiss LSM 510 software package. Biofilms were classified as heavy (>50-μm thick, with dense and nearly total surface coverage), moderate (<50-μm thick with less dense but nearly total surface coverage), or weak (small adherent microcolonies or scattered individual cells).
Microtiter plate biofilm assays.
Bacteria grown in TMH medium (38) for 48 h at room temperature were diluted in fresh media to 105 bacteria/ml, and 100 μl was added to four replicate wells in a 96-well polystyrene microtiter plate, which was then incubated at room temperature with shaking at 200 rpm. After 48 h, the media and planktonic cells were removed, and the wells were washed four times with 200 μl of water. The adherent bacteria were stained with 200 μl of a 0.05% safranine solution in water for 10 min. The staining solution was removed, and the wells were washed four times with water and air dried. Bound dye was solubilized with 200 μl of 30% acetic acid, and the absorbance at 450 nm was measured for each well, using a microtiter plate reader. Background staining was corrected by subtracting the safranine binding to uninoculated control wells. The ratio of the A450 reading for each strain relative to that of Y. pestis KIM6+ was calculated.
RESULTS
Effect of serotype on survival of Y. pseudotuberculosis in the flea digestive tract.
To test the ability of Y. pseudotuberculosis to colonize the digestive tract of fleas, we infected X. cheopis fleas with 18 different Y. pseudotuberculosis strains representing 11 O-antigen serotypes (Table 1). Different Y. pseudotuberculosis serotypes varied greatly in their ability to produce a chronic infection. All serotype O1b, O2, O5, and O9 strains tested maintained the infection throughout the entire 28-day period. In contrast, all three serotype O3 strains, one of three serotype O4 strains, and the single serotype O6 strain tested were not recovered from any fleas 28 days after infection. Serotype O3, O4, and O6 strains that failed to stably colonize persisted in only 5 to 10% of fleas at 7 days and were completely eliminated from all fleas by day 28.
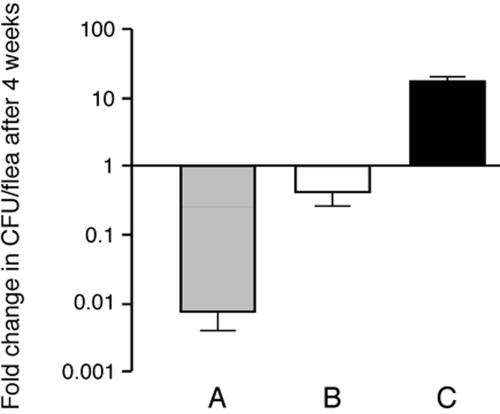
The average number of Y. pseudotuberculosis CFU per infected flea was also determined (Table 1). Numbers of both Y. pseudotuberculosis and Y. pestis in fleas decreased during the first 24 h. Following this initial decrease, most of the strains maintained fairly constant infection levels of about 103 to 104 CFU/flea over the 28 days. Other strains (K163, K174, and IP32821) decreased in numbers so that at day 28 there were only 60 to 550 CFU per flea. Thus, most Y. pseudotuberculosis strains persisted in the flea digestive tract, but their numbers decreased 1- to 10-fold over 4 weeks from the initial infectious dose of 104 bacteria/flea, and the decrease was not serotype dependent (Fig. 1). In contrast, the average number of Y. pestis increased 10-fold over 28 days.
FIG. 1.
The number of Y. pseudotuberculosis in infected fleas decreases over time. The change (n-fold) in CFU/flea (mean CFU/flea on day 28 divided by mean CFU/flea immediately after infection) was determined separately for Y. pseudotuberculosis strains K163, K174, and K177 (A) and IP32953, PB1, PB1 ΔddhD-wzz, PB1 ΔddhD-wzz(pHMS1.2), IP32951, K170, K175, K177, K186, and K199 (B) and for Y. pestis KIM6+ (C). The mean and standard error of the mean of the change (n-fold) in CFU are shown for each group; the mean CFU/flea for each time point was determined from samples of 20 fleas per strain except where noted in Table 1.
Effect of the Y. pestis ymt gene on survival and replication of Y. pseudotuberculosis serotype O3 in fleas.
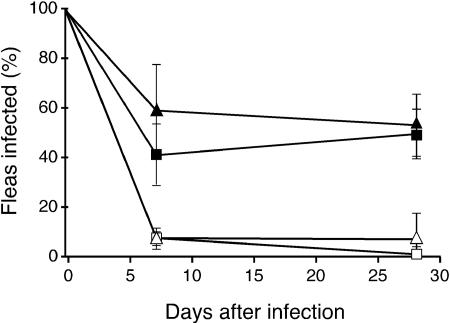
Notably, all three of the serotype O3 strains tested failed to establish a chronic infection in X. cheopis fleas (Table 1), similar to Δymt strains of Y. pestis (23). To determine whether the Y. pestis ymt gene is sufficient to allow serotype O3 strains to survive in fleas, strains K171 and YPIII were provided a copy of the ymt gene on plasmid pCH16. The presence of the Y. pestis ymt gene greatly increased the infectivity of Y. pseudotuberculosis O3 strains for fleas. Between 40 and 60% of fleas infected with the ymt-transformed strains remained infected at days 7 and 28 (Fig. 2), with an average bacterial load of 1 × 104 to 2 × 104 CFU per infected flea (Table 1).
FIG. 2.
Effect of the Y. pestis ymt gene on the ability of serotype O3 Y. pseudotuberculosis strains to persist in the flea digestive tract. Percentages of fleas that remained infected during a 4-week period following a single infectious blood meal containing the YPIII (□), K171 (▵), YPIII(pCH16) (▪), or K171(pCH16) (▴) strain of serotype O3. The average and standard deviation of the results of three independent infection experiments for each strain are shown.
Y. pseudotuberculosis does not form a biofilm or cause proventricular blockage in fleas.
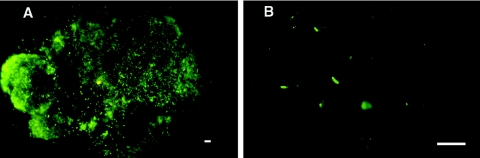
Y. pestis forms dense, biofilm-like aggregates in the flea midgut composed of bacteria surrounded by an extracellular matrix (27). Y. pestis also colonizes the proventriculus, where the aggregates may become lodged and lead to blockage. Since most Y. pseudotuberculosis strains were able to colonize the flea digestive tract, we examined midguts dissected from infected fleas for evidence of biofilm-like growth. Rather than forming clumps in the midgut, however, Y. pseudotuberculosis bacteria occurred only as individual cells (Fig. 3). Furthermore, colonization by Y. pseudotuberculosis was restricted to the midgut; bacteria were never observed in the proventriculus.
FIG. 3.
Y. pseudotuberculosis does not form a biofilm in the flea digestive tract. The digestive tracts of fleas infected with Y. pestis KIM6+(pGFP) (A) or Y. pseudotuberculosis PB1 (B) were dissected, and the bacteria were visualized with fluorescence microscopy, either directly (Y. pestis) or after indirect fluorescent antibody (Y. pseudotuberculosis). The bar corresponds to 100 μm.
To verify this, we monitored fleas infected with Y. pseudotuberculosis for development of foregut blockage (Table 2). None of the fleas infected with any Y. pseudotuberculosis strain ever developed foregut blockage during the 28-day observation period, including strain PB1 ΔddhD-wzz(pHMS1.2), which, like Y. pestis, lacks O antigen and is strongly pigmented on Congo red agar. In contrast, between 25 and 50% of fleas infected with Y. pestis became blocked during the 28-day period.
TABLE 2.
Comparison of biofilm formation by Y. pseudotuberculosis and Y. pestis in different in vitro and in vivo environments
| Species | Strain | Serotype | Pgm phenotypea | Biofilm formation
|
|||
|---|---|---|---|---|---|---|---|
| In vitro
|
In vivo
|
||||||
| Microtiter wellb | Flow cellc | C. elegans nematodesd | X. cheopis flease | ||||
| Y. pseudotuberculosis | K163 | O1b | − | 0.20 ± .03 | Moderate | None | − |
| K170 | O2c | − | 1.11 ± .12 | Heavy | Intermediate | − | |
| K171 | O3 | − | 0.59 ± .10 | Weak | Severe | − | |
| YPIII(K1) | O3 | − | 0.49 ± .03 | Moderate | Severe | − | |
| K174 | O4a | − | 0.27 ± .03 | Heavy | Weak | − | |
| K175 | O4b | − | 0.25 ± .05 | Heavy | None | − | |
| K177 | O5a | − | 0.17 ± .04 | Weak | Intermediate | − | |
| K186 | O9 | − | 0.34 ± .07 | Moderate | None | − | |
| K199 | Unknown | − | 0.09 ± .05 | Heavy | Weak | − | |
| PB1 ΔddhD-wzz | Rough | − | 1.16 ± .06 | Moderate | NDg | − | |
| PB1 ΔddhD-wzz(pHMS1.2) | Rough | + | 1.37 ± .10 | Heavy | NDg | − | |
| Y. pestis | KIM6+ | Rough | + | 1.00 | Heavy | NDg | + |
| KIM6 | Rough | − | 0.01 ± .01 | Weak | NDg | +/−f | |
Shows pigmentation on Congo red agar.
Means ± standard deviations of the results of three safranine-binding assays expressed relative to the results for Y. pestis KIM6+.
Biofilms were classed as heavy, moderate, or weak based on surface area coverage and depth as explained in Materials and Methods.
Scoring for infection of C. elegans was based on accumulation of bacterial biofilm on the mouthparts of the nematodes and the effect on feeding and movement (data are from reference 29).
+, has the ability to form aggregates in the digestive tract and block fleas; −, does not have the ability to form aggregates in the digestive tract and block fleas.
Forms aggregates in the midgut but does not block fleas (22).
ND, not determined.
In vitro biofilm formation by Y. pseudotuberculosis.
The ability of Y. pestis to block the proventriculus of fleas correlates with hms-dependent pigmentation on Congo red agar (22) and biofilm formation in vitro at 21°C (27, 35). We tested the ability of nine Y. pseudotuberculosis strains, none of which was pigmented on Congo red agar, to form biofilms on two different substrates in vitro. In glass flow cells, the nine strains varied considerably in their patterns of biofilm formation, with no obvious correlation to serotype. Similar to Y. pestis, some of the strains showed evidence of the formation of microcolonies and thin biofilms after 24 h of growth. By 48 h, Y. pseudotuberculosis strains YPIII, K163, K171, K177, and K186 appeared as scattered microcolonies or very thin biofilms, while strains K170, K174, K175, and K199 had formed more-substantial, thicker biofilms (Fig. 4). Although thick biofilms were produced by some Y. pseudotuberculosis strains, they had a greater tendency to slough off the glass surface than biofilms produced by Y. pestis.
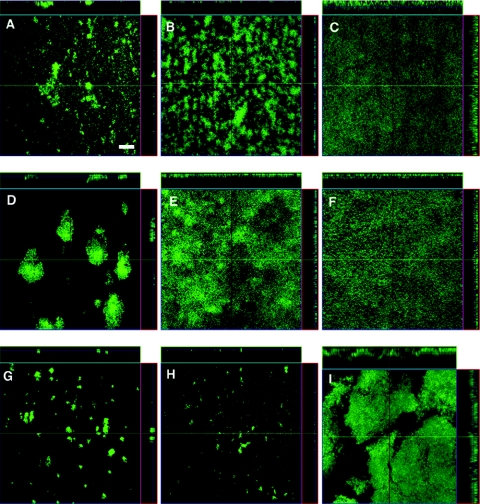
FIG. 4.
Y. pseudotuberculosis biofilms in glass flow cells. Confocal scanning laser microscopy images of biofilms produced after 48 h at 21°C for Y. pseudotuberculosis serotype O3 strain YPIII (A), serotype O1b strain K163 (B), serotype O2c strain K170 (C), serotype O3 strain K171 (D), serotype O4a strain K174 (E), serotype O4b strain K175 (F), serotype O5a strain K177 (G), serotype O9 strain K186 (H), and strain K199 (serotype unknown) (I). The right side and top of each panel are reconstructed vertical cross sections of the biofilm. The bar corresponds to 100 μm.
The amount of biofilm formed in polystyrene microtiter plates by the different Y. pseudotuberculosis strains relative to that of Y. pestis was quantified by means of a safranine-binding assay. More biofilm was formed by some of the Y. pseudotuberculosis strains in this system than was produced by Y. pestis (Table 2). The growth rates of the strains in the liquid medium were equivalent, and thus did not account for the differences in biofilm formation.
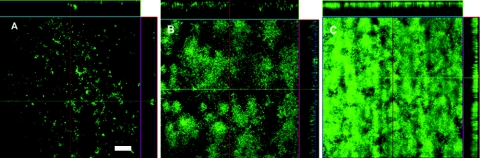
We also tested the effect of the loss of O polysaccharide and hms-dependent pigmentation on Y. pseudotuberculosis by comparing the biofilms formed by a Y. pseudotuberculosis O1b rough mutant (PB1 ΔddhD-wzz), which, like Y. pestis, produces lipopolysaccharide that lacks O antigen (S. Kiljunen, E. Pinta, J.-A. Bengoechea, O. Holst, and M. Skurnik, submitted for publication) and PB1 ΔddhD-wzz transformed with pHMS1.2, a recombinant plasmid that contains the Y. pestis hmsHFRS operon (34). The presence of this plasmid resulted in a strongly pigmented colony phenotype (Pgm+) on Congo red agar and the formation of a thicker biofilm (Fig. 5). However, this biofilm was less dense than a Y. pestis biofilm produced under the same conditions, and transformation with pHMS1.2 did not affect the tendency of the biofilm to slough off the surface. The O-antigen-negative Pgm+ Y. pseudotuberculosis strain also failed to colonize or block the proventriculus of fleas (Table 2).
FIG. 5.
The Pgm+ phenotype correlates with enhanced biofilm formation by Y. pseudotuberculosis in vitro. Confocal scanning laser microscopy images of 48-h, 21°C biofilms produced by the Hms− strain Y. pseudotuberculosis PB1 ΔddhD-wzz (A), the Hms+ strain PB1 ΔddhD-wzz(pHMS1.2) (B), and the Hms+ strain Y. pestis KIM6+ (C). The bar corresponds to 100 μm.
DISCUSSION
Comparison of Y. pseudotuberculosis and Y. pestis flea infection phenotypes.
During the evolution of Y. pestis from Y. pseudotuberculosis in the relatively recent past, Y. pestis adopted a dramatically different lifestyle, which relies on transmission by rodent fleas. As a starting point in identifying the evolutionary steps that led to arthropod-borne transmission of plague, we compared the ability of Y. pseudotuberculosis and Y. pestis to infect the rat flea X. cheopis.
Both species were able to survive for long periods of time in the flea digestive tract. Two marked differences were observed, however. Unlike Y. pestis, Y. pseudotuberculosis did not increase in numbers during the 28-day observation period (Fig. 1), and the ability of Y. pseudotuberculosis to establish a chronic infection was serotype dependent (Table 1). All three serotype O3 strains and one serotype O4 and O6 strain were quickly eliminated from the flea midgut, whereas other serotypes maintained a steady infection over 28 days (Table 1). Interestingly, transformation of Y. pseudotuberculosis with the Y. pestis ymt gene could eliminate both differences. For example, serotype O3 strains that were normally unable to infect fleas achieved 40 to 60% infection rates when transformed with ymt (Fig. 2). Additionally, for Y. pseudotuberculosis strains such as PB1 that are able to infect fleas, the number of bacteria per infected flea is significantly increased by the presence of ymt (23).
Survival of Y. pestis in the flea midgut depends on the phospholipase D activity encoded by ymt, which is present on a Y. pestis-specific plasmid. Ymt− Y. pestis experience outer membrane perturbation in the flea midgut and are almost entirely eliminated from the flea within 24 h (23). Even though Y. pseudotuberculosis does not possess a ymt homolog, most strains could persist in the flea midgut for weeks, suggesting that they are innately more resistant to membrane-perturbing agents in the flea gut. Although it is possible that Y. pseudotuberculosis serotypes able to persist in fleas encode an alternative phospholipase D, the genome sequence of the serotype O1 Y. pseudotuberculosis strain IP32953 does not suggest this (9). Instead, Y. pseudotuberculosis serotypes appear to be differentially susceptible to antibacterial factors in the flea gut, and transformation with ymt is able to eliminate these differences. The differential susceptibility might be related to the serotype-specific O antigen itself. However, the O antigen of serotype 3 strains, which were rapidly eliminated from fleas, is structurally most similar to those of serotype O1b, O2c, O4a, O5a and O7 strains (37), which were able to survive in fleas. Furthermore, an O-antigen-negative (rough) mutant of Y. pseudotuberculosis O1b did not have reduced ability to colonize fleas (Table 1). Most of the genomic differences among Y. pseudotuberculosis serotypes can be attributed to genes unrelated to O-antigen structure (9, 20), and these may be more important for the observed relationship between serotype and flea colonization. The phoP gene of Y. pseudotuberculosis O3 YPIII strains is known to be nonfunctional, and other O3 strains have a 9-kb deletion in the high-pathogenicity island (14, 17). Although a defect in phoP could conceivably reduce Y. pseudotuberculosis survival in the flea midgut because phoP affects resistance to cationic antimicrobial peptides commonly produced by insects (31), neither phoP nor the high-pathogenicity island affects the survival of Y. pestis in fleas (22; B. J. Hinnebusch, unpublished data).
Y. pseudotuberculosis biofilm formation in vitro or on the mouthparts of C. elegans is not predictive of flea infection or blockage.
The most striking difference between Y. pestis and Y. pseudotuberculosis flea infections is that Y. pseudotuberculosis is unable to cause the characteristic foregut blockage typical of Y. pestis that is required for efficient transmission (2, 3). Fundamentally, flea blockage is a bacterial biofilm phenomenon. Y. pestis and Y. pseudotuberculosis can accumulate as biofilms on the external head and mouthparts and prevent feeding of C. elegans nematodes, suggesting the possibility that this invertebrate might be a useful surrogate model to identify both host and bacterial factors required for flea infection and blockage (10, 13, 25, 29, 39). To assess the correlation between the various models, we systematically compared the in vitro biofilm and flea infection phenotypes of 9 Y. pseudotuberculosis strains for which C. elegans infection phenotypes had previously been reported (29). There was little correlation between biofilm-forming characteristics in the two in vitro systems and on the surface of C. elegans. None of the Y. pseudotuberculosis strains caused blockage in fleas, regardless of the ability to form biofilms in vitro or on C. elegans. Furthermore, there was no correlation between worm infection phenotype and survival in the flea digestive tract (Table 2). Interestingly, some strains that failed to form biofilms in polystyrene microtiter plates formed thick biofilms in glass flow cells, and vice versa (Table 2). Thus, the ability of Y. pseudotuberculosis to form biofilms in fleas and on nematodes and in microtiter dishes and in flow cells may not be predictive of one another and may depend on specific factors unique to each environment. Therefore, although biofilm formation in vitro or on C. elegans are relevant models of proventricular blockage in fleas, proposed roles for genes identified in these artificial systems should be tested with fleas before extrapolating results from one system to another (12).
Y. pestis Pgm− strains with reduced biofilm-forming capabilities in vitro are unable to block fleas (27). Consistent with this model, we found that Y. pseudotuberculosis biofilms under flow are typically weaker and more easily disrupted than Y. pestis, which correlates with their inability to block fleas (Fig. 4, 5). Thus far, few factors have been identified that influence Yersinia biofilms in any environment. Changes to the lipopolysaccharide (LPS) core affect Y. pseudotuberculosis biofilms on C. elegans (29). In contrast, the O-antigen portion of LPS did not have any consistent effect on biofilm formation either in microtiter wells or on C. elegans (29). In flow cells, both of the serotype O4 strains tested formed relatively thick biofilms, while both serotype O3 strains formed poor biofilms (Fig. 4). However, more strains must be tested before any relationship between serotype and biofilm in flow cells can be determined.
We have shown here that a Y. pseudotuberuculosis Pgm+ phenotype enhances biofilm formation on glass flow cells (Fig. 5). This is also true in microtiter wells and on C. elegans worms (29). However, even a Y. pseudotuberculosis strain with a strong, constitutive Pgm+ phenotype on Congo red agar forms a weaker in vitro biofilm than Y. pestis and is unable to block fleas (Fig. 5; Table 2). It is likely that even if Y. pseudotuberculosis initially colonizes the surface of the proventricular spines, it does not form a biofilm that is cohesive enough to withstand the rapid contractions of the proventricular valve and hydrodynamic forces generated when the flea feeds. Consequently, the bacteria are washed back into the midgut. Y. pseudotuberculosis is unlikely to face such forces on the head and mouthparts of C. elegans, and this could explain why it is able to prevent nematode feeding but is unable to block fleas.
The lack of any correlation between Y. pseudotuberculosis biofilm formation in the flea and on C. elegans could also be due to differences in surface characteristics. The flea proventriculus and C. elegans are covered with cuticle, but insect and nematode cuticles differ substantially in composition and ultrastructure. Nematode cuticle is composed primarily of collagen (33). The external surface coat of C. elegans is a thin (5- to 20-nm), negatively charged, carbohydrate-rich layer that can bind lectins and can be sloughed off (6). In contrast, the cuticle lining of the flea proventriculus is composed primarily of chitin filaments embedded in a complex, highly sclerotized protein matrix (19, 28). The nature of this protein matrix has not been determined for fleas, but the cuticle lining the proventriculus of bees contains abundant cysteine-rich proteins that confer an overall acidic character (32).
The Y. pseudotuberculosis genome (9) contains all of the identified hms genes, which are >99% identical to the Y. pestis homologues, yet most Y. pseudotuberculosis strains have a Pgm− phenotype on Congo red agar (7). Such differences in the structure or composition of cell envelope or extracellular structures, as well as the substrate of the biofilm and its surrounding medium, can have profound effects on biofilm formation. For example, mutations in Salmonella enterica that result in either incomplete (rough) LPS structure or loss of cellulose production abolish biofilm development on glass, but not on gallstones (36). The effects of changes in regulatory functions on biofilm development are also unique to different environments and perhaps even more difficult to predict (reviewed in reference 4). Thus, the molecular mechanisms that explain how Y. pestis evolved from Y. pseudotuberculosis to block fleas may be complex and due to subtle genetic differences that have yet to be identified. Our results imply that one or more genetic changes occurred in the Y. pseudotuberculosis progenitor of Y. pestis that increased the production or stability of the hms-dependent extracellular matrix in the flea gut environment, enhancing biofilm formation in the proventriculus and resulting in efficient arthropod-borne transmission.
Acknowledgments
We thank Mikael Skurnik, J.-A. Bengoechea, Elizabeth Carniel, Robert Perry, and Creg Darby for providing strains and plasmids. We also thank Florent Sebbane and Cuong Vuong for critical readings of the manuscript.
This work was supported by the Division of Intramural Research, NIAID, NIH and in part by the Ellison Medical Foundation (New Scholars Award in Global Infectious Diseases to B.J.H.).
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacot, A. W. 1915. Further notes on the mechanism of the transmission of plague by fleas. J. Hyg. Plague 14(Suppl. 4):774-776. [PMC free article] [PubMed] [Google Scholar]
- 3.Bacot, A. W., and C. J. Martin. 1914. Observations on the mechanism of the transmission of plague by fleas. J. Hyg. Plague 13(Suppl. 3):423-439. [PMC free article] [PubMed] [Google Scholar]
- 4.Beloin, C., and J. M. Ghigo. 2005. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 13:16-19. [DOI] [PubMed] [Google Scholar]
- 5.Blanc, G., and M. Balthazard. 1944. Contribution a l'etude du comportement de microbes pathogenes chez la puce du rat Xenopyslla cheopis. Le bacille de la pseudo-tuberculose des rongeurs. C. R. Soc. Biol. Paris 138:811-812. [Google Scholar]
- 6.Blaxter, M. L., A. P. Page, W. Rudin, and R. M. Maizels. 1992. Nematode surface coats: actively evading immunity. Parasitol. Today 8:243-247. [DOI] [PubMed] [Google Scholar]
- 7.Brubaker, R. R. 1991. Factors promoting acute and chronic diseases caused by yersiniae. Clin. Microbiol. Rev. 4:309-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrows, T. W., and G. A. Bacon. 1960. V and W antigens in strains of Pasteurella pseudotuberculosis. Br. J. Exp. Pathol. 41:38-44. [PMC free article] [PubMed] [Google Scholar]
- 9.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cipollo, J. F., A. M. Awad, C. E. Costello, and C. B. Hirschberg. 2004. Srf-3, a mutant of Caenorhabditis elegans, resistant to bacterial infection and to biofilm binding, is deficient in glycoconjugates. J. Biol. Chem. 279:52893-52903. [DOI] [PubMed] [Google Scholar]
- 11.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darby, C., S. L. Ananth, L. Tan, and B. J. Hinnebusch. 2005. Identification of gmhA, a Yersinia pestis gene required for flea blockage, by using a Caenorhabditis elegans biofilm system. Infect. Immun. 73:7236-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243-244. [DOI] [PubMed] [Google Scholar]
- 14.Fukushima, H., Y. Matsuda, R. Seki, M. Tsubokura, N. Takeda, F. N. Shubin, I. K. Paik, and X. B. Zheng. 2001. Geographical heterogeneity between Far Eastern and Western countries in prevalence of the virulence plasmid, the superantigen Yersinia pseudotuberculosis-derived mitogen, and the high-pathogenicity island among Yersinia pseudotuberculosis strains. J. Clin. Microbiol. 39:3541-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia Vescovi, E., F. C. Soncini, and E. A. Groisman. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165-174. [DOI] [PubMed] [Google Scholar]
- 16.Gemski, P., J. R. Lazere, T. Casey, and J. A. Wohlhieter. 1980. Presence of a virulence-associated plasmid in Yersinia pseudotuberculosis. Infect. Immun. 28:1044-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grabenstein, J. P., M. Marceau, C. Pujol, M. Simonet, and J. B. Bliska. 2004. The response regulator PhoP of Yersinia pseudotuberculosis is important for replication in macrophages and for virulence. Infect. Immun. 72:4973-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 19.Hepburn, H. R. 1885. Structure of the integument, p. 1-58. In G. A. Kerkut and L. I. Gilbert (ed.), Comprehensive insect physiology, biochemistry, and pharmacology, vol. 3. Integument, respiration, and circulation. Permagon Press Inc., Elmsford, N.Y. [Google Scholar]
- 20.Hinchliffe, S. J., K. E. Isherwood, R. A. Stabler, M. B. Prentice, A. Rakin, R. A. Nichols, P. C. Oyston, J. Hinds, R. W. Titball, and B. W. Wren. 2003. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 13:2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinnebusch, B. J., E. R. Fischer, and T. G. Schwan. 1998. Evaluation of the role of the Yersinia pestis plasminogen activator and other plasmid-encoded factors in temperature-dependent blockage of the flea. J. Infect. Dis. 178:1406-1415. [DOI] [PubMed] [Google Scholar]
- 22.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 23.Hinnebusch, B. J., A. E. Rudolph, P. Cherepanov, J. E. Dixon, T. G. Schwan, and A. Forsberg. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296:733-735. [DOI] [PubMed] [Google Scholar]
- 24.Hinnebusch, J., P. Cherepanov, Y. Du, A. Rudolph, J. D. Dixon, T. Schwan, and A. Forsberg. 2000. Murine toxin of Yersinia pestis shows phospholipase D activity but is not required for virulence in mice. Int. J. Med. Microbiol. 290:483-487. [DOI] [PubMed] [Google Scholar]
- 25.Hoflich, J., P. Berninsone, C. Gobel, M. J. Gravato-Nobre, B. J. Libby, C. Darby, S. M. Politz, J. Hodgkin, C. B. Hirschberg, and R. Baumeister. 2004. Loss of srf-3-encoded nucleotide sugar transporter activity in Caenorhabditis elegans alters surface antigenicity and prevents bacterial adherence. J. Biol. Chem. 279:30440-30448. [DOI] [PubMed] [Google Scholar]
- 26.Itoh, Y., X. Wang, B. J. Hinnebusch, J. F. Preston III, and T. Romeo. 2005. Depolymerization of β-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarrett, C. O., E. Deak, K. E. Isherwood, P. C. Oyston, E. R. Fischer, A. R. Whitney, S. D. Kobayashi, F. R. DeLeo, and B. J. Hinnebusch. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190:783-792. [DOI] [PubMed] [Google Scholar]
- 28.Jin, P. 1994. Studies on the digestive system of fleas: structures of the proventriculi of fourteen flea species. Acta Entomol. Sin. 37:51-58. [Google Scholar]
- 29.Joshua, G. W., A. V. Karlyshev, M. P. Smith, K. E. Isherwood, R. W. Titball, and B. W. Wren. 2003. A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology 149:3221-3229. [DOI] [PubMed] [Google Scholar]
- 30.Lillard, J. W., Jr., J. D. Fetherston, L. Pedersen, M. L. Pendrak, and R. D. Perry. 1997. Sequence and genetic analysis of the hemin storage (hms) system of Yersinia pestis. Gene 193:13-21. [DOI] [PubMed] [Google Scholar]
- 31.Marceau, M., F. Sebbane, F. Ewann, F. Collyn, B. Lindner, M. A. Campos, J. A. Bengoechea, and M. Simonet. 2004. The pmrF polymyxin-resistance operon of Yersinia pseudotuberculosis is upregulated by the PhoP-PhoQ two-component system but not by PmrA-PmrB, and is not required for virulence. Microbiology 150:3947-3957. [DOI] [PubMed] [Google Scholar]
- 32.Neves, C. A., E. B. M. I. Peixoto, and J. E. Serrao. 2000. Histochemistry of the cuticle from proventriculus in stingless bee, Melipona quadrifasciata anthidioides (Hymenoptera, Apidae). Folia Histochem. Cytobiol. 38:193-196. [PubMed] [Google Scholar]
- 33.Page, A. P., and A. D. Winter. 2003. Enzymes involved in the biogenesis of the nematode cuticle. Adv. Parasitol. 53:85-148. [DOI] [PubMed] [Google Scholar]
- 34.Pendrak, M. L., and R. D. Perry. 1993. Proteins essential for expression of the Hms+ phenotype of Yersinia pestis. Mol. Microbiol. 8:857-864. [DOI] [PubMed] [Google Scholar]
- 35.Perry, R. D., M. L. Pendrak, and P. Schuetze. 1990. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J. Bacteriol. 172:5929-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prouty, A. M., and J. S. Gunn. 2003. Comparative analysis of Salmonella enterica serovar Typhimurium biofilm formation on gallstones and on glass. Infect. Immun. 71:7154-7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skurnik, M. 2004. Lipopolysaccharides of Yersinia, p. 215-241. In E. Carniel and B. J. Hinnebusch (ed.), Yersinia: molecular and cellular biology. Horizon Bioscience, Wymondham, U.K.
- 38.Straley, S. C., and W. S. Bowmer. 1986. Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect. Immun. 51:445-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan, L., and C. Darby. 2004. A movable surface: formation of Yersinia sp. biofilms on motile Caenorhabditis elegans. J. Bacteriol. 186:5087-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trebesius, K., D. Harmsen, A. Rakin, J. Schmelz, and J. Heesemann. 1998. Development of rRNA-targeted PCR and in situ hybridization with fluorescently labeled oligonucleotides for detection of Yersinia species. J. Clin. Microbiol. 36:2557-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vuong, C., S. Kocianova, J. M. Voyich, Y. Yao, E. R. Fischer, F. R. DeLeo, and M. Otto. 2004. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 279:54881-54886. [DOI] [PubMed] [Google Scholar]
- 42.Wang, X., J. F. Preston III, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]