Abstract
The Bacillus subtilis transcriptional regulator Fnr is an integral part of the regulatory cascade required for the adaptation of the bacterium to low oxygen tension. The B. subtilis Fnr regulon was defined via transcriptomic analysis in combination with bioinformatic-based binding site prediction. Four distinct groups of Fnr-dependent genes were observed. Group 1 genes (narKfnr, narGHJI, and arfM) are generally induced by Fnr under anaerobic conditions. All corresponding promoters contain an essential Fnr-binding site centered −41.5/−40.5 bp upstream of the transcriptional start point, suggesting their induction by direct Fnr interaction. Group 2 genes (alsSD, ldh lctP, ywcJ, and cydABCD) are characterized by anaerobic repression in the presence of nitrate. Mutational analysis of the Fnr-binding sites found in three of the corresponding promoters excluded their function in Fnr-mediated repression. Genetic evidence showing that group 2 genes are anaerobically repressed by nitrate reductase formation was accumulated. A possible role of the redox regulator YdiH in the regulation of group 2 genes was initially investigated. Group 3 genes are characterized by their Fnr-dependent activation in the presence of nitrate and the lack of an Fnr-binding site in their promoters. The analysis of Group 3 gene transcription (ykuNOP and ydbN) indicated that Fnr induces nitrate reductase production, which leads to the formation of the regulatory compound nitrite from nitrate. Finally, the group 4 operon acoABCL, lacking an Fnr-binding site, requires Fnr-dependent nitrate reductase formation for its general anaerobic induction. A regulatory model for the observed complex Fnr-mediated gene expression was deduced.
The gram-positive model organism Bacillus subtilis adapts to an anaerobic environment by changing its metabolic activity (26). Under anaerobic conditions, B. subtilis performs a mixed-acid fermentation with lactate, acetate, and acetoin as the major products (7, 24). In the presence of nitrate, B. subtilis performs the respiratory process of ammonification (8, 13, 14, 22).
The regulatory network underlying anaerobic adaptation has been extensively studied during the last decade. A regulatory cascade describing the coordinated regulation of genes involved in anaerobiosis was established (7). One major regulatory switch in the adaptation to anaerobiosis is the two-component system ResDE (32, 35). While the mechanism of signal perception by ResDE is still unknown, the downstream regulatory network is elucidated to a significant depth. Activated ResD binds to promoter regions of nasDE, encoding the nitrite reductase, the flavohemoglobin gene hmp, and the gene encoding the redox regulator Fnr (23, 25). Fnr in turn is responsible for the induction of the narGHJI operon and narK, encoding the respiratory nitrate reductase and a potential nitrite extrusion protein, respectively (6, 24). Mutation of fnr strongly affects anaerobic growth of B. subtilis on nitrate (6, 24). Furthermore, Fnr activates the expression of the arfM gene encoding an anaerobic respiration and fermentation modulator protein by direct interaction with the arfM promoter region (16). The promoter regions of all three Fnr-regulated genes carry the highly conserved potential B. subtilis Fnr-binding site (TGTGA-N6-TCACA) centered 41.5/40.5 bp from the transcriptional start point. Complementation experiments using an Escherichia coli crp mutant revealed that the DNA-binding domain of Fnr of B. subtilis is similar to that of Crp from E. coli, the well-studied cyclic AMP receptor protein (6). In B. subtilis, additional potential Fnr-binding sites were found in the promoter regions of a second potential nitrite transporter gene, ywcJ, as well as the fermentation operons ldh-lctP and alsSD (6, 7, 34). The latter operons encode lactate dehydrogenase, lactate permease, and acetolactate synthase and acetolactate decarboxylase, respectively. Transcription of alsSD and ldh-lctP was found to be anaerobically induced and repressed by the presence of nitrate (7). Nitrate repression was related to nitrate reductase activity (7).
Global transcriptional profiling was used to analyze changes in the mRNA population after adaptation to anaerobic growth conditions (35). Several hundred genes were observed to be induced or repressed under anaerobic conditions. Additionally, changes in gene expression patterns were measured for an anaerobically grown resDE mutant strain. About 50 genes, including 15 operons, were found to be ResDE dependent (35). However, these results did not distinguish between genes that are directly regulated by ResDE and indirect effects of ResDE resulting from the induction of fnr transcription.
In this study, the Fnr regulon was intensively characterized by an experimental approach combining DNA macroarrays and a bioinformatic investigation based on transcription factor binding site predictions. Identified promoters of interest were further investigated using genetic and molecular biology methods. Finally, a model of the Fnr regulon was proposed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All B. subtilis strains used in this study are listed in Table 1. B. subtilis strains were grown in Spizizen's minimal medium (10) supplemented with 10 mM sodium nitrate or 10 mM sodium nitrite where indicated. For aerobic growth, bacteria were inoculated with a culture grown overnight to a final optical density at 578 nm of 0.05 and grown in shake flasks with vigorous agitation (250 rpm) for the indicated times. For anaerobic growth, bacteria were inoculated with a culture grown overnight to a final optical density at 578 nm of 0.25 and incubated in transfusion flasks filled up to the top under conditions detailed above. For RNA preparations, bacteria were harvested at mid-log phase. For β-galactosidase assays, bacteria were harvested at late mid-log phase.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| JH642 | trpC2 pheA1 | BGSCa |
| THB1 | trpC2 pheA1 narGH::tet | 14 |
| THB2 | trpC2 pheA1 fnr::spec | 16 |
| LUW48 | trpC2 amyE::cydA′-lacZ | 33 |
| LUW273 | ydiHΩ pYDIH1 spec | 15 |
| QB700 | trpC2 amyE::acoA-lacZ cat | 1 |
| TDB2 | trpC2 pheA1 amyE::narG lacZ cat | This work |
| HRB5 | trpC2 pheA1 fnr::spec amyE::narG lacZ cat | This work |
| HRB6 | trpC2 pheA1 narG::tet amyE::narG lacZ cat | This work |
| TDB8 | trpC2 pheA1 amyE::Δfnr(−49/−48 TG→CC) narG lacZ cat | This work |
| HRB7 | trpC2 pheA1 amyE::alsS lacZ cat | This work |
| HRB8 | trpC2 pheA1 fnr::spec amyE::alsS lacZ cat | This work |
| HRB9 | trpC2 pheA1 narGH::tet amyE::alsS lacZ cat | This work |
| HRB10 | trpC2 pheA1 amyE::Δfnr(−4/−5/−6 ACA→CGG) alsS lacZ cat | This work |
| HRB11 | trpC2 pheA1 fnr::spec amyE::Δfnr(−4/−5/−6 ACA→CGG) alsS lacZ cat | This work |
| HRB19 | trpC2 pheA1 amyE::ywcJ lacZ cat | This work |
| HRB20 | trpC2 pheA1 fnr::spec amyE::ywcJ lacZ cat | This work |
| HRB21 | trpC2 pheA1 amyE::Δfnr(−37/−36 TG→CC) ywcJ lacZ cat | This work |
| HRB22 | trpC2 pheA1 narGH::tet amyE::ywcJ lacZ cat | This work |
| HRB23 | trpC2 pheA1 amyE::cydA′-lacZ | This work |
| HRB24 | trpC2 pheA1 fnr::spec amyE::cydA′-lacZ | This work |
| HRB25 | trpC2 pheA1 narGH::tet amyE::cydA′-lacZ | This work |
| HRB26 | trpC2 pheA1 amyE::acoA-lacZ | This work |
| HRB27 | trpC2 pheA1 narGH::tet amyE::acoA-lacZ | This work |
| HRB28 | trpC2 pheA1 narGH::tet amyE::acoA-lacZ | This work |
| HRB29 | ydiHΩ pYDIH1 spec amyE::ywcJ lacZ cat | This work |
BGSC, Bacillus Genetic Stock Center.
Preparation of RNA and Northern blot analysis.
Preparation of RNA and Northern blot analysis were performed as described elsewhere previously (11).
DNA macroarray analysis.
For the synthesis of labeled cDNA, 2 μg of total cellular RNA prepared from B. subtilis strains JH642 (wild type) and THB2 (fnr mutant) was mixed with 4 μl of primer labeling mix (Sigma-Genosys Ltd., The Woodlands, Tex.) and adjusted with water to a final volume of 12 μl. The samples were heated to 95°C for 10 min and subsequently incubated for 30 min at 42°C to allow annealing of the primers. For reverse transcription, a solution containing 10 μl first-strand buffer (supplied with the reverse transcriptase), 10 mM dithiothreitol, 400 mM dATP, 400 mM dGTP, 400 mM dTTP, 45 μCi [α-33P]dCTP (Amersham Biosciences, Freiburg, Germany), and 300 U SuperScript II reverse transcriptase (Invitrogen Life Technologies GmbH, Karlsruhe, Germany) was mixed, and cDNA was synthesized for 1.5 h at 42°C in a total volume of 50 μl. Further steps were performed as described elsewhere previously (31).
Macroarray data analysis.
Hybridized DNA arrays were read out using a phosphorimager (Molecular Imager FX; Bio-Rad Laboratories, Hercules, Calif.), and obtained data were processed with ArrayVision software (version 6.0; Imaging Research, St. Catherine's, Ontario, Canada). Background levels were deduced from the area surrounding all spots. A quality score was calculated from these values and used to distinguish absent, median, and present signals. Data normalization was done using GeneSpring software (version 4.2; Silicon Genetics, Redwood City, Calif.). Three independent measurements for each analyzed condition and mutant were performed. Results obtained were averaged accordingly. Finally, data were exported to Microsoft Excel, and expression ratios were calculated (raw data are presented in the supplemental material).
β-Galactosidase assays.
Crude cell extracts were prepared as described previously (11). β-Galactosidase assays were performed as described elsewhere previously (17).
Construction of the reporter gene fusions and site-directed mutagenesis of potential Fnr-binding sites.
A transcriptional fusion between the E. coli lacZ gene and the alsS upstream region was constructed. An 894-bp PCR fragment spanning the region from positions −341 to +540 relative to the translational start point of alsS was amplified with the primers EH46 (5′-AGTTGAATTCCTTGTCCGATTTG-3′) and EH47 (5′-GTGGATCCTGCCCTGCTGACGCTAT-3′). Using the restriction sites for EcoRI and BamHI created by the primers (underlined), we cloned the promoter region of alsS into plasmid pDIA5322 (6), resulting in plasmid palsS-lacZ. This plasmid was transformed into B. subtilis JH642 (wild type), THB2 (fnr mutant), and THB1 (narG mutant) strains. Transformants were screened for double-crossover integration at the amyE locus, resulting in strains HRB7, HRB8, and HRB9, respectively.
A similar cloning strategy was used to create narG-lacZ and ywcJ-lacZ transcriptional fusions. A 456-bp PCR fragment spanning the region from positions −243 to +197 relative to the translational start point of narG was amplified with the primers EH18 (5′-GCGGATCCAATATTCCAGCTGCAAGA-3′) and EH19 (5′-CGGAATTCGGTATCTGCATACATCAC-3′) (restriction sites are underlined). Cloning of the PCR fragment into pDIA5322 resulted in plasmid pnarG-lacZ. Transformation into B. subtilis strains JH642 (wild type), THB2 (fnr mutant), and THB1 (narG mutant) resulted in strains TDB2, HRB5, and HRB6, respectively. A 395-bp PCR fragment spanning the region from positions −203 to +176 relative to the translational start point of ywcJ was amplified with the primers EH20 (5′-CGGAATTCGCCTGCTTTACCAGTCAC-3′) and EH21 (5′-GCGGATCCAACGGAGAATCAGCCATA-3′). Cloning of the PCR fragment into pDIA5322 resulted in plasmid pywcJ-lacZ. Transformation into B. subtilis JH642 (wild type), THB2 (fnr mutant), and THB1 (narG mutant) strains resulted in strains HRB19, HRB20, and HRB22, respectively.
To analyze cydA-lacZ, genomic DNA from strain LUW48 (33) was transformed into B. subtilis JH642 (wild type), THB2 (fnr mutant), and THB1 (narG mutant) strains. After selection for homologous recombination at the amyE locus, strains HRB23, HRB24, and HRB25 were obtained. The acoA-lacZ fusion of B. subtilis strain QB700 (1) was transformed into the amyE locus of B. subtilis JH642 (wild type), THB2 (fnr mutant), and THB1 (narG mutant), which resulted in strains HRB26, HRB27, and HRB28, respectively. To obtain strain HRB29, we transformed strain LUW273 (ydiH mutant) with plasmid pywcJ-lacZ (15).
The potential Fnr-binding sites in the promoter regions of alsS, narG, and ywcJ were mutated using primers for crossover PCR (12). The potential Fnr-binding site of alsS was changed from AGTGA-CT-TCACA to AGTGA-CT-ATCCGG (exchanged bases are shown in boldface type). Crossover PCRs were performed with the following two primers containing the desired base exchanges (in boldface type): EH66 (5′-AGAGTGTATAGTGAAACTTATCCGGAGATA-3′) and EH67 (5′-TATCTCCGGATAAGTTTCCGGATACACTCT-3′). Two PCR products were generated with primer pairs EH46-EH67 (356 bp) and EH66-EH47 (568 bp). In a second PCR, we used the first two PCR products as templates and amplified the whole promoter region with the primer pair EH46-EH47. The complete promoter fragments were cloned into the plasmid pDIA5322 as described above for the wild-type sequence, resulting in the plasmid palsS(Δfnr)-lacZ. After transformation into B. subtilis strain JH642 and selection for double-crossover integration at the amyE locus, strain HRB10 was obtained.
The potential Fnr-binding site of narG was changed from 5′-TGTGA-TA-TCACA-3′ to CCTGA-TA-TCACA (exchanged bases are shown in boldface type). Crossover PCRs were performed with the following two primers containing the desired base exchanges (in boldface type): EH108 (5′-AAAAGCAGAGTGCCTGACATAGTT-3′) and EH109 (5′-ACTATGTCAGGCACTCTGCTTTT-3′). Two PCR products were generated with primer pairs EH19-EH109 (241 bp) and EH18-EH108 (291 bp). In a second PCR, we used the first two PCR products as templates, amplified the whole narG promoter region with the primer pair EH18-EH19, and cloned it into the plasmid pDIA5322 to create plasmid pnarG(Δfnr)-lacZ. Transformation of B. subtilis strain JH642 yielded strain TDB8.
The potential Fnr-binding site of ywcJ was changed from TGTGA-TA-TCACA to CCTGA-TA-TCACA (exchanged bases are shown in boldface type). Crossover PCRs were performed with the following two primers containing the desired base exchanges (in boldface type): EH24 (5′-TATTTCAGGATTAATTTTTACGAAT-3′) and EH25 (5′-ATTCGTAAAAATTAATCCTGAAATA-3′). Two PCR products were generated with primer pairs EH20-EH24 (156 bp) and EH21-EH25 (264 bp). The whole ywcJ promoter region was generated in a second PCR with the primer pair EH20-EH21 using the two PCR products as templates and cloned into the plasmid pDIA5322 to create plasmid pywcJ(Δfnr)-lacZ. Transformation of B. subtilis strain JH642 resulted in strain HRB21. In general, all transformants were tested for amylase activity to ensure integration of the promoter-lacZ construct at the amyE locus. In addition, two independently obtained clones of each newly constructed strain were used for β-galactosidase assays. All cloned fragments made by PCR were sequenced to check for PCR-induced errors.
Primer extension.
For each primer extension analysis, 50 μg of total cellular RNA was used. Reverse transcription was initiated from the γ-32P-end-labeled primer EH41 (5′-GGCCAAAATGGACCGAAGCACATAACG-3′) for ywcJ and primer EH113 (5′-TTATTTGAATGGTGTTCGATAGGAGAG-3′) for narG according to a standard procedure (2). The sequencing reactions were performed with the same primers used for reverse transcription. The primer extension products and the sequencing reactions were analyzed on a 6% denaturing polyacrylamide gel in Tris-borate buffer. The dried gel was analyzed by phosphorimaging.
Prediction of Fnr-binding sites.
A position weight matrix (PWM) model of the Fnr-binding site was created by use of an aligned training set consisting of three sequences from the PRODORIC database (19). The PWM was computed using the widely accepted information theoretical approach, with some modifications (28). At first, the information vector Rsequence(l) was generated from the alignment where f(b,l) is the frequency of the base b at position l and was calculated as follows: 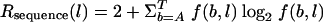 . We additionally considered the nucleotide bias in genomes by using a linear correction of noise (30). The position weight matrix m(b,l) values were calculated afterwards as follows: m(b,l) = f(b,l) · Rsequence(l). For the case where f(b,l) = 0, we introduced a penalty function in dependence of the sample size n as follows:
. We additionally considered the nucleotide bias in genomes by using a linear correction of noise (30). The position weight matrix m(b,l) values were calculated afterwards as follows: m(b,l) = f(b,l) · Rsequence(l). For the case where f(b,l) = 0, we introduced a penalty function in dependence of the sample size n as follows: 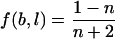 . Using the PWM as a scoring function, a genome-wide search for potential binding sites was performed using the program “Virtual Footprint” (20). Theprogram is interconnected with the PRODORIC database (accessible at http://www.prodoric.de/vfp). The sequence logo was created using WebLogo software (http://weblogo.berkeley.edu) (29).
. Using the PWM as a scoring function, a genome-wide search for potential binding sites was performed using the program “Virtual Footprint” (20). Theprogram is interconnected with the PRODORIC database (accessible at http://www.prodoric.de/vfp). The sequence logo was created using WebLogo software (http://weblogo.berkeley.edu) (29).
RESULTS AND DISCUSSION
Expression profiling using DNA macroarrays for definition of the Fnr regulon in B. subtilis.
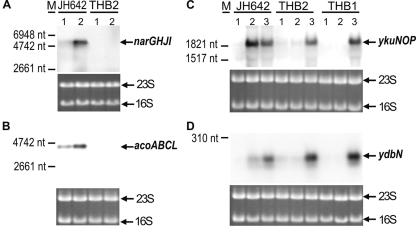
The global transcription profiles of fnr mutant strain THB2 and B. subtilis wild-type strain JH642 grown under fermentative conditions or nitrate-respiratory conditions were compared. To establish appropriate anaerobic growth conditions for RNA preparation, we monitored the anaerobic and Fnr-dependent expression of the narGHJI operon by Northern blot analysis. A single transcript of about 6,400 nucleotides (nt), which corresponds to the size of the narGHJI operon, was detected using a narG-specific RNA probe in total cellular RNA isolated from fermentatively grown cells. The amount of narGHJI transcript was found to be fourfold higher when nitrate was present in the growth medium (Fig. 1A). The narGHJI transcript was not present in RNA prepared from B. subtilis fnr mutant strain THB2. These results indicated that proper growth conditions were used for the RNA preparation.
FIG. 1.
Influence of anaerobiosis on transcription of narGHJI, acoABCL, ykuNOP, and ydbN in B. subtilis wild-type (JH642), narG mutant (THB1), and fnr mutant (THB2) strains. Northern blot hybridizations were performed with RNA extracted from cells grown under the following anaerobic conditions: fermentative (1), nitrate respiratory (2), and with the addition of nitrite (3). A narG-specific RNA probe detected a single transcript of 6.4 kb (A), and an acoA-specific probe detected a single transcript of 4,637 nt (B). Both transcripts were only found in wild-type cells. A threefold nitrate-dependent induction of both transcripts is visible when lanes 1 and 2 are compared. A single transcript of 1,900 nt was detected with the ykuNOP-specific probe (C), while the ydbN-specific probe detected an approximately 200-nt transcript, which corresponds to a single ydbN transcript (D). No obvious transcripts were detected for conditions of anaerobic fermentation in the wild-type strain and for fermentative and nitrate-respiratory conditions in the mutant strains. Ethidium bromide staining of the gels showed that equal amounts of RNA were analyzed. The size standard was RNA molecular weight marker no. 1 (Roche Diagnostics GmbH, Mannheim, Germany) and the 16S and 23S rRNA species.
Fnr-dependent transcription of B. subtilis genes was subsequently investigated by hybridization of labeled cDNAs prepared from mRNA isolated from the anaerobically grown B. subtilis wild type and a corresponding fnr mutant to DNA macroarrays representing about 4,100 of the genes of the organism. In total, we found the expression of 37 genes, including 10 operons, affected by the fnr mutation (Table 2).
TABLE 2.
Transcriptome analysis for definition of the Fnr regulona
| Gene | Description | Putative Fnr box | Induction/repression (fold)
|
|
|---|---|---|---|---|
| Fermentation | Nitrate respiration | |||
| Group 1 | ||||
| narG | Nitrate reductase (alpha subunit) | ++ | 1.3 | 1.0 |
| narH | Nitrate reductase (beta subunit) | 2.0 | 1.8 | |
| narI | Assembling factor | 2.1 | 5.6 | |
| narJ | Nitrate reductase (gamma subunit) | 3.1 | 1.9 | |
| narK | Nitrite extrusion protein | ++ | 1.6 | 1.4 |
| fnr | Anaerobic redox regulator | 1.6 | 4.1 | |
| arfM | Anaerobic modulator | ++ | 1.7 | 2.2 |
| Group 2 | ||||
| alsS | Alpha-acetolactate decarboxylase | + | (1.3) | (1.9) |
| alsD | Alpha-acetolactate synthase | (2.8) | (2.2) | |
| cydA | Cytochrome bd ubiquinol oxidase (I) | − | (1.8) | (1.3) |
| cydB | Cytochrome bd ubiquinol oxidase (II) | (1.4) | (3.3) | |
| cydC | ABC transporter | (1.3) | (2.5) | |
| cydD | ABC transporter | (1.4) | (2.0) | |
| ldh | l-Lactate dehydrogenase | + | (1.4) | (17.6) |
| lctP | l-Lactate permease | (0.9) | (4.0) | |
| ywcJ | Similar to formate/nitrite transporter | + | (1.2) | (1.4) |
| Group 3 | ||||
| dhbA | 2,3-Dihydro-2,3-DHB dehydrogenase | 1.6 | 1.8 | |
| dhbB | Isochorismatase | 1.3 | 5.5 | |
| dhbC | Isochorismate synthase | − | 1.5 | 5.1 |
| dhbE | 2,3-DHB-AMP ligase | 1.1 | 2.7 | |
| dhbF | Similar to E. coli EntF | 1.1 | 4.5 | |
| hmp | Flavohemoglobin | − | 1.2 | 33.3 |
| nasD | Nitrite reductase; subunit | 1.0 | 12.4 | |
| nasE | Nitrite reductase; subunit | − | 1.1 | 4.6 |
| nasF | Uropophyrin-III C-methyltransferase | 0.9 | 6.9 | |
| ycgT | Similar to thioredoxin reductase | − | 1.2 | 2.5 |
| ydbN | Unknown | − | 1.3 | 11.4 |
| ykjA | Unknown | − | 0.9 | 3.5 |
| ykuN | Unknown; similar to flavodoxin | 1.5 | 5.5 | |
| ykuO | Unknown | − | 1.9 | 19.5 |
| ykuP | Unknown; similar to flavodoxin | 1.3 | 29.8 | |
| Group 4 | ||||
| acoA | Acetoin dehydrogenase; E1 subunit | − | 7.1 | 8.6 |
| acoB | Acetoin dehydrogenase; E1 subunit | 2.5 | 2.8 | |
| acoC | Acetoin dehydrogenase; E2 component | 5.3 | 3.4 | |
| acoL | Acetoin dehydrogenase; E3 component | 4.1 | 4.5 | |
Total cellular RNA was prepared from wild-type B. subtilis and a corresponding fnr mutant strain grown anaerobically under the indicated conditions. Numbers give the severalfold induction and, when shown in parentheses, the severalfold repression mediated by Fnr. ++, Fnr boxes in the promoter region of the indicated genes responsible for a direct Fnr effect; +, found Fnr boxes without direct function; −, no Fnr box found.
The Fnr regulon consists of four groups of differentially expressed genes.
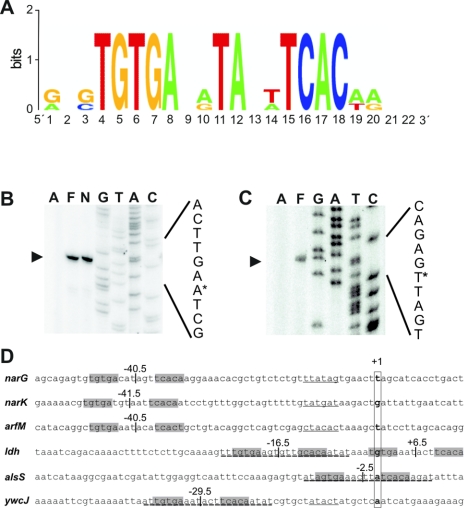
Four different groups of Fnr-dependent genes were distinguished by their expression patterns. The first group consists of genes that were induced by Fnr under conditions of fermentative growth and anaerobic nitrate respiration. This group included arfM, narKfnr, and narGHJI. The arfM gene encodes an anaerobic modulator protein (16), and narGHJI encodes the respiratory nitrate reductase (13). The narK gene for a potential nitrate/nitrite transporter protein forms an operon with the fnr gene, and Fnr is required for transcription from the narK promoter (6). Next, we analyzed the promoter regions of these genes for potential Fnr-binding sites using a bioinformatic approach. For this purpose, a position weight matrix for the Fnr-binding site was created. Two lines of evidence are currently available for functional B. subtilis Fnr-binding sites. First, the role of a proposed Fnr-binding site was proven indirectly by an in vivo complementation analysis. Here, a narK promoter-lacZ reporter gene fusion from B. subtilis was activated by the catabolite regulatory protein Crp of E. coli (6). Protein sequence alignments revealed that the amino acid residue RE-R motif within the helix-turn-helix DNA-binding domain of E. coli Crp is conserved in the potential DNA-binding domain of B. subtilis Fnr. Consequently, Fnr of B. subtilis was supposed to bind to DNA sequence motifs similar to those of Crp from E. coli (TGTGA-TA-TCACA). Second, Marino et al. (16) identified such a motif (TGTGAAATACATCACT) upstream of the Fnr-dependent gene coding for the anaerobic modulator ArfM (restriction sites are underlined). Mutations of this potential Fnr box centered −40.5 bp upstream of the transcriptional start site of arfM resulted in a complete loss of transcriptional activation under anaerobic growth conditions (16). Moreover, it was previously shown by Nakano et al. (25) that the transcription of narGHJI and narK is completely Fnr dependent. Consequently, within the promoter regions of both genes, a potential Fnr-binding sequence representing the E. coli Crp binding site motif was identified (6). Primer extension experiments performed previously by Cruz Ramos et al. revealed that an Fnr box was centered −41.5 bp from the transcriptional start point of narK (6). To prove whether the position of the Fnr-binding site upstream of the narGHJI operon also belongs to the same type of promoter, we determined the transcriptional start point of the narGHJI transcript by primer extension analysis (Fig. 2B). We found the Fnr-binding site located −40.5 bp upstream of the 5′ end of the narGHJI transcript (Fig. 2D). Therefore, a position weight matrix model for the B. subtilis Fnr-binding site was created, representing the three Fnr boxes found in the narG, narK, and arfM promoters and visualized as a sequence logo in Fig. 2A (29). Using the Virtual Footprint tool of the PRODORIC database and the corresponding B. subtilis Fnr position weight matrix, we screened the B. subtilis genome for potential Fnr boxes (20). All genes and operons found in group 1 of the macroarray analysis possess an Fnr-binding site. These promoters resemble class II catabolite activator protein-dependent promoters (5).
FIG. 2.
Promoter analysis of group 1 and group 2 Fnr-dependent promoters. (A) Sequence logo of the Fnr-binding site of B. subtilis based on the information weight matrix model. The height of each stack of letters represents the degree of sequence conservation measured in bits of information according to the equation given in Materials and Methods. The height of each letter within a stack is proportional to its frequency at that position in the binding site. The letters are sorted with the most frequent on top. (B, C) Determination of transcription start points of narG and ywcJ by primer extension analysis. Total cellular RNA was extracted from wild-type strain JH642 grown aerobically (A) and anaerobically under fermentative (F) and nitrate-respiratory (N) conditions. The same primer used for the primer extension analysis was used for sequencing reactions (lanes G, A, T, and C). Arrows indicate the primer extension products, and asterisks mark the 5′ end of the narG (B) and ywcJ (C) mRNA in the sequence. (D) Localization of putative Fnr-binding sites in promoter regions of group 1 and group 2 Fnr-dependent genes. Putative Fnr-binding sites are marked by gray boxes, and their positions with respect to the transcriptional start points are given. Potential YdiH-binding sites are marked by dashed lines. The −10 promoter regions are underlined. Transcriptional start sites are boxed and indicated by boldface type.
The second group of Fnr-dependent genes was characterized by the unaffected expression under anaerobic fermentative conditions but with Fnr-dependent repression under nitrate-respiring conditions. The genes ldh-lctP for lactate utilization, alsSD for acetoin formation, cydABCD for a potential high-affinity oxygen-dependent terminal cytochrome bd oxidase, and ywcJ, encoding a second potential nitrate/nitrite transport protein, belong to this group. The expression of group 2 genes is repressed in an Fnr-dependent manner. Putative Fnr-binding sites were found by Virtual Footprint centered at positions −16.5 and +6.5 relative to the transcriptional start point of the ldh promoter and at position −2.5 of the alsS promoter (Fig. 2D) (7). Obviously, the position of these Fnr boxes is different from those in Fnr-activated promoters. Comparable results were found for E. coli Fnr, where repression of transcription is mediated by Fnr binding to sites differently located compared to activating sites (9). To determine the position of the putative Fnr-binding site upstream of the ywcJ gene relative to the transcriptional start site, we performed a primer extension analysis (Fig. 2C). The putative Fnr-binding site was then mapped to a position centered at position −29.5 upstream of the transcriptional start point in the ywcJ promoter region (Fig. 2D). All genes of the Fnr-repressed group 2, with the exception of the cydABCD operon, possess Fnr-binding sites that are located closer to the transcriptional start point compared to the Fnr boxes of group 1 genes.
The third class of Fnr-dependent genes whose expression is not altered under fermentative conditions but strongly induced under nitrate-respiring conditions contains the dhbABCEF operon for synthesis of 2,3-dihydroxybenzoate (DHB), its modification and esterification to the iron siderophore corynebactin, the hmp gene for the flavohemoglobin involved in nitric oxide (NO)/nitrite detoxification, and the nasDEF operon coding for nitrite reductase. Furthermore, open reading frames of unknown function, ycgT, ydbN, ykjA, and ykuNOP, belong to this group (Table 2). Interestingly, this cluster included genes of the Fur (dhbABCEF, ydbN, and ykuNOP) and ResDE (hmp and nasDEF) regulons (3, 35). Promoters of genes of the Fnr-dependent expression group 3 do not contain Fnr-binding sites at all. For those genes, we postulate an indirect effect of Fnr.
The fourth class of Fnr-dependent genes consists only of the acoABCL operon involved in acetoin utilization (1). While its anaerobic expression was completely dependent on the presence of the fnr gene, no obvious Fnr-binding site was detected in the corresponding promoter region (Table 2).
Furthermore, four genes, ydbL, yceB, yceC, and ywiC, containing a putative Fnr box in their promoter region were identified, but their expression was not influenced by Fnr under the growth conditions we tested.
Functional analysis of group 1 Fnr-dependent genes.
The group 1 Fnr-dependent genes (narKfnr, narGHJI, and arfM) are characterized by their Fnr-dependent induction under all anaerobic conditions tested and by the presence of a conserved Fnr box in their upstream region. However, only the functionality of the Fnr box of the arfM promoter was characterized by a mutagenesis approach (16). Fnr-dependent regulation of the narK promoter via the Fnr box found −41.5 bp upstream of the transcriptional start point was concluded by its glucose-dependent induction via Crp in E. coli (6). Finally, the narGHJI promoter was only characterized via reporter gene fusion and fnr regulator mutants (25). In order to elucidate the detailed nature of the Fnr-dependent narGHJI promoter, we constructed new lacZ reporter gene fusions and integrated them into the amyE locus of B. subtilis. All previously published investigations of the narGHJI promoter were performed using a lacZ fusion located at the nar locus. Testing of narG-lacZ integrated at the amyE locus allowed the mutation of the Fnr-binding site via single base exchanges and then subsequent chromosomal integration and selection of appropriate strains. Furthermore, using this approach, it was also possible to test the expression of a narG-lacZ fusion in narG mutant strain THB1.
The expression of the newly constructed reporter gene fusion was analyzed in the wild type and an fnr mutant strain (THB2). Expression of narG-lacZ in wild-type cells was found to be induced fourfold under anaerobic growth conditions. Anaerobic expression was totally abolished in the fnr mutant strain (HRB5) (Table 3). To connect the observed regulation with the Fnr box of the narG promoter, we mutated the corresponding sequence upstream of narG. The mutation was performed according to the strategy used as described above for the arfM promoter region, where the 5′-TGTGA-3′ half-site was changed to 5′CCTGA-3′ (exchanged bases are shown in boldface type), destroying the originally palindromic sequence (16). The mutation of the putative Fnr box (strain TDB8) resulted in a total loss of anaerobic narG-lacZ induction, indicating the essential role of the Fnr box for Fnr-dependent gene activation. Surprisingly, in the presence of nitrate under anaerobic conditions, narG promoter expression was further increased by a factor of 4, indicating a further nitrate-dependent induction of the narG promoter. Very similar results were obtained during Northern blot analysis of the transcript derived from the nar operon (Fig. 1A). This nitrate-dependent induction was also observed for the narJ gene; however, it was not clearly visible for narGH and narI during the array analysis. Since the detected amount for all transcripts of the narGHJI operon was weak compared to the RNA amounts measured via Northern blot analysis, one can only speculate whether the missing induction of narG and narH is due to failure in reverse transcription during macroarray testing.
TABLE 3.
Influence of anaerobiosis on transcription of narG-lacZ, alsS-lacZ, ywcJ-lacZ, cydA-lacZ, and acoA-lacZ in B. subtilis wild-type, narG mutant, fnr mutant, and ydiH mutant strainsa
| Strain | Genotype | Fusion | β-Galactosidase activity (Miller units)
|
|||
|---|---|---|---|---|---|---|
| Aerobic | Anaerobic
|
|||||
| Fermentative | With nitrate | With nitrite | ||||
| TDB2 | Wild type | narG-lacZ | <5 | 19 | 81 | NDb |
| HRB5 | fnr mutant | narG-lacZ | <5 | <5 | <5 | ND |
| TDB8 | Wild type | narG (ΔFnr box)-lacZ | <5 | <5 | <5 | ND |
| HRB6 | narG mutant | narG-lacZ | <5 | 22 | 103 | ND |
| HRB7 | Wild type | alsS-lacZ | 20 | 689 | 266 | 835 |
| HRB8 | fnr mutant | alsS-lacZ | 31 | 720 | 718 | ND |
| HRB10 | wild type | alsS (ΔFnr box)-lacZ | 30 | 714 | 304 | ND |
| HRB11 | fnr mutant | alsS (ΔFnr box)-lacZ | 37 | 618 | 608 | ND |
| HRB9 | narG mutant | alsS-lacZ | 30 | 567 | 597 | ND |
| HRB19 | Wild type | ywcJ-lacZ | <5 | 76 | 23 | 66 |
| HRB20 | fnr mutant | ywcJ-lacZ | <5 | 71 | 63 | ND |
| HRB21 | wild type | ywcJ (ΔFnr-box)-lacZ | 10 | 248 | 248 | ND |
| HRB22 | narG mutant | ywcJ-lacZ | <5 | 73 | 79 | ND |
| HRB29 | ydiH mutant | ywcJ-lacZ | 33 | 237 | 240 | ND |
| HRB23 | Wild type | cydA-lacZ | <5 | 519 | 262 | 570 |
| HRB24 | fnr mutant | cydA-lacZ | <5 | 580 | 596 | ND |
| HRB25 | narG mutant | cydA-lacZ | <5 | 619 | 655 | ND |
| HRB26 | Wild type | acoA-lacZ | 93 | 1,289 | 1,703 | 2,663 |
| HRB27 | fnr mutant | acoA-lacZ | 28 | 39 | 38 | ND |
| HRB28 | narG mutant | acoA-lacZ | 35 | 26 | 30 | ND |
Strains were grown anaerobically using 50 mM glucose as a carbon source and ammonia as the nitrogen source, with the indicated additions (10 mM nitrate or nitrite) to the mid-exponential phase of growth. Results represent the average of at least three independent experiments performed in duplicate, with a standard error of less than 10%.
ND, not determined.
To prove whether nitrate itself or a product of the ammonification process is responsible for the observed additional induction, narG-lacZ was analyzed in a narG mutant strain (HRB6). Nitrate induction of narG-lacZ was still observed in the narG mutant, indicating its independence of the metabolization of nitrate. Nitrate induction was only observed when analyzed cells were harvested in the exponential phase and when nitrate was still present in the medium. After 6 h of cultivation, when the cells entered the stationary phase, we no longer detected the nitrate-dependent narG-lacZ induction. Determination of the nitrate concentration in the medium revealed that nitrate is almost completely utilized and therefore can no longer act as an inducer. This observation might explain previously published results where no obvious nitrate induction of narG expression was observed (16).
Functional analysis of group 2 Fnr-dependent genes.
The genes of group 2 (alsSD, ldh-lctP, ywcJ, and cydABCD) were characterized by their Fnr-dependent anaerobic repression in the presence of nitrate. However, only alsSD, ldh-lctP, and ywcJ carry an Fnr box-like sequence upstream of their coding region. The cydABCD promoter did not contain an obvious Fnr box. The potential Fnr boxes of the alsS and ldh promoters were centered at positions −16.5 and +6.5 for ldh and at position −2.5 for alsS with respect to the transcriptional start point. Primer extension analysis revealed that the 5′ end of the ywcJ mRNA is localized 27 bp upstream of the translational start, −29.5 bp downstream of the center of the Fnr box palindrome (Fig. 2D). In order to test general promoter and Fnr box function in vivo, alsS-lacZ, ywcJ-lacZ, and cydA-lacZ were integrated into the amyE locus and analyzed for their anaerobic expression. Due to the previously determined almost-identical expression behavior of the alsS and ldh promoters, we refrained from a detailed ldh promoter analysis (16).
First, the expression of alsS-lacZ, ywcJ-lacZ, and cydA-lacZ was found to be induced under fermentative conditions. But when nitrate was present, a consistent two- to threefold repression occurred for all three tested gene fusions (Table 3). Similar observations were previously made for alsSD and ldh-lctP expression (6). These findings demonstrate that essential genes for fermentation and for a high-affinity cytochrome oxidase required for microaerophilic growth are repressed during anaerobic nitrate-respiratory conditions. Clearly, B. subtilis adapts this way to the most efficient energy-generating pathway under anaerobic growth conditions. Interestingly, for the fnr mutant strain, we consistently found that the fermentative expression of alsS-lacZ, ywcJ-lacZ, and cydA-lacZ remained unaffected, indicating that fnr is not required for strong anaerobic expression (Table 3). However, nitrate-dependent repression of alsS-lacZ, ywcJ-lacZ, and cydA-lacZ gene expression was abolished in the fnr mutant strain, pointing towards a role of Fnr in mediating nitrate-dependent repression. These findings are in agreement with previous findings for the alsS and ldh promoters tested at their original chromosomal location (6). Next, the question arose whether Fnr directly interacts with the putative Fnr-binding sites found in the promoter regions of alsS and ywcJ to mediate the observed repression. To test this hypothesis, we mutated the corresponding putative Fnr-binding sites in both promoters. The putative Fnr box in the alsS promoter was changed from AGTGA-CT-TCACA to AGTGA-CT-TCCGG, while the ywcJ promoter-localized Fnr box was changed from TGTGA-TA-TCACA to CCTGA-TA-TCACA (exchanged bases are shown in boldface type). The mutation in the palindromic sequence did not result in a nitrate-dependent derepression of alsS-lacZ in the wild-type strain (Table 3). When the mutated promoter was analyzed in the fnr mutant strain (HRB11), derepression was still visible (Table 3). As previously demonstrated for ldh expression by Cruz Ramos etal. (7), these results indicate that the nitrate-dependent repression of alsS-lacZ is not mediated by the putative Fnr boxes in its upstream region.
In contrast, the expression of ywcJ-lacZ was found to be derepressed by about 3-fold under fermentative conditions and even 10-fold under nitrate-respiratory conditions, resulting in comparable levels of ywcJ expression under both conditions. However, this behavior was not observed in the fnr mutant strain with the ywcJ wild-type promoter, demonstrating that Fnr is not responsible for the dramatic derepression of ywcJ expression when the putative Fnr-binding site is mutated. These results suggest that ywcJ expression might be regulated by another yet-unknown transcription factor that recognizes the same or an overlapping consensus sequence like Fnr. In agreement with previous findings for ldh-lctP (7), these results demonstrate that expression of alsSD and ywcJ is not directly influenced by the trans-acting factor Fnr via a cis-acting element. The expression behavior mediated by the Fnr box-free cydA promoter perfectly fits the Fnr- and Fnr box-independent expression of the other group 2 genes.
Nevertheless, Fnr influences the transcription of alsSD, ldh- lctP, ywcJ, and cydABCD during anaerobic nitrate respiration. Next, we investigated the possible indirect Fnr effect on the nitrate-dependent repression of group 2 genes via the regulation of narGHJI expression. For this reason, we analyzed the expression of alsS-lacZ, ywcJ-lacZ, and cydA-lacZ reporter gene fusions in nitrate reductase mutant strains (HRB9, HRB22, and HRB25) (Table 3). Interestingly, for all three reporter gene fusions tested, a derepression similar to that of the fnr mutant strains was detected. Since Fnr is the major regulator of the nitrate reductase operon under anaerobic growth conditions, these results indicate that Fnr influences the transcription of alsSD, ywcJ, and cydABCD indirectly by controlling nitrate reductase function. The dissimilatory nitrate reductase converts nitrate to nitrite under anaerobic conditions. To test whether the product of the nitrate reductase causes the derepression, we analyzed gene expression in the wild-type strains in the presence of nitrite. Interestingly, nitrite showed no inhibitory effect on the transcription of alsS-lacZ, ywcJ-lacZ, or cydA-lacZ (Table 3). Similar results were previously observed for ldh expression (7).
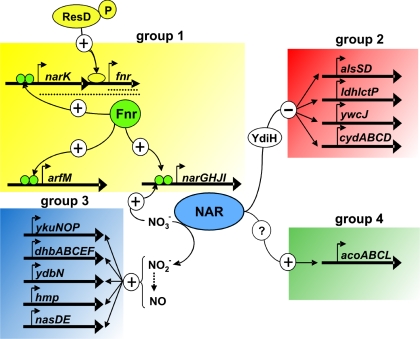
These results demonstrate that Fnr mediates the nitrate-dependent repression of alsSD, cydABCD, ldh lctP, and ywcJ by anaerobic induction of nitrate reductase production. The observed effect is not mediated by the substrate and the product, nitrate and nitrite, of the nitrate reductase reaction. Nevertheless, active nitrate reductase is required for the observed regulation (Fig. 3). Possibly, parts of the electron transport system or the NADH-to-NAD+ ratio is involved in the repression of alsSD, cydABCD, ldh lctP, and ywcJ. In Streptomyces coelicolor, expression of cydABCD is regulated by Rex, a novel redox-sensing repressor. The DNA-binding activity of Rex appears to be controlled by the redox poise of the NADH/NAD+ pool (4). Rex homologues exist in most gram-positive bacteria. In B. subtilis, a Rex homolog, YdiH, encoded by the ydiH gene acts as a repressor for cydABCD transcription under aerobic growth conditions (27). Very recently, it was demonstrated that YdiH of B. subtilis acts as a negative regulator of cydABCD, ldh-lctP, and ywcJ and coordinates the expression of these genes during the transition from aerobic to anaerobic growth (15). DNase I footprinting analysis revealed three binding sites of YdiH in the cydA promoter region, and a consensus sequence was proposed. Interestingly, the YdiH-binding sequence is also present in the ywcJ and ldh promoters overlapping the potential, nonfunctional Fnr-binding site. We already postulated the binding of a repressor at the ywcJ promoter, which is abolished by mutating the potential Fnr-binding site. To test whether YdiH functionally represses ywcJ expression under nitrate-respiratory conditions, we analyzed ywcJ-lacZ expression in a ydiH mutant strain (HRB29). A derepression of ywcJ-lacZ expression comparable to those when the potential Fnr-binding site of the ywcJ promoter was mutated was measured (Table 3). These results suggest that repression of ywcJ expression is mediated by YdiH and that the binding site of the repressor overlaps the deduced nonfunctional Fnr-binding site. The results previously reported by Larsson et al. (15) in combination with our findings provide the first insight into the redox regulatory network of B. subtilis. In the presence of nitrate under anaerobic growth conditions, this alternative electron acceptor is used to reoxidize NADH to NAD+. With this NADH-to-NAD+ ratio, the YdiH repressor is active and represses expression of the group2 genes cydABCD, ldh-lctP, alsSD, and ywcJ. After nitrate is used up, NADH accumulates and YdiH gets inactivated, which in turn leads to the derepression of group 2 genes. Thus, YdiH fills the missing link in how nitrate respiration and fermentation are coordinated at the transcriptional level (Fig. 3).
FIG. 3.
Regulatory model for Fnr function during the transition to anaerobic growth conditions. Under anaerobic conditions, Fnr directly induces transcription of narGHJI, arfM, and narKfnr via a cis-acting Fnr box in the corresponding promoter regions (group 1 genes). By regulation of nitrate reductase formation, Fnr most likely mediates the repression of alsSD, ldh lctP, ywcJ, and cydABCD via YdiH (group 2 genes). Nitrite-derived nitric oxide serves as a second messenger and induces transcription of a subset of genes that overlap with the Fur, ResDE, and other unknown regulons (group 3 genes). Finally, acoABCL is the only member of expression group 4. Anaerobic expression of the operon is completely dependent on Fnr-dependent nitrate reductase formation.
Functional analysis of group 3 Fnr-dependent genes.
The third group of genes, ydbN, ykuNOP, dhbABCEF, ykjA, ycgT, hmp, and nasDE, identified by transcriptional profiling were found to be anaerobically induced in the presence of nitrate, and all lack Fnr-binding sites. To test how Fnr is involved in group 3 gene regulation, we analyzed the expression of ykuNOP and ydbN, encoding proteins of unknown functions, using the Northern blot technique. Using the ykuN-specific probe, a single transcript of about 1,900 nt, which corresponded to the size of the ykuNOP operon, was detected in equal amounts of RNA isolated from anaerobically grown cells in the presence of nitrate and nitrite (Fig. 1C). The transcript was not detected in RNA from fermentatively grown cells. By analyzing an fnr mutant strain, we found the ykuNOP transcript only in RNA prepared from cells that were grown anaerobically in the presence of nitrite (Fig. 1C). A specific ykuNOP transcript was only detected in RNA prepared from a narG mutant strain (strain THB1) anaerobically grown in the presence of nitrite (Fig. 1C). Since this expression pattern was identical to those observed for the fnr mutant strain, an indirect Fnr function was obvious. The anaerobic expression of ykuNOP was only observed when nitrate was reduced to nitrite or when nitrite was directly added to the medium. Therefore, we conclude that ykuNOP expression is solely dependent on nitrite in the medium. The almost-identical expression pattern was found for ydbN, where we detected a specific transcript of about 200 nt in the Northern blot analysis (Fig. 1D). This size corresponds nicely to a single transcript of the predicted ydbN open reading frame of 177 bp. Consequently, Fnr anaerobically induces narGHJI transcription and formed nitrate reductase converts nitrate to nitrite, which in turn acts as an inducer for a yet-unknown N-oxide regulatory system (Fig. 3).
Some of the genes found in group 3 of our transcriptional profiling analysis have previously been described to be members of the Fur regulon, i.e., dhbABCEF, ykuNOP, and ydbN (4). NO can lead to nitrosylation of the ion center of Fur and thereby trigger derepression of the Fur regulon (18). Furthermore, members of the Fur regulon were found to be induced under nitrate-respiratory conditions (35). Moreover, hmp and nasDE belong to the ResDE regulon (35). Recent studies showed that the expression of most genes of this cluster (hmp, nasD, dhbA, ykuN, and ycgT) is dependent on the presence of NO (18, 21). Under cellular conditions, nitrite is spontaneously converted to nitric oxide (21). Thus, it is possible that the observed induction of group 3 genes upon the addition of nitrite is actually mediated by nitric oxide instead of nitrite.
Functional analysis of group 4 Fnr-dependent genes.
The group 4 Fnr-dependent operon acoABCL was characterized by its strict Fnr-dependent expression under all anaerobic conditions tested and by the absence of an obvious Fnr box in its upstream region. Northern blot analysis revealed the strict dependence of acoABCL expression under fermentative and nitrate-respiratory conditions on the presence of an intact fnr gene (Fig. 1B). A slight nitrate induction was visible. Similar observations were made for the analysis of acoA-lacZ reporter gene fusions (Table 3). Again, a slight nitrate-dependent induction was observed. The total loss of reporter gene activity of the same fusion tested under anaerobic and nitrate-respiratory conditions in a narG mutant clearly demonstrated the indirect effect of Fnr via narGHJI induction. Therefore, the anaerobic expression of the acoABCL operon is, even under fermentative conditions, strictly dependent on the production of nitrate reductase mediated by Fnr.
Finally, a summary of the complex Fnr regulation pattern during the onset of anaerobic metabolism in B. subtilis is given in Fig. 3.
Supplementary Material
Acknowledgments
We thank G. Homuth, T. Koburger, J. Mosterz, and L. Steil for their help with the macroarray techniques and evaluation. We thank Anja Hartmann for her excellent technical assistance. Strains LUW48 and LUW273 were a generous gift from C. von Wachenfeldt, Lund University, Sweden, and we thank M. Debarbouille for supplying us with strain QB700.
This study was funded by grants from the Deutsche Forschungsgemeinschaft (Ha3456-1/1).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ali, N. O., J. Bignon, G. Rapoport, and M. Debarbouille. 2001. Regulation of the acetoin catabolic pathway is controlled by sigma L in Bacillus subtilis. J. Bacteriol. 183:2497-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Baichoo, N., T. Wang, R. W. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 4.Brekasis, D., and M. S. B. Paget. 2003. A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). EMBO J. 22:4856-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 6.Cruz Ramos, H., L. Boursier, I. Moszer, F. Kunst, A. Danchin, and P. Glaser. 1995. Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 14:5984-5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz Ramos, H., T. Hoffmann, M. Marino, H. Nedjari, E. Presecan-Siedel, O. Dreesen, P. Glaser, and D. Jahn. 2000. Fermentative metabolism of Bacillus subtilis: physiology and regulation of gene expression. J. Bacteriol. 182:3072-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaser, P., A. Danchin, F. Kunst, P. Zuber, and M. M. Nakano. 1995. Identification and isolation of a gene required for nitrate assimilation and anaerobic growth of Bacillus subtilis. J. Bacteriol. 177:1112-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guest, J. R., J. Green, A. S. Irvine, and S. Spiro. 1996. The FNR modulon and FNR-regulated gene expression, p. 317-342. In E. C. C. Lin and A. Simon (ed.), Regulation of gene expression in Escherichia coli. R.G. Landes Co., Austin, Tex.
- 10.Harwood, C. R., and S. M. Cutting (ed.). 1990. Modern microbiological methods: molecular biological methods for Bacillus subtilis, p. 548. Wiley & Sons, Chichester, United Kingdom.
- 11.Härtig, E., H. Geng, A. Hartmann, A. Hubacek, R. Münch, R. W. Ye, and M. M. Nakano. 2004. Bacillus subtilis ResD induces expression of the potential regulatory genes yclJK upon oxygen limitation. J. Bacteriol. 186:6477-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann, T., N. Frankenberg, M. Marino, and D. Jahn. 1998. Ammonification in Bacillus subtilis utilizing dissimilatory nitrite reductase is dependent on ResDE. J. Bacteriol. 180:186-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann, T., B. Troup, A. Szabo, C. Hungerer, and D. Jahn. 1995. The anaerobic life of Bacillus subtilis. Cloning and characterization of the genes encoding the respiratory nitrate reductase system. FEMS Microbiol. Lett. 131:219-225. [DOI] [PubMed] [Google Scholar]
- 15.Larsson, J. T., A. Rogstam, and C. von Wachenfeldt. 2005. Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis. Microbiology 151:3323-3335. [DOI] [PubMed] [Google Scholar]
- 16.Marino, M., H. Cruz Ramos, T. Hoffmann, P. Glaser, and D. Jahn. 2001. Modulation of anaerobic energy metabolism of Bacillus subtilis by arfM (ywiD). J. Bacteriol. 183:6815-6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics, p. 54. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Moore, C. M., M. M. Nakano, T. Wang, R. W. Ye, and J. D. Helmann. 2004. Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside. J. Bacteriol. 186:4655-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Münch, R., K. Hiller, H. Barg, D. Heldt, S. Linz, E. Wingender, and D. Jahn. 2003. PRODORIC: prokaryotic database of gene regulation. Nucleic Acids Res. 31:266-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Münch, R., K. Hiller, A. Grote, M. Scheer, J. Klein, M. Schobert, and D. Jahn. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187-4189. [Online.] [DOI] [PubMed] [Google Scholar]
- 21.Nakano, M. M. 2002. Induction of ResDE-dependent gene expression in Bacillus subtilis in response to nitric oxide and nitrosative stress. J. Bacteriol. 184:1783-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano, M. M., Y. P. Dailly, P. Zuber, and D. P. Clark. 1997. Characterization of anaerobic fermentative growth in Bacillus subtilis: identification of fermentation end products and genes required for the growth. J. Bacteriol. 179:6749-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakano, M. M., and Y. Zhu. 2001. Involvement of ResE phosphatase activity in down-regulation of ResD-controlled genes in Bacillus subtilis during aerobic growth. J. Bacteriol. 183:1938-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52:165-190. [DOI] [PubMed] [Google Scholar]
- 25.Nakano, M. M., P. Zuber, P. Glaser, A. Danchin, and F. M. Hulett. 1996. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J. Bacteriol. 178:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priest, F. G. 1993. Systematics and ecology of Bacillus, p. 3-16. In A. L. Sonnenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 27.Schau, M., Y. Chen, and F. M. Hulett. 2004. Bacillus subtilis YdiH is a direct negative regulator of the cydABCD operon. J. Bacteriol. 186:4585-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider, T. D. 1997. Information content of individual genetic sequences. J. Theor. Biol. 189:427-441. [DOI] [PubMed] [Google Scholar]
- 29.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schreiber, M., and C. Brown. 2002. Compensation for nucleotide bias in a genome by representation as a discrete channel with noise. Bioinformatics 18:507-512. [DOI] [PubMed] [Google Scholar]
- 31.Steil, L., T. Hoffmann, I. Budde, U. Völker, and E. Bremer. 2003. Genome-wide transcriptional profiling analysis of the adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185:6358-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, G., E. Sharkova, R. Chesnut, S. Birkey, M. F. Duggan, A. Sorokin, P. Pujic, S. D. Ehrlich, and F. M. Hulett. 1996. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J. Bacteriol. 178:1374-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winstedt, L., K. I. Yoshida, Y. Fujita, and C. von Wachenfeldt. 1998. Cytochrome bd biosynthesis in Bacillus subtilis: characterization of the cydABCD operon. J. Bacteriol. 180:6571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yashphe, J., J. A. Hoch, and N. O. Kaplan. 1978. Regulation of the lactate dehydrogenase synthesis in Bacillus subtilis. Biochim. Biophys. Acta 544:1-7. [DOI] [PubMed] [Google Scholar]
- 35.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.