Abstract
Genome duplication and segregation normally are completed before cell division in all organisms. The temporal relation of duplication and segregation, however, can vary in bacteria. Chromosomal regions can segregate towards opposite poles as they are replicated or can stay cohered for a considerable period before segregation. The bacterium Vibrio cholerae has two differently sized circular chromosomes, chromosome I (chrI) and chrII, of about 3 and 1 Mbp, respectively. The two chromosomes initiate replication synchronously, and the shorter chrII is expected to complete replication earlier than the longer chrI. A question arises as to whether the segregation of chrII also is completed before that of chrI. We fluorescently labeled the terminus regions of chrI and chrII and followed their movements during the bacterial cell cycle. The chrI terminus behaved similarly to that of the Escherichia coli chromosome in that it segregated at the very end of the cell division cycle: cells showed a single fluorescent focus even when the division septum was nearly complete. In contrast, the single focus representing the chrII terminus could divide at the midcell position well before cell septation was conspicuous. There were also cells where the single focus for chrII lingered at midcell until the end of a division cycle, like the terminus of chrI. The single focus in these cells overlapped with the terminus focus for chrI in all cases. It appears that there could be coordination between the two chromosomes through the replication and/or segregation of the terminus region to ensure their segregation to daughter cells.
In the past few years, the ability to visualize DNA sites and proteins in bacteria has advanced our knowledge of chromosome replication and segregation, particularly on how the two processes are related to each other with respect to cell growth and division (12, 36). These studies have revealed several principles governing chromosome segregation. It is clear that bacterial chromosomes can segregate in an orderly fashion, but the pattern of segregation can vary depending on the bacterium and its developmental state. The segregation can occur in conjunction with replication (coreplicational segregation) (24), or the replicated regions can stay together for a considerable period before separation (sister chromosome cohesion) (1). There are special sequences close to the origin of replication that serve as a centromere and are used to actively mobilize origin-proximal daughter DNA towards opposite cell poles (9, 11, 39) or to anchor them to specific locations on the cell membrane (2). It also appears that molecular motors, common in eukaryotic chromosome segregation, are not involved in bacteria. The motive force may come from the act of replication itself (extrusion-capture model [21]) or in a centromere-mediated process through the involvement of a cytoskeletal element, as in eukaryotes (11, 19). In the latter case, only the centromeric DNA is segregated actively and the rest of the chromosome, including the terminus region, is segregated in a sequence-nonspecific manner, using a mechanism such as chromosome condensation (14, 29).
In addition to the centromere, the replication terminus region is of special importance in the segregation process. First, cell division must not occur until a round of chromosome replication has been completed. In bacteria, it is believed that this temporal order is ensured by the presence of the replicating DNA in midcell. This causes steric hindrance to septum formation, although the process could be more involved (28, 38). Second, chromosome dimers are produced by homologous recombination between sister chromosomes in about 15% of the cells of a growing culture of Escherichia coli (32). A dimer cannot be distributed to daughter cells unless they are first converted to monomers. A site-specific recombination system, acting near the terminus region, accomplishes this. Dimer resolution also requires FtsK, a division septum protein essential for cell division (16) as well as proper positioning of the resolution site (dif), which is found in the terminus region (3, 23). Finally, the multiply catenated daughter chromosomes that arise at the completion of replication need to be unlinked. This requires the action of topoisomerases and FtsK (8). Localization of enzymatic machineries responsible for both dimer resolution and decatenation at the division septum implies that the process of cell division may contribute to chromosome segregation directly. However, septal ring formation does not appear to be essential for proper chromosome segregation. When septum formation is conditionally prevented, separate nucleoids are still seen during filamentous growth of rod-shaped bacteria. It appears that, for segregation of unlinked monomeric daughter chromosomes, active segregation of the origin region is the key event.
The connection of termination of DNA replication to cell division is particularly intriguing in Vibrio cholerae, which has two chromosomes (chromosome I [chrI] and chrII) of very different sizes (13, 35). The origins of the two chromosomes are different, but they share some replication initiation factors and initiate replication synchronously (6, 7). It follows then that the shorter chrII should finish replication before the larger chrI, assuming similar elongation rates for the two chromosomes. The question arises as to when the dimer resolution and decatenation of chrII occur. If these processes require FtsK and formation of the septal ring, as is currently believed for E. coli, then the segregation of the two chromosomal termini is likely to be temporally linked to the formation of the septal ring. Orthologues of the well-studied XerCD recombinases for dimer resolution in E. coli are present in V. cholerae, and the resolution site, dif, is present antipodal to the origin of both the chromosomes (27).
Here we have followed the position of the terminus region of the two chromosomes in growing V. cholerae cells using fluorescent tags to determine whether their segregation is temporally linked or independent. We found only a single focus for the terminus region of chrI in the cell center, even when cell septation was apparent. The results were mixed for the focus representing the chrII terminus region. In half of the cells, it split at the cell center and the sister foci migrated away from the cell center towards cell-quarter positions before any septum formation was apparent. In the other half, the focus did not split until the end of the division cycle. Moreover, in these cells, the focus overlapped with the one for the chrI terminus. This suggests that, although not as a rule, the two terminus regions could be coupled for simultaneous segregation at the time of cell septation.
MATERIALS AND METHODS
A general strategy for integrating foreign genes into V. cholerae chromosomes.
The genes of interest were transferred from a plasmid vector to the chromosome by two homologous recombination steps. To provide homology for integration, a 2-kb region spanning the point of insertion was first amplified from chromosomal DNA by PCR using primers containing restriction enzyme sites at their ends. The amplified fragments were digested with those enzymes and ligated to a similarly digested R6K γ-ori-based suicide vector, pDS132 (31). The foreign genes of interest were then cloned into a restriction enzyme site in the middle of the chromosomal fragment. For cloning purposes, DH5α(λpir) was used as a plasmid host. For conjugal transfer of plasmids to V. cholerae strains, E. coli SM10(λpir) was used as the donor. Since pDS132 cannot replicate in V. cholerae, selection of the plasmid-borne drug marker usually resulted in integration of the entire plasmid in the chromosome by a single crossover. When desired, the plasmid backbone was eliminated in a second recombination step by sucrose selection, due to the presence of the sacB gene in pDS132 (31). The integration of the genes of interest was confirmed by PCR.
Construction of a V. cholerae strain with a constitutively expressing araE transporter gene and an inducible operon carrying λcI-ecfp and lacI-eyfp fusion genes under PBAD control.
The full-length araE gene of E. coli with a linked Kanr cassette was amplified from strain BR8701 (Table 1) by PCR using primers PS33 and PS34 (Table 2) with HindIII and StuI sites. To get the araE gene only, the PCR product was digested with XhoI and StuI enzymes and ligated to similarly digested plasmid pZC496 that contained the λcI-ecfp and lacI-eyfp fusion genes (Table 1). The resultant plasmid was called pPS17. A locus between VCA0293 and VCA0294, both coding for hypothetical proteins, was selected for inserting the araE transporter and the fusion genes of pPS17. To provide homology for integration, a 2-kb region, with the point of insertion (chrII coordinate 312511) in its middle, was amplified using primers PS35 and PS36 with SphI and PstI sites. The product was digested and cloned in similarly digested pDS132 to generate pPS18. Next pPS17 was digested with PciI and XhoI, and the larger of the two products, after the ends were blunted with Klenow polymerase, was ligated to SpeI-digested and blunt-ended pPS18. The resultant plasmid, pPS19, was then conjugally transferred to a V. cholerae strain, N16961Strr (CVC209, Table 1), carrying pCP20 (pCP20 is a temperature-sensitive plasmid that confers Ampr on the recipient and was used here to counterselect the donor). The transconjugants were selected at 30°C on plates containing chloramphenicol (Cm) (25 μg/ml), spectinomycin (Spc) (40 μg/ml), and ampicillin (Amp) (50 μg/ml). Single colonies of transconjugants were inoculated in LB medium without Cm and grown at 37°C and plated on sucrose plates. Single colonies were patched on appropriate antibiotic-containing media to check for the loss of pDS132 backbone (Cm sensitivity) and pCP20 (Amp sensitivity) and the presence of the Spcr marker in the transconjugant. The integration of the PBADλcI-ecfp lacI-eyfp PCP18araE cassette was verified by PCR using primers RAF76 and RAF58. The resulting strain, CVC239, was used as the recipient in all other strains constructed for cytological studies. The growth rate of CVC239 was identical to that of the parent strain N16961Strr even when the fusion proteins were expressed.
TABLE 1.
Bacterial strains and plasmids used in the present study
| Strain or plasmid | Relevant description | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α(λpir) (= CVC23) | Supplies R6K π protein | M. Waldor |
| SM10(λpir) (= CVC201) | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu (λpir) pro endA hsdA hsdR supF | 7 |
| BW25113 | lacIqrrnB3ΔlacZ4787 hsdR514 ΔaraBAD567 ΔrhaBAD568 | 18 |
| BW27750 (= BR8701) | BW25113 ΔaraFGH φ(ΔaraEp kan PCP18-araE): araE promoter replaced with constitutive promoter CP18 | 18 |
| V. cholerae strains | ||
| CVC209 | N16961Strr | M. Waldor |
| CVC239 | N16961StrrPBADlacI-eyfp λcI-ecfp araE tetR Spcr | This study |
| CVC243 | Same as CVC239 except for 64× lacO in chrII | This study |
| CVC244 | Same as CVC239 except for 64× λOL1 in chrI | This study |
| CVC269 | Same as CVC243 except for 64× λOL1 in chrI | This study |
| CVC270 | Same as CVC239 except for 64× lacO in chrI | This study |
| CVC271 | Same as CVC239 except for 128× lacO in chrI | This study |
| CVC272 | Same as CVC239 except for 64× λOL1 in chrII | This study |
| CVC300 | N16961StrrPCP18araE kan | This study |
| CVC301 | N16961StrrPCP18araE | This study |
| CVC250 | N16961recA::kan | D. Pal |
| Plasmids | ||
| pCP20 | pSC101rep(Ts) Ampr; source of FLP recombinase | 4 |
| pDS132 | R6K γori, mobRP4 sacB Cmr; suicide vector for conjugal transfer and integration | 31 |
| pLAU85 | pBAD24 encoding LacI-ECFP and FtsZ-EYFP; Ampr | 20 |
| pPS9 | Coordinates 1512931-1516681 of chrI cloned in pDS132 | This study |
| pPS10 | Coordinates 462660-464883 of chrII cloned in pDS132 | This study |
| pPS11 | pDS132 + vca0527::64× lacO Kanr | This study |
| pPS17 | pZC496 + araE Spcr | This study |
| pPS18 | Coordinates 311001-313800 of chrII cloned in pDS132 | This study |
| pPS19 | pDS132 PBADlacI-eyfp λcI-ecfp PCP18araE tetR Spcr | This study |
| pPS20 | pDS132 PCP18araE Kanr | This study |
| pPS30 | pUC19 + 128× lacO Kanr | This study |
| pPS33 | pDS132 + vca0527::128× lacO Kanr | This study |
| pPS41 | pDS132 + vc1418::64× λOL1 Ampr | This study |
| pPS42 | pDS132 + vca0527::64× λOL1 Ampr | This study |
| pPS50 | pDS132 + vc1418::64× lacO Kanr | This study |
| pRFB110 | pUC19 + 64× lacO Kanr | 9 |
| pRFB122 | pUC19 + 64× λOL1 Cmr | 9 |
| pZC496 | pSC101 PBADlacI-eyfp λcI-ecfp tetR Spcr | Don Biek |
| pZE12-luc | Source of Ampr cassette | 26 |
TABLE 2.
Primers used in this study
| Primer name | Location | Primer sequence |
|---|---|---|
| PS25 | chrI | CTGCTCTAGACAACGAAGAAGCTCAGCTCGCTCTGCATGC |
| PS26 | chrI | ACTACGAGCTCGGACTCATCAGGTTCATCACCCC |
| PS27 | chrI | CGAGTACTTAATTCAAGGGCGTGATATCGC |
| PS31 | chrI | CCACTATACAATTTTGTGAGTTCTTATACG |
| PS28 | chrII | CTGCTCTAGAGACACGGAAAATATCGGAGATCATCCGCCC |
| PS29 | chrII | ACTACGAGCTCCCTACTGCATGAGCTGCGTCAAGGTGTGC |
| PS30 | chrII | GGCCGCTACTGCAGAGATGATCACCGCACGGTACCG |
| PS32 | chrII | CACTACCCAGAAAGATTGAAAGAGCGTTACC |
| PS33 | BR8701 | CGCCCAAGCTTGGCGACCAACAATACTCAACAACTACG |
| PS34 | BR8701 | TAGAAGGCCTCCCAGCTCATTCCTCCCAGCAAACC |
| PS35 | chrII | AAAACTGCAGGGCGGGCGTTAGCCATTTGGGAGAAC |
| PS36 | chrII | ACATGCATGCCCTAACGGCGAAACGGTTGTGCTCG |
| PS37 | Kan | CAGTCATAGCCGAATAGCCT |
| PS38 | Kan | CGGTGCCCTGAATGAACTGC |
| PS43 | Amp | GTTCTGCTATGTGGCGCGGTATTATCCCGTGTTGACGCCGGGCAAG |
| PS44 | Amp | GGAGGGCTTACCATCTGGCCCCAGTGCTGCAATGATACCGCGAGACC |
| RAF76 | lacI | CGAGGGCTAGCAGGAAACAGCTATGAAACCAGTAACGTTATACGATG |
| RAF58 | λcI | TGCATTGATGCCATTAAATAAAGCGCTAACGCCTGACTGC |
| RAF70 | araE | TTCCGCCTCAATATGACG |
Insertion of operators near the dif site of chrI.
A gene coding for hypothetical protein VC1418 near the dif site of chrI was selected for inserting an array of 64 λOL1 operators. A 2-kb region surrounding the point of insertion was amplified by PCR using primers PS25 and PS26 with SacI and XbaI sites. After digestion of the PCR product with the cognate enzymes, it was cloned in pDS132 digested with the same enzymes. The resultant plasmid was named pPS9. To clone the λOL1 array, the Cmr cassette of the operator carrying plasmid pRFB122 (9) was replaced by the Ampr cassette of pZE12-luc (26). pZE12-luc was digested with SacI and XhoI, and after the ends were blunted, the Ampr cassette was cloned in BsaI- and ScaI-digested pRFB122. The resultant plasmid, pPS15, was digested with SacII and XbaI, and the larger fragment containing the Ampr cassette linked to λ operators was gel purified, blunted, and cloned in FspI-digested pPS9. The plasmid so constructed, pPS41, was conjugally transferred to CVC239 and subjected to sucrose selection. The integration of the genes of interest was confirmed by PCR using primers PS27 and PS31, and those internal to the Ampr cassette, PS43 and PS44. This strain, CVC244, was used for cytological mapping of the dif region of chrI.
Instead of λ operators, we also integrated lac operators at the same locus. The plasmid pRFB110 (9) was digested with NdeI and blunt ended. The fragment that carried the lac operators and the linked Kanr cassette (4.2 kb) was cloned in the FspI-digested pPS9, as before. The resultant plasmid, pPS50, was conjugally transferred to CVC239, and the transconjugant, called CVC270, was directly used for localizing dif of chrI.
A strain was also made with an array of 128 lac operators. pRFB110 was digested with HincII and SacI, and the smaller 2.2-kb fragment containing lac operators was gel eluted and cloned between the SmaI and SacI sites of pRFB110. The resultant plasmid, pPS30, thus had two tandem arrays of 64 operators and a Kanr cassette. The plasmid was digested with NdeI, and the larger fragment was blunt ended and cloned in pPS9 digested with FspI. The resultant plasmid, pPS33, was conjugally transferred to CVC239, and a transconjugant, called CVC271, was directly used for localizing dif of chrI.
Insertion of operators near the dif site of chrII.
The gene coding for hypothetical protein VCA0527 near the dif site of chrII was selected for inserting the lac operator arrays. PCR primers PS28 and PS29 with SacI and XbaI sites were used to amplify a 2-kb region encompassing VCA0527. The product was digested with SacI and XbaI and cloned between the SacI and XbaI sites in pDS132 to get the plasmid pPS10. The 64 lac operators and the linked Kanr cassette, obtained from plasmid pRFB110 as previously described, were ligated to pPS10 linearized with NheI and blunted with Klenow polymerase to give the plasmid pPS11. The plasmid was transferred conjugally to CVC239. The integration of the plasmid was verified by PCR using primers PS30 and PS32 and two primers internal to the Kanr cassette, PS37 and PS38. The resultant strain, CVC243, was used directly for localizing dif of chrII.
We also labeled chrII at the same locus with λOL1 operators. For this purpose plasmid pPS15 was used as the source of the operators and the cassette was cloned in pPS10 digested with NheI and blunted with Klenow polymerase. The resultant strain was named CVC272.
Insertion of operators near dif of both chrI and chrII.
The strain CVC243, containing lac operators near dif of chrII, was used as the recipient for conjugal transfer of pPS41 carrying one operator with flanking DNA homologous to the dif region of chrI. The integration of the operators in the transconjugant was verified by PCR using primers described earlier for CVC244. The transconjugant, CVC269, was used for localizing the two dif loci simultaneously.
Insertion of araE in chrII.
To visualize FtsZ-yellow fluorescent protein (YFP) and a terminus region in the same cell, we used pLAU85 (20) encoding LacI-cyan fluorescent protein (CFP) and FtsZ-YFP. The full-length araE gene and the linked Kanr cassette were amplified by PCR from BR8701 using primers PS33 and PS34 with HindIII and StuI sites. After the ends were blunted with Klenow polymerase, the PCR product was cloned in plasmid pPS18 as before to get plasmid pPS20. This plasmid was electroporated to SM10(λpir) and then conjugally transferred to N16961Strr. The resulting strain, CVC300, was transformed with plasmid pCP20 to remove the Kanr cassette flanked by FRT sites. The loss of the cassette and the integration of araE in the resultant strain, CVC301,were confirmed by PCR using primers PS33 and RAF70.
Flow cytometry.
To determine the number of origins per cell, the replication runoff method was followed (33). Cultures of log-phase cells at an optical density at 600 nm (OD600) of 0.3 were treated with rifampin at 150 μg/ml and cephalexin at 30 μg/ml to prevent replication initiation and cell division and were allowed to grow for 3 to 5 h to complete DNA replication that had already begun (replication runoff). The culture was chilled, and all subsequent operations were done between 0 and 4°C. Typically, cells were pelleted from 1.8 ml culture, washed twice with 1× phosphate-buffered saline (PBS) buffer containing 1 mM EDTA (PBSE). The cells were resuspended in 0.1 ml of PBSE to which 0.9 ml of 77% ethanol was added to fix the cells. The cell suspension was stored at −20°C. Before flow cytometric analysis, the cells were resuspended in PBSE twice and OD600 of the cell suspension was determined. An aliquot representing 107 cells (assuming that an OD600 of 1 equals 109 cells) was removed, and the volume was adjusted to 0.9 ml with PBSE. To this 0.1 ml of Hoechst 33342 stain (Molecular Probes) was added to a final concentration of 0.5 μg/ml and the staining was continued for 2 to 3 h in the dark before being analyzed by the flow cytometer BD FACS DIVA LSR II (Becton Dickinson).
For quantitating the fluorescence due to enhanced YFP (EYFP) or enhanced CFP (ECFP), the cells from log-phase cultures were centrifuged and resuspended in 1× PBS. An aliquot representing 2 × 107 cells was directly used for flow cytometry.
Light microscopy.
Cells were grown either in L broth or in 1× M63 glucose medium supplemented with 2 mM MgSO4, 0.1 mM CaCl2, 1 μg/ml thiamine, and 0.2% glucose to an OD600 of 0.3 to 0.4, at which time they were induced with arabinose (0.005 to 0.0005% in different experiments). After an hour of induction the cells were examined in the microscope either directly or after being concentrated 50-fold by centrifugation at 2,500 rpm for 5 min and resuspended in 50 mM HEPES, pH 7.6. A 3-μl aliquot of cell suspension was placed on a microscope slide and covered with a polylysine-treated coverslip. Fluorescence and phase images were obtained to locate fluorescent foci relative to the cell poles. Differential interference contrast (DIC) images were also obtained to locate cell invagination more precisely.
For determining pole-to-focus or pole-to-pole distance for V. cholerae, which is shaped like a curved rod, the length along the curved longitudinal axis between two points (focus to pole or pole to pole) was approximated by breaking the curve into multiple segments and using NIH Image software to measure the linear distance of the individual segments. The individual segments were then summed to give the total point-to-point distance along the curved axis.
For time-lapse microscopy, an agarose pad was first made by pipetting 90 μl of 1% agarose containing 30% LB or 1× M63 glucose medium onto the slide and covering it with a silanized coverslip. After the agarose solidified, the coverslip was removed and 3 μl of cell suspension in the same medium was placed on the surface of the pad and covered again with a new coverslip. The slide was placed on a microscope stage heated to 37°C, and the cells were observed at intervals of approximately 15 min for a total of 165 min.
RESULTS
Strategy for subcellular localization of replication terminus by fluorescent repressor-operator system.
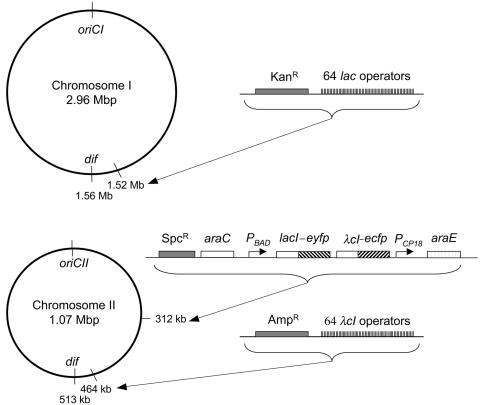
In order to visualize the terminus region of the two V. cholerae chromosomes, we used the fluorescent repressor-operator system with either lac repressor fused to EYFP or the λcI repressor fused to ECFP and the corresponding binding site arrays of 64 lac or λOL1 operators (9, 34). We have employed these reagents previously in E. coli to follow the cellular positions of several chromosomal markers including the terminus of replication (9). In the present study, the operators were integrated near the dif site of V. cholerae chrI and chrII (Fig. 1) (15). The site is located diametrically opposite to the origin of replication for both the chromosomes, and, as in E. coli, it is likely that the replication termini are located near the dif site. With this assumption, we chose to integrate the operators near dif at an FspI site (coordinate 1514551) in chrI and at an NheI site (coordinate 463720) in chrII (Fig. 1) (Materials and Methods).
FIG. 1.
Maps of chrI and chrII of V. cholerae showing the sites of integration of lac and λcI operator cassettes, the araE transporter gene, and the synthetic operon expressing lacI-eyfp λcI-ecfp repressors. The repressor genes are inducible under the PBAD/araC control. The transporter gene is expressed constitutively from the PCP18 promoter.
We initially employed a pSC101-based plasmid containing an artificial operon comprising lacI-eyfp and λcI-ecfp genes under the PBAD promoter to express the fluorescent proteins. This resulted in nonhomogenous expression of the fluorescent proteins in the population of cells of the culture, either due to an all-or-none response of the PBAD promoter at suboptimal inducer concentrations (18) or due to copy number fluctuations of the pSC101-derived plasmid in V. cholerae. To achieve uniform expression of fusion proteins in the cell population, the araE transporter gene of E. coli was placed under control of a constitutive promoter, CP18 (18). The transporter and the PBADlacI-eyfp λcI-ecfp operon were both integrated in the intergenic region between two hypothetical proteins in chrII at coordinate 312511. The cell-to-cell uniformity of fluorescent protein expression at various suboptimal arabinose concentrations was confirmed by flow cytometry (data not shown).
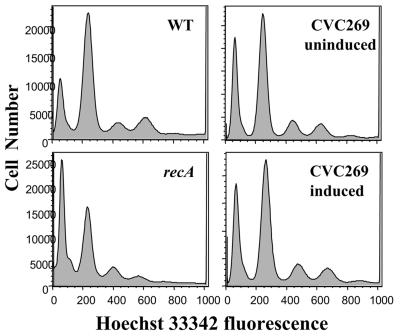
It has been reported that cell growth is impaired when DNA binding proteins fused to fluorescent proteins are expressed in cells that contain their binding site arrays (20). At the level of expression of the fusion proteins used in the present study, no significant changes in cell generation times were observed (data not shown). In fact no effect on growth was seen even when the inducer concentration was 100× the highest concentration used in this study. The replication initiation was also unaffected after induction of fusion proteins for 1 hour as determined by flow cytometry (Fig. 2). Finally, no foci were observed when the fluorescence proteins were expressed without the operator cassettes present in the same cell (data not shown).
FIG. 2.
Flow cytometric analysis of origin content of wild-type V. cholerae (CVC209), of an isogenic recA mutant (CVC250), and of CVC269 (Table 1) before and after induction with 0.2% arabinose, all in M63-plus-glucose medium. Before analysis, log-phase cells were treated with rifampin and cephalexin, which allowed distinct peaks of DNA contents to be seen, representing cells with integral numbers of chromosomes, which equate to number of origins per cell at the time of drug addition. The recA mutant was used to represent asynchronous replication, as is the case in E. coli (33).
Localization of the chrI terminus.
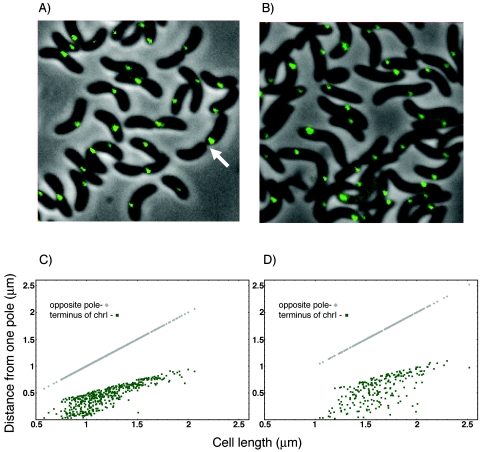
To visualize the operator arrays, fluorescent foci were monitored after exponentially growing cells were transferred to a microscope slide. To locate the foci with respect to cell boundaries, fluorescence, DIC, and phase pictures were taken of the same field. The fluorescence pictures were overlaid onto the phase pictures, and focus position was measured from the pole with a focus nearest to it and plotted with respect to the pole-to-pole distance of the cell (cell length, Fig. 3). DIC pictures were used to examine the initiation of cell septation. In both L broth and M63-glucose media, there was only one focus in most cells (Table 3). The focus position was distributed between the pole and the midcell, but there was a bias of the focus to be at the midcell. Cells with two foci were rare (Table 3) and were seen only in very large cells. It appears that the terminus segregates near the very end of the cell division process (arrow, Fig. 3A).
FIG. 3.
Localization of the terminus region of chrI marked with λOL1 operators. Shown are representative fields of L broth (A)- and M63-plus-glucose (B)-grown cells (CVC244) showing a single fluorescent focus even when invagination is well advanced (arrow). Distribution of focus positions in cells under the two growth conditions is shown in panels C and D, respectively. The focal distance was measured from the cell pole, which was closer to the focus.
TABLE 3.
Distribution of terminus foci in exponentially growing cells
| Terminus region | Medium | No. of cells (% of total no.) with no. of foci:
|
|||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| chrI | L broth | 458 (97) | 14 (3) | 0 | 0 |
| chrI | M63 + glucose | 225 (98) | 4 (2) | 0 | 0 |
| chrII | L broth | 549 (82) | 117 (17) | 2 (0.3) | 4 (0.6) |
| chrII | M63 + glucose | 291 (82) | 65 (18) | 0 | 0 |
Localization of the chrII terminus.
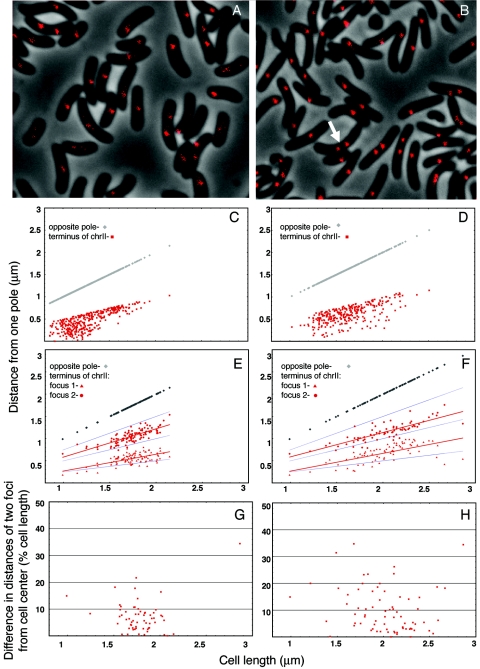
Mapping the position of the chrII terminus showed two differences from that seen for the chrI terminus. The foci were not located as close to the pole in small cells, and cells with two foci were significantly more abundant (arrow, Fig. 4; Table 3). Cells with two foci were most frequent when the cell length exceeded 1.5 μm (Fig. 4E). The cell lengths in these experiments varied from about 0.8 μm to about 2.2 μm. In other words, the segregation of the chrII terminus could occur when the cells reached about middle age, much before cell division. But this was true only in about half of the cells. In the remaining cells larger than 1.5 μm, only one focus was seen that was generally located at midcell. Considering synchrony of replication initiation, we believe the chrII terminus duplicates at a particular cell age, which corresponds to a cell size of ≤1.5 μm, and then either segregates immediately or waits until the end of the division cycle.
FIG. 4.
Localization of the terminus region of chrII marked with 64 lac operators. (A to F) The cells were CVC243; otherwise the details are as in Fig. 3. Note cells with two foci prior to septum formation (arrow). In two-focus cells, focal distances were measured from the pole from which the nearby focus was closer. L-broth-grown cells are represented in panels A, C, and E, and M63-plus-glucose-grown cells are in panels B, D, and F. Distribution of focus positions in cells with one focus is shown in panels C and D and in cells with two foci in panels E and F. The mean focus positions are shown by red lines, and cell half and quarter positions are shown by blue lines (E and F). (G and H) The difference plots show the degree of asymmetry of the two foci with respect to the cell center.
When seen, the two foci were symmetrically distributed with respect to the midcell position (Fig. 4E and H). It seems that these foci separated soon after replication and their separation then increased in proportion to the cell length. The separation, however, did not proceed as far as quarter-cell positions. Although far fewer in number, two foci were also seen in cells smaller than 1.5 μm. This is expected if L broth cells had four origins and hence could have up to four copies of the terminus for chrII.
Switching the operator arrays between lac and λOL1 or their linked drug cassettes between Amp and Kan did not affect the localization patterns for either terminus (data not shown).
Localization of the terminus of chrI and chrII in the same cell.
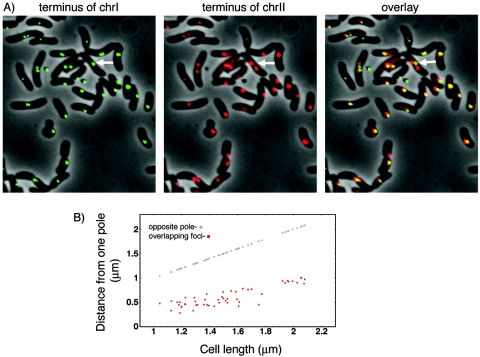
Mapping of the terminus of chrI and chrII simultaneously in the same cells (CVC269) confirmed and extended the results obtained from singly tagged cells. As expected, cells mostly had one focus for the chrI terminus but could have two foci for the chrII terminus (Fig. 5). This supports the view that the chrII terminus can segregate earlier in the cell cycle than the chrI terminus. In larger cells when two chrII foci were present, they were symmetrically disposed with respect to the center of the cell (arrow, Fig. 5). As before, when the terminus regions of chrI and chrII were localized separately the focus positions were broadly distributed in cells with a single focus. However, the overlay showed that they overlap (overlay, Fig. 5A). At least some overlap was seen in all cells with a single focus for each of the two chromosomes. This implies some coordination between the two chromosomes at their terminus region in a cell-length-independent fashion (Fig. 5B).
FIG. 5.
Simultaneous localization of the terminus region of chrI (green) and chrII (red). (A) In cells with two foci for chrII, they are found in either side of the focus for chrI (arrow). In cells with one focus for each chromosome, they were often found to overlap (yellow foci in the overlay). (B) Distribution of the positions of overlapping foci from the cell pole closer to them.
Terminus movement in growing cells.
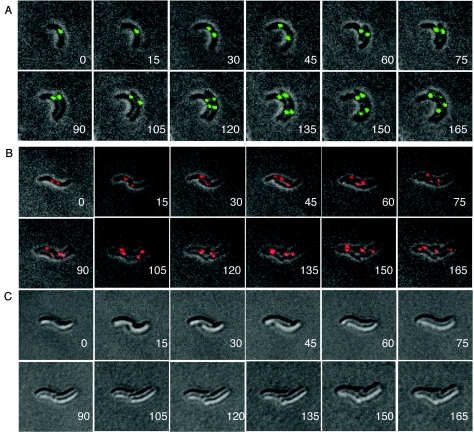
We performed time-lapse microscopy to visualize terminus dynamics in cells placed on a nutrient-containing agarose pad. Our experimental conditions allowed cell growth and periodic fluorescence imaging for about two generations. The 64 operator foci, although adequate for single images, bleached during time-lapse photography. To overcome this, we used a cassette with 128 operators that allowed us to record up to 12 pictures without bleaching.
The focus for the chrI terminus appeared to split only when cell septation was almost complete (30-min panel, Fig. 6A). This resulted in the focus being at the new poles in both the daughter cells. The foci then moved asynchronously away from the poles. Both foci then split again at the division plane, at the end of the second cell cycle (120-min panel, Fig. 6A). This sequence of events is consistent with the inferences drawn from static pictures of a random population and allows us to conclude that in newborn cells the chrI terminus is present at the new pole. Under these conditions, the doubling time initially was approximately 45 min. The growth of the cells slowed as the experiment progressed, extending the doubling time to over 90 min. Growth was observed in more than 90% of the cells of the population.
FIG. 6.
Time-lapse microscopy of cells marked with 128 lac operators at the terminus region of chrI (A) and with 64 λOL1 operators at the terminus region of chrII (B and C). (A) CVC271 cells were grown to log phase in L broth and then transferred to a microscope slide layered with an agarose pad containing threefold-diluted L broth to reduce autofluorescence during imaging. (B) The cells were CVC243; otherwise they were treated as for panel A. (C) DIC pictures of the same cells as in panel B. The number in each panel represents the time interval (min) since the initiation of photography. Note that, in the two daughters of the same cell, segregation of the chrII terminus is apparent by 45 min in one and by 75 min in the other.
In the case of the chrII terminus, cells with two foci but without an intervening cell septum were common (0-min panel, Fig. 6B). Even though after cell division the single focus was not localized, it separated into two only at the midcell (by 45 min in one daughter and by 75 min in the other; Fig. 6B). From DIC pictures it was clear that the splitting occurred before the appearance of cell septation (0-, 45-, and 75-min panels, Fig. 6C). The example of Fig. 6B also highlights the fact that chrII terminus segregation can happen at two different times in the daughters of the same cell. In summary, the terminus of chrII is proximal to the new poles in newborn cells, as is the case for chrI, but its segregation is less intimately connected to the cell septation event.
Localization of terminus relative to the division septum.
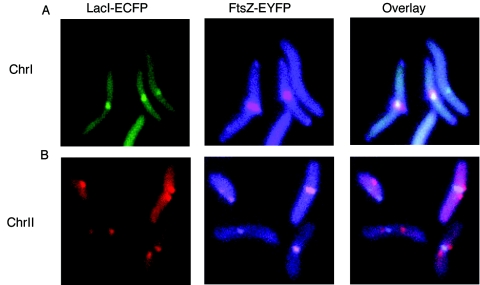
To visualize the relative position of the two terminus regions with respect to the division septum, we first integrated the lac operators near the dif site of chrI or chrII in CVC301. These cells were transformed with pLAU85 to supply LacI-ECFP and FtsZ-EYFP. Although ftsZ in pLAU85 was derived from E. coli, the gene product is 75% identical to the FtsZ of V. cholerae. An FtsZ ring was found at the cell center in the majority of the cases. However in some cells it was also found at the poles as observed during the Caulobacter crescentus cell cycle (30). Expressing FtsZ-EYFP in CVC301 without the operators also gave similar localization of the FtsZ ring (data not shown). In cells where both the focus for the chrI terminus and the FtsZ ring were seen, they colocalized at the cell center in all cases (Fig. 7). For chrII, when both a terminus focus and the Z ring were seen in the same cell, in 85% of cases there were two terminus foci on opposite side of the Z ring. In the remaining 15% of cases, a single terminus focus was seen overlapping the Z ring. Apparently, by the time the Z ring forms, the terminus regions of both chrI and chrII have already arrived at the cell center. There were also cells with two terminus foci but no Z ring (these represent 24% of the two-focus cells). This suggests that completion of chrII segregation can be independent of the FtsZ ring formation.
FIG. 7.
Simultaneous localization of the FtsZ ring and the terminus focus. (A) Focus for the chrI terminus (green) and the ring of FtsZ (purple) and the overlay of the two panels. (B) Same as panel A except that the foci (red) represent the terminus region of chrII.
DISCUSSION
Bacterial chromosomes can segregate either coreplicationally (24) or in stages because of sister chromosome cohesion (1). In the coreplicational segregation model, the prediction in the case of V. cholerae with two unequally sized chromosomes would be that the terminus of the shorter one should segregate earlier than the longer one, replication initiation being synchronous for the two. In the case of cohesion, depending upon the nature and duration of cohesion, the difference in segregation timing of the two terminus regions may not be that different or may even vanish. For example, in eukaryotes, activation of separase at the metaphase-to-anaphase transition allows segregation of all chromosomes at the same time (40). It is conceivable that, in multichromosome bacteria, there could be control at the stage of termination of replication, such that terminus segregation waits until replication of all chromosomes is completed.
We find that the terminus of the larger chrI segregates when cell septation is about to be completed, as has been found for the E. coli chromosome (20, 25). The results for the shorter chrII are different. Its terminus region segregates well before cell septation in about half of cells. In the rest, only a single terminus focus is seen at the midcell in mother cells, as is the case for chrI. Thus, chrII terminus can segregate both early and late in the cell cycle. We suggest that a nonspecific process, like chromosome condensation, suffices for completing segregation of chrII, but in case this type of mechanism proves inadequate, cell septation triggers a separase function, such as dimer resolution at dif. We note, however, that although terminus segregation appears to coincide with cell septation, the two processes may not be related. In cells treated with cephalexin, a drug that inhibits cell septation, the terminus focus split for both chrI and chrII in all elongated cells (data not shown).
In cells with two chrII terminus foci, the foci were well separated but did not quite reach the quarter-cell positions (Fig. 4). The origin of V. cholerae chrII was found either in mid- or in quarter-cell positions (10). The terminus could follow the origin, as would be expected from coreplicational segregation. We note, however, that the terminus focus was not randomly distributed around the quarter-cell positions but biased more towards the midcell. The reason for this bias remains to be understood but could be a consequence of mechanisms like oriented transcription and transertion that could restrict free diffusion of the terminus region around the cell-quarter positions (5, 37). Since the terminus region of the two chromosomes often overlapped (Fig. 5B), another mechanism could have also constrained their positioning.
As alluded to earlier, a reason for the heterogeneous behavior of the chrII terminus could be cohesion of the sister chromosomes. Although the origin content is expected to increase in L broth compared to M63-plus-glucose-grown cells, we find that the number of terminus foci per cell remained the same in both the media (Table 3). This is expected for the chrI terminus from the E. coli paradigm, where the terminus number does not increase irrespective of the growth rate. But there might have been four terminus foci for chrII, since it is expected to complete replication much earlier than chrI. Although rarely, in young cells (smaller than 1.5 μm) two foci were seen for the chrII terminus (Fig. 4E). Thus, in L-broth-grown cells, a focus could represent two chrII terminus regions or four regions in large cells that remain conjoined until acted upon by cell septation.
A common finding between the foci for the terminus of chrI and chrII was that, although they can occupy apparently random positions, they arrived in midcell before segregation (Fig. 3 to 7). Both termini are at midcell by the time the cell reaches 1.5 μm (Fig. 3C and 4C). In fact, between 1- and 1.5-μm cell length, the distribution of the focus positions of the two chromosomes looks quite similar (Fig. 3C and 4C), and when seen together in the same cell, the foci for the two chromosomes almost always overlapped (overlay, Fig. 5). We cannot speculate at the moment what could be the purpose, if any, of this apparent togetherness.
The arrival of the two terminus regions at the midcell and splitting of chrII terminus there could mean that the terminus region of both the chromosomes duplicates in the midcell, which in turn predicts that replication factories involved are also in the cell center at the time of terminus duplication. This is expected from our current knowledge of replication factory localization in E. coli (20), Bacillus subtilis (22), and C. crescentus (17). For chromosomal replication, the factory could be stationary at the midcell, as is believed to be the case in E. coli and B. subtilis, or could be mobile, being at a pole during initiation of replication but then moving gradually to midcell at the end of replication, as in C. crescentus. Interestingly, studies of origin localization in V. cholerae suggest that its chromosomes are replicated differently: chrI is replicated by a mobile factory and chrII by a stationary factory (10). Since in either mode the terminus should replicate at the midcell, our data, although consistent, do not substantiate either model.
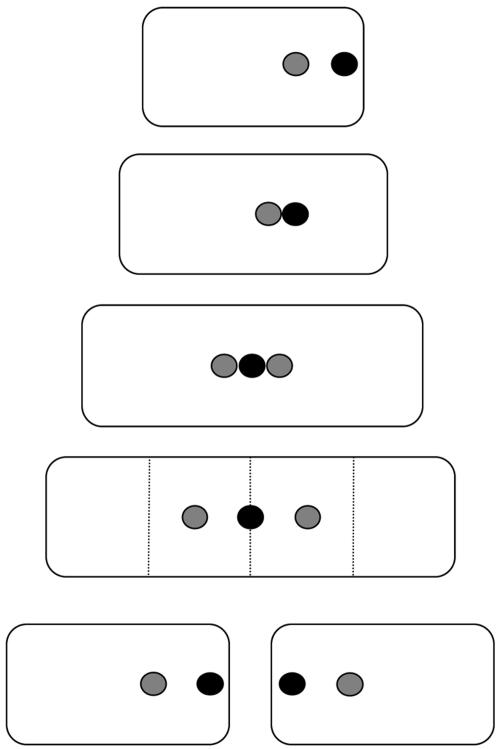
Our data are summarized in the model shown in Fig. 8. It shows that the chrI terminus is at the new pole of a newborn cell and it segregates when the mother cell divides. The focus representing the chrII terminus is on an average more centrally located in newborn cells and splits at the midcell before cell division. The separation of the daughter foci increases in proportion to cell length but does not reach quarter-cell positions. Although not shown, the focus for chrII terminus can also behave like the chrI focus, in which case it does not divide until the time of cell septation. The basis for this heterogeneous behavior remains to be studied. The synchronous segregation of the terminus region of chrI and chrII in at least half of the cells suggests that there could be coordination between the two chromosomes at the stage of replication termination as has been found for replication initiation (6).
FIG. 8.
A model showing positions of terminus region for chrI (black dots) and chrII (gray dots) during a bacterial division cycle. The vertical dotted lines represent cell half or quarter positions. In this model, the terminus of chrI traverses from the new pole to the midcell (i.e., spans only the new-pole half of the cell). The terminus region of both the chromosomes duplicates in midcell position. The terminus of chrII is seldom polar and preferentially stays close to the midcell position overlapping with the terminus for chrI.
Acknowledgments
We are grateful to Matt Waldor for strains, David Sherratt and Christophe Possoz for the plasmid pLAU85, Joe Pogliano for instructions on time-lapse microscopy, and Michael Lichten and Don Biek for critical reading of the manuscript.
REFERENCES
- 1.Bates, D., and N. Kleckner. 2005. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121:899-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Yehuda, S., M. Fujita, X. S. Liu, B. Gorbatyuk, D. Skoko, J. Yan, J. F. Marko, J. S. Liu, P. Eichenberger, D. Z. Rudner, and R. Losick. 2005. Defining a centromere-like element in Bacillus subtilis by identifying the binding sites for the chromosome-anchoring protein RacA. Mol. Cell 17:773-782. [DOI] [PubMed] [Google Scholar]
- 3.Corre, J., and J. M. Louarn. 2002. Evidence from terminal recombination gradients that FtsK uses replichore polarity to control chromosome terminus positioning at division in Escherichia coli. J. Bacteriol. 184:3801-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dworkin, J., and R. Losick. 2002. Does RNA polymerase help drive chromosome segregation in bacteria? Proc. Natl. Acad. Sci. USA 99:14089-14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egan, E. S., A. Lobner-Olesen, and M. K. Waldor. 2004. Synchronous replication initiation of the two Vibrio cholerae chromosomes. Curr. Biol. 14:R501-R502. [DOI] [PubMed] [Google Scholar]
- 7.Egan, E. S., and M. K. Waldor. 2003. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell 114:521-530. [DOI] [PubMed] [Google Scholar]
- 8.Espeli, O., and K. J. Marians. 2004. Untangling intracellular DNA topology. Mol. Microbiol. 52:925-931. [DOI] [PubMed] [Google Scholar]
- 9.Fekete, R. A., and D. K. Chattoraj. 2005. A cis-acting sequence involved in chromosome segregation in Escherichia coli. Mol. Microbiol. 55:175-183. [DOI] [PubMed] [Google Scholar]
- 10.Fogel, M. A., and M. K. Waldor. 2005. Distinct segregation dynamics of the two Vibrio cholerae chromosomes. Mol. Microbiol. 55:125-136. [DOI] [PubMed] [Google Scholar]
- 11.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329-341. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, G. S., D. Sitnikov, C. D. Webb, A. Teleman, A. Straight, R. Losick, A. W. Murray, and A. Wright. 1997. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell 90:1113-1121. [DOI] [PubMed] [Google Scholar]
- 13.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes, V. F., and N. R. Cozzarelli. 2000. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc. Natl. Acad. Sci. USA 97:1322-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber, K. E., and M. K. Waldor. 2002. Filamentous phage integration requires the host recombinases XerC and XerD. Nature 417:656-659. [DOI] [PubMed] [Google Scholar]
- 16.Ip, S. C., M. Bregu, F. X. Barre, and D. J. Sherratt. 2003. Decatenation of DNA circles by FtsK-dependent Xer site-specific recombination. EMBO J. 22:6399-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen, R. B., S. C. Wang, and L. Shapiro. 2001. A moving DNA replication factory in Caulobacter crescentus. EMBO J. 20:4952-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khlebnikov, A., K. A. Datsenko, T. Skaug, B. L. Wanner, and J. D. Keasling. 2001. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147:3241-3247. [DOI] [PubMed] [Google Scholar]
- 19.Kruse, T., and K. Gerdes. 2005. Bacterial DNA segregation by the actin-like MreB protein. Trends Cell Biol. 15:343-345. [DOI] [PubMed] [Google Scholar]
- 20.Lau, I. F., S. R. Filipe, B. Soballe, O. A. Okstad, F. X. Barre, and D. J. Sherratt. 2003. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol. 49:731-743. [DOI] [PubMed] [Google Scholar]
- 21.Lemon, K. P., and A. D. Grossman. 2001. The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev. 15:2031-2041. [DOI] [PubMed] [Google Scholar]
- 22.Lemon, K. P., and A. D. Grossman. 2000. Movement of replicating DNA through a stationary replisome. Mol. Cell 6:1321-1330. [DOI] [PubMed] [Google Scholar]
- 23.Lesterlin, C., F. X. Barre, and F. Cornet. 2004. Genetic recombination and the cell cycle: what we have learned from chromosome dimers. Mol. Microbiol. 54:1151-1160. [DOI] [PubMed] [Google Scholar]
- 24.Li, Y., K. Sergueev, and S. Austin. 2002. The segregation of the Escherichia coli origin and terminus of replication. Mol. Microbiol. 46:985-996. [DOI] [PubMed] [Google Scholar]
- 25.Li, Y., B. Youngren, K. Sergueev, and S. Austin. 2003. Segregation of the Escherichia coli chromosome terminus. Mol. Microbiol. 50:825-834. [DOI] [PubMed] [Google Scholar]
- 26.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLeod, S. M., and M. K. Waldor. 2004. Characterization of XerC- and XerD-dependent CTX phage integration in Vibrio cholerae. Mol. Microbiol. 54:935-947. [DOI] [PubMed] [Google Scholar]
- 28.Migocki, M. D., P. J. Lewis, R. G. Wake, and E. J. Harry. 2004. The midcell replication factory in Bacillus subtilis is highly mobile: implications for coordinating chromosome replication with other cell cycle events. Mol. Microbiol. 54:452-463. [DOI] [PubMed] [Google Scholar]
- 29.Moller-Jensen, J., R. B. Jensen, and K. Gerdes. 2000. Plasmid and chromosome segregation in prokaryotes. Trends Microbiol. 8:313-320. [DOI] [PubMed] [Google Scholar]
- 30.Moller-Jensen, J., R. B. Jensen, J. Lowe, and K. Gerdes. 2002. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 21:3119-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philippe, N., J. P. Alcaraz, E. Coursange, J. Geiselmann, and D. Schneider. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246-255. [DOI] [PubMed] [Google Scholar]
- 32.Sherratt, D. J. 2003. Bacterial chromosome dynamics. Science 301:780-785. [DOI] [PubMed] [Google Scholar]
- 33.Skarstad, K., R. Bernander, and E. Boye. 1995. Analysis of DNA replication in vivo by flow cytometry. Methods Enzymol. 262:604-613. [DOI] [PubMed] [Google Scholar]
- 34.Viollier, P. H., M. Thanbichler, P. T. McGrath, L. West, M. Meewan, H. H. McAdams, and L. Shapiro. 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. USA 101:9257-9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waldor, M. K., and D. RayChaudhuri. 2000. Treasure trove for cholera research. Nature 406:469-470. [DOI] [PubMed] [Google Scholar]
- 36.Webb, C. D., A. Teleman, S. Gordon, A. Straight, A. Belmont, D. C. Lin, A. D. Grossman, A. Wright, and R. Losick. 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88:667-674. [DOI] [PubMed] [Google Scholar]
- 37.Woldringh, C. L. 2002. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol. Microbiol. 45:17-29. [DOI] [PubMed] [Google Scholar]
- 38.Wu, L. J., and J. Errington. 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117:915-925. [DOI] [PubMed] [Google Scholar]
- 39.Yamaichi, Y., and H. Niki. 2004. migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J. 23:221-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanagida, M. 2005. Basic mechanism of eukaryotic chromosome segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:609-621. [DOI] [PMC free article] [PubMed] [Google Scholar]