Abstract
The complete nucleotide sequence and genetic organization of a new genomic island (AGI-3) isolated from the extraintestinal avian pathogenic Escherichia coli strain BEN2908 is reported. This 49,600-bp island is inserted at the selC locus and contains putative mobile genetic elements such as a phage-related integrase gene, transposase genes, and direct repeats. AGI-3 shows a mosaic structure of five modules. Some of these modules are present in other E. coli strains and in other pathogenic bacterial species. The gene cluster aec-35 to aec-37 of module 1 encodes proteins associated with carbohydrates assimilation such as a major facilitator superfamily transporter (Aec-36), a glycosidase (Aec-37), and a putative transcriptional regulator of the LacI family (Aec-35). The aec-35 to aec-37 cluster was found in 11.6% of the tested pathogenic and nonpathogenic E. coli strains. When present, the aec-35 to aec-37 cluster is strongly associated with the selC locus (97%). Deletion of the aec-35-aec-37 region affects the assimilation of seven carbohydrates, decreases the growth rate of the strain in minimal medium containing galacturonate or trehalose, and attenuates the virulence of E. coli BEN2908 for chickens.
Escherichia coli, a commensal inhabitant of the gastrointestinal tract of mammals and birds, is also the causative agent of several diseases in animals and human worldwide. Pathogenic E. coli strains have been divided into intestinal pathogenic E. coli and extraintestinal pathogenic E. coli (ExPEC) depending on the location of the infection they are causing. ExPEC strains are responsible for a variety of infections, including bacteremia, urinary tract infections, neonatal meningitis, pneumonia, deep surgical wound infections, endovascular infections, vertebral osteomyelitis, and septicemia (35, 54).
Avian pathogenic Escherichia coli (APEC) strains belong to the ExPEC group. They are mainly responsible for a respiratory disease in poultry usually followed by a systemic infection and a fatal septicemia. Characteristic fibrinopurulent lesions are aerosacculitis, pericarditis, and perihepatitis. APEC strains can also be involved in localized infections such as omphalitis, salpingitis, swollen head syndrome, and cellulitis (2, 17). APEC isolates commonly belong to serogroups O1, O2, O5, O8, O18, O35, and O78 (4, 17). Various virulence factors of APEC strains, such as adhesins (F1 and P fimbriae and curli), anti-host defense factors (OmpA, Iss, lipopolysaccharide, and K1), iron acquisition systems (aerobactin, Iro proteins, yersiniabactin, and the Sit iron acquisition locus), autotransporters (Tsh and Vat), and the IbeA protein have been identified (20, 21, 25, 36-38, 42, 46, 50). Using various genomic approaches, several putative virulence factors of APEC strains have been identified (9, 20, 38, 58). However, the above components cannot explain all the disease process, suggesting the existence of other, unidentified components.
It is well described that pathogenicity factors can be encoded by mobile genetic elements (transposons, phages, plasmids, integrons, and genomic islands) which are capable of horizontal gene transfer. Genomic islands (GEIs) are clusters of chromosomal genes that are often associated with tRNA genes (28). GEIs often possess genes or cryptic pseudogenes coding for mobility-related elements such as phage genes, insertion sequence elements, transposases, and origins of replication (28). They have a modular organization and carry gene clusters that encode a wide range of functions that contribute to the adaptation of bacteria to the environment and/or to the development of symbiosis, antibiotic resistance, or virulence. In the last case, GEIs are designated pathogenicity islands (PAIs).
GEIs can be unstable due to homologous recombination between their flanking direct repeats. These direct repeats are often homologous to phage attachment sites. The selC tRNA locus has been shown to be a hot spot for integration of foreign DNA, and particularly of PAIs: PAI-1 in the uropathogenic E. coli strain 536, the locus of enterocyte effacement (LEE) in enteropathogenic and enterohemorrhagic E. coli strains, the toxigenic invasion locus A in the enterotoxigenic E. coli strain H10407, the locus of proteolysis activity in the Shiga toxin-producing E. coli strain 4797/97, SPI-3 in Salmonella enterica, and SHI-2 in Shigella flexneri (5, 7, 23, 24, 44, 57). The selC locus is also the attachment site for the E. coli retronphage ΦR73 (61).
As selC is a hot spot for insertion of PAIs, we searched for the presence of foreign DNA inserted at the selC locus of the avian ExPEC strain BEN2908. We thus identified and characterized a new genomic island containing genes involved in carbohydrate uptake and virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The main bacterial strains and plasmids used in this study are described in Table 1. E. coli strain BEN2908, O2:K1:H5 (fim+ iut+ ibeA+), is a nalidixic acid-resistant derivative of strain MT78 that was isolated from the trachea of a chicken with a respiratory infection (16, 25). E. coli strain BEN2269 was isolated from the intestine of a healthy chicken and is nonpathogenic (16). For prevalence analysis a total of 285 E. coli strains were also used, including 240 avian isolates (205 pathogenic and 35 nonpathogenic), 37 strains isolated from humans with extraintestinal diseases, 7 isolates from animals (other than poultry) with extraintestinal syndrome, and the nonpathogenic K-12 MG1655 strain as a negative control.
TABLE 1.
Relevant strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| BEN2908 | Pathogenic avian isolate O2:K1:H5 fim+iut+ibeA+ Nalr | 16 |
| BEN2269 | Nonpathogenic avian isolate, O2:K1−:H−, formerly EC79 | 16 |
| BEN2929 | BEN2908 Δ(aec-35-36-37)::Kanr | This study |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pKD4 | Plasmid carrying a kanamycin resistance cassette and oriRγ origin | 14 |
| pKD46 | Red recombinase expression plasmid, temperature-conditional replicon, Ampr | 14 |
| pCR2.1 | TA cloning vector, Kanr Ampr, high copy number | Invitrogen |
| pBEN182 | pCR2.1 containing the aec-35 to aec-37 genes of AGI-3 | This study |
Strains were grown in Luria-Bertani broth (LB) at 37°C. When necessary, ampicillin at 100 μg ml−1, kanamycin at 50 μg ml−1 or nalidixic acid at 30 μg ml−1 were added. For growth rate experiments, overnight LB cultures were centrifuged, washed twice, and resuspended in the same volume in a minimal medium (100 mM NaCl, 30 mM triethanolamine HCP [pH 7.5], 5.0 mM NH4Cl, 2.0 mM NaH2PO4, 0.25 mM Na2SO4, 0.05 mM MgCl2, 1.0 mM KCl) (66). The strains were then cultured in triplicate at 37°C in 100-well, sterile, covered microplates (Labsystems, Helsinki, Finland). Each well contained 300 μl of the above minimal medium supplemented with 5 mM Na-d-galacturonate or 5 mM trehalose and 3 μl of the bacterial suspension. The plates were incubated in a Microbiology Reader Bioscreen C apparatus (Labsystems, Helsinki, Finland) and the optical density was measured at 450 nm every 15 min, after shaking. The generation time was calculated as described by Miller (43).
DNA preparation.
Plasmids and cosmids were purified using Nucleobond AX100 or AX500 columns (Macherey-Nagel, Düren, Germany). Genomic DNA was prepared using a NucleoSpin tissue kit from Macherey-Nagel (Düren, Germany) or by the boiling method (56).
PCR and primers.
The primers used in this study are listed in Table 2. PCRs were performed in 25 μl containing 500 nM of the forward and reverse primers, 200 μM of each deoxynucleoside triphosphate (Finzyme, Ozyme France), 1 U of Taq DNA polymerase (New England Biolabs Inc.), and 2 mM MgCl2 in a PCR buffer containing 1 mM KCl, 1 mM (NH4)2SO4, 200 μM MgSO4, 0.1% Triton X-100, 2 mM Tris-HCl, pH 8.8 (New England Biolabs Inc.). The PCR conditions were as follows: initial denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min/kb. A final extension step at 72°C for 7 min was included. Reactions were performed in a Perking Elmer thermocycler (GeneAmp 9700, Applied Biosystems).
TABLE 2.
Oligonucleotide primers used for PCR
| Primer | Sequencea | Reference |
|---|---|---|
| K260 | GAGCGAATATTCCGATATCTGGTT | 41 |
| K261 | CCTGCAAATAAACACGGCGCAT | 41 |
| K255 | GGTTGAGTCGATTGATCTCTGG | 41 |
| CA8R | CCACCTTCACCGGCATAAC | This study |
| CA8F | CATCGGTGATACCTGTCGCC | This study |
| cat75 | ACATTACGCTTATTTTCCCTTGACCGG AAGAATCAGAGGCTGTGGTTTCAgtgt aggctggagctgcttc | This study |
| cat76 | ACTCTTACAGTGCCTTTATTTGGCCAT GATTTACCCTCTACAATAAATTAcata tgaatatcctccttag | This study |
| cat75′ | ACATTACGCTTATTTTCCCTTGACCGG AAGAATCAGAGGCTGTGGTTTCA | This study |
| cat76′ | ACTCTTACAGTGCCTTTATTTGGCCAT GATTTACCCTCTACAATAAATTA | This study |
| cat47 | CTTTGAAGTGCACGATAACC | This study |
| cat82 | GGTGGAGCTGCATGACAAGG | This study |
| cat51 | GTGTAGGCTGGAGCTGCTTC | This study |
| cat52 | CATATGAATATCCTCCTTAG | This study |
| IC79 | AATCGGAGTTGGCTCTTTCC | This study |
| IC36 | CGTGCCCGCGAGAAAGTATTGG | This study |
| IC39 | CATCCGGTGGGGTGATTATGAGTCA | This study |
| IC64 | AAGCCTATTTTGTCAGCAGTAACGCCA | This study |
| IC45 | GGATAATGCTGGTCAGTGGCAGGAA | This study |
| IC50 | GCCCTGCTTCCACGACACTTGC | This study |
| IC37 | TGCGATAATGCCTTGCTGATGCTTT | This study |
| IC60 | TGTCGCAGAAGCCTCAGCACCTC | This study |
| IC43 | GGTTCAGCCGCATGGATTGCC | This study |
| IC44 | CAAGTTCCTGCCACTGACCAGCATT | This study |
| IC80 | CAAGTGGGGACTTCATTGCT | This study |
| SelC1 | GCGTGTATTAGGCGGAAAAAAC | This study |
For gene inactivation, sequences complementary to the kanamycin resistance cassette are in lowercase letters.
DNA sequencing and sequence analysis.
Cosmid clones were sequenced by primer walking. Nucleotide sequencing was performed by Genome Express (Meylan, France). Sequences were assembled and analyzed using the VectorNTI package (InforMax, Inc., Bethesda, Md.). Coding sequences were predicted using GeneMark (39). Homology searches were performed by using the BLAST server from the National Center for Biotechnology Information.
Construction of the mutant strain BEN2929 (BEN2908Δaec-35-37::Kanr) and plasmid pBEN182.
The aec-35 to aec-37 chromosomal DNA region was deleted by recombination with the PCR product as described by Datsenko and Wanner (14). Briefly, the method of Tung et al. was used to electroporate E. coli BEN2908 cells with pKD46, a temperature-sensitive plasmid carrying the Red recombinase system from the λ bacteriophage under the control of the arabinose-inducible PBAD promoter (14, 62). The Red recombinase system allowed the replacement of the aec-35 to aec-37 DNA region by a kanamycin resistance cassette obtained by amplifying pKD4 template plasmid using primers cat75 and cat76 (Table 2). The 5′ ends of primers cat75 and cat76 are homologous to the 5′ and 3′ ends of the aec-35-aec-37 region, respectively. The amplified product was then inserted into E. coli BEN 2908 carrying pKD46 by electroporation and a kanamycin-resistant and ampicillin-sensitive clone was isolated. The replacement of the aec-35-aec-37 gene cluster was confirmed by PCR using primers cat47 and cat82 (flanking the deleted region) (Table 2) and by Southern blot with a kanamycin probe generated by PCR amplification of pKD4 using primers cat51 and cat52 (Table 2). The mutant was named BEN2929 and stored at −70°C.
Cloning of the aec-35 to aec-37 DNA region was carried out after amplification of this region from BEN2908 chromosomal DNA using primers cat75′ and cat76′ (Table 2). The amplified region was cloned into pCR2.1-TOPO (Invitrogen), leading to plasmid pBEN182. The construction was verified by sequencing the inserted region.
Hybridization.
For Southern blot hybridization, DNA restriction fragments generated by BlnI digestion were submitted to electrophoresis and transferred to a Hybond-N+ membrane (Amersham Pharmacia Biotech). The 1.5-kb fragment carrying the kanamycin resistance gene obtained by PCR amplification from plasmid pKD4 using primers cat51 and cat52 (Table 2) was labeled by using enhanced chemiluminescence with the ECL RPN3000 kit (Amersham Pharmacia Biotech) according to the manufacturer's protocol. Hybridization, washes, and detection were performed as instructed by the kit.
Experimental colibacillosis.
An in vivo virulence assay was conducted as described previously with some modifications (25). Briefly, 25-day-old White Leghorn specific-pathogen-free chickens (line PA12) from the Institut National de la Recherche Agronomique experimental farm were inoculated in the right thoracic air sac with a 0.1-ml suspension containing 5 × 106 CFU. The inocula were prepared from bacteria cultivated in LB at 37°C with agitation to an optical density at 600 nm of 0.6.
Groups of 20 and 19 animals were inoculated with strain BEN2908 and mutant strain BEN2929, respectively. One control group of three animals was inoculated with the avian nonpathogenic strain BEN2269. Blood samples were collected 24 and 48 h postinoculation and serial dilutions were plated onto Drigalski agar plates for bacterial quantification. Animals were euthanized 48 h postinoculation by injection of Nesdonal (Rhône-Mérieux, Lyon, France), and necropsied. A piece of liver was collected and after homogenization in saline, serial dilutions were plated onto Drigalski agar plates supplemented when needed with nalidixic acid (30 μg ml−1) or kanamycin (50 μg ml−1). Macroscopic fibrinous lesions were observed and scored (air sacs, 0 to 4; heart, 0 to 2; and liver, 0 to 2) as previously described (42).
Bacterial growth assay in 90% normal chicken serum.
Blood from 2-week-old specific-pathogen-free chickens was collected and allowed to clot for 1 h at room temperature. Sera were then pooled and stored at 4°C until used. Fifty milliliters of LB was inoculated with 100 μl of bacteria that had been grown for 24 h in LB and then incubated at 37°C with agitation to an optical density at 600 nm of 0.3. After centrifugation, the bacterial pellet was washed twice and resuspended in 1 ml of sterile Tris buffer (50 mM Tris-HCl, 2 mM MgCl2, 0.4 mM CaCl2, 40 mM NaCl, pH 8.4); 100 μl of this bacterial inoculum was incubated at 37°C in 900 μl of fresh serum. Bacterial counts at 0, 1, 2, and 3 h were obtained by plating 10-fold serial dilutions onto LB agar plates. Slide agglutination did not reveal the presence of antibodies specifically directed against O2 E. coli in the serum used.
Phenotype microarray tests.
The effect of the aec-35-aec-37 gene cluster on the carbon metabolism of strain BEN2908 was tested in Phenotype microarray plates (Biolog Inc., Hayward, California) (8). These are 96-well microtiter plates containing different carbohydrates dried on the bottom of each well. When a carbohydrate is assimilated, it allows the respiration of the bacteria and the electrons produced are transferred to an indicator dye (tetrazolium violet), resulting in a purple color. Cells were streaked onto LB agar plates supplemented with the appropriate antibiotic (nalidixic acid, kanamycin, or ampicillin) and grown overnight at 37°C. Individual colonies were then picked up from the surface of the plates using a cotton swab and resuspended (at an optical density at 450 nm of 0.08) in the minimal medium containing the indicator dye provided by the supplier (IF0 inoculating fluid). The suspensions were then distributed (100 μl per well) into PM1 and PM2 microplates containing a panel of 190 different carbon sources. The plates were incubated aerobically at 37°C for 24 h and the reduction of the tetrazolium dye was then quantified by measuring the optical density at 590 nm (enzyme-linked immunosorbent assay reader Multiskan Ascent).
Statistical analysis.
Statistical analysis of the data from the in vivo virulence assay was done by applying a Mann-Whitney test. Values were arranged in increasing numbers, dead animals being given the highest rank. Exact P values were calculated with the StatXact software (version 5.0; Cytel Software, Cambridge, MA) and P < 0.05 was considered significant. The prevalence data were analyzed by using the chi-square test. Exact P values were calculated with the software StatXact and P < 0.05 was considered significant. The phenotype microarray data were analyzed online (http://home.clara.net/sisa/t test.htm) by using Student's t test for statistical significance.
Nucleotide sequence accession number.
The AGI-3 nucleotide sequence has been assigned GenBank accession no. AY857617.
RESULTS
Identification and sequencing of a genomic island inserted at the selC locus of the APEC strain BEN2908.
To search for the presence of foreign DNA inserted at the selC locus of the avian ExPEC strain BEN2908, we first checked the integrity of the locus. For that aim we tried to amplify the selC locus (527 bp in strain MG1655) with primers K260 and K261 (Table 2) located upstream and downstream of selC, respectively, in E. coli strain MG1655 (41). No PCR product was obtained with strain BEN2908, suggesting the insertion of foreign DNA at the selC locus.
As the regions flanking PAIs inserted at selC are relatively well conserved, we then used the K255 primer (matching the left end of the locus of enterocyte effacement) and the K260 primer (located upstream of the selC locus) to screen by PCR a previously made genomic cosmid library of the strain (Table 2) (40). One of the cosmids (CA8), which yielded a PCR product of 418 bp, was sequenced by primer walking. As the insert in CA8 did not cover the entire DNA region inserted at selC, the cosmid library was screened again by PCR with primers CA8R and CA8F matching the 216-bp end of the insert of CA8 (Table 2). Cosmid DG6, which yielded the predicted PCR product of 216 bp, was identified. After sequencing of the DG6 insert by primer walking, the right end of the DNA region inserted at the selC locus was identified.
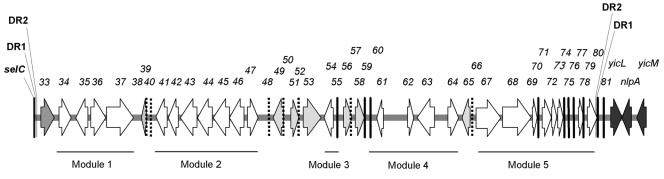
The DNA region inserted at selC was named AGI-3 for APEC Genomic Island 3. AGI-3 is inserted at the 3′ end of selC (Fig. 1). By comparison with the genome of the nonpathogenic E. coli strain K-12 MG1655, the insertion leads to a deletion of 1,903 bp comprising the yicK gene (6). AGI-3 is 49,600 bp long and contains two direct repeats: DR1 of 16 bp, TTCGACTCCTGTGATC, and DR2 of 21 bp,TTTGGGGGT(A/T)CTT(T/A)(A/T)GGGGGT. One DR1 is located at one end of AGI-3 (3′ extremity of selC) and the second DR1 is located inside aec-80. DR1 of AGI-3 was found to also flank PAI-I of E. coli 536 and the toxigenic invasion locus A PAI of E. coli H10407, and corresponds to half of the attP site of phage φR73 (18, 24, 32). One DR2 is located between DR1 and aec-33, and the second DR2 is located inside aec-80. AGI-3 was predicted to contain 49 open reading frames (ORFs) numbered sequentially from aec-33 to aec-81 (aec for avian E. coli) (Table 3, Fig. 1). The overall G+C content of AGI-3 is 49.25%, which is close to the average of the E. coli genome (51%). Nevertheless, the G+C content shows variability across the AGI-3 genomic island (Table 3).
FIG. 1.
Genetic organization of the AGI-3 genomic island. Open reading frames larger than 360 bp are indicated as dark grey arrows for core genome genes, as a grey arrow for the phage-like integrase gene aec-33, as light grey arrows for mobility genes, and as white arrows for genomic island-associated genes. ORFs smaller than 360 bp (120 amino acids) are indicated in bold black lines or in dotted black lines (mobility genes). ORFs are labeled consecutively from 33 to 81 and correspond to aec-33 to aec-81. DR, direct repeat.
TABLE 3.
Genetic features of the identified AGI-3 open reading frames
| ORF | G+C content (%) | Size (aa) | Positiona (bp) | Similarities with representative sequence in database (Blastp E value)b | Accession no. | Conserved domain | Predicated function |
|---|---|---|---|---|---|---|---|
| aec-33 | 45 | 394 | 598-1782 | Putative prophage integrase, E. coli CFT073 (0.0) | NP_756353.1 | cd00801 | Integrase |
| aec-34 | 46 | 347 | 2194-3237 | ShiA homolog, E. coli CFT073 (0.0) | NP_756354.1 | None | Reduction of inflammation |
| aec-35 | 44 | 356 | C 3635-4705 | Putative transcriptional regulator, E. coli CFT073 (0.0) | NP_756356.1 | smart00354, pfam00532 | Transcriptional regulator LacI family |
| aec-36 | 42 | 452 | 4910-6268 | Hexuronate transporter, E. coli CFT073 (0.0) | NP_756357.1 | pfam00083 | Sugar (and other) transport |
| aec-37 | 43 | 795 | 6277-8664 | Putative glucosidase, E. coli CFT073 (0.0) | NP_756359.1 | pfam01055 | α-Glycosidase |
| aec-38 | 55 | 151 | C 9306-9761 | IS1 protein InsB, E. coli EDL933 (3e-61) | AAG55747.1 | pfam03400 | Transposase |
| aec-39 | 53 | 91 | C 9680-9955 | IS1 ORFB, Shigella flexneri 2a str. 2457T (7e-42) | NP_836645.1 | pfam03811 | Transposase |
| aec-40 | 51 | 70 | C 9772-9984 | Putative iso-IS1 ORF, Shigella flexneri (5e-25) | AAK18551.1 | None | Unknown |
| aec-41 | 44 | 320 | C 10588-11550 | Putative fructokinase, E. coli EDL933 (9e-61) | NP_311269.1 | cd01167 | Fructokinase |
| aec-42 | 50 | 302 | C 11664-2572 | No significant similarities | |||
| aec-43 | 49 | 497 | C 12691-14184 | Glycosylhydrolase, Bacteroides thetaiotaomicron VPI-5482 (9e-37) | AAO76867.1 | smart00640 | Glycosylhydrolase |
| aec-44 | 46 | 420 | C 14199-15461 | No significant similarities | |||
| aec-45 | 56 | 477 | C 15558-16991 | Sucrose hydrolase, E. coli (e-152) | AAP79503.1 | smart00640 | Glycosylhydrolase |
| aec-46 | 49 | 412 | C 17011-18249 | Sugar transporter FruP, Bacillus megaterium (6e-98) | AAM19070.1 | pfam01306 | Sugar transporter |
| aec-47 | 52 | 311 | 18551-19486 | Putative sucrose-specific transcriptional regulator, Yersinia pestis KIM (2e-86) | NP_669120.1 | smart00354, pfam00532 | Transcriptional regulator |
| aec-48 | 49 | 127 | C 20425-20808 | (ORF truncated) Possible transposase remnant, Yersinia pseudotuberculosis IP 32953 (1e-60) | CAF25366.1 | pfam00665 | Transposase |
| aec-49 | 56 | 301 | C 20793-21698 | Hypothetical ORF in IS2, Klebsiella pneumoniae (e-162) | AAR07888.1 | pfam00665 | Transposase |
| aec-50 | 51 | 136 | C 21656-22066 | IS2 ORFA, Shigella flexneri 2a str. 2457T (3e-60) | NP_838126.1 | pfam01527 | Transposase |
| aec-51 | 53 | 225 | 22338-23015 | Unknown protein encoded by ISEc8, E. coli CFT073 (e-115) | NP_755534.1 | pfam01527 | Transposase |
| aec-52 | 55 | 115 | 23015-23362 | Unknown protein encoded by ISEc8, E. coli CFT073 (4e-60) | NP_755535.1 | pfam05717 | Transposase |
| aec-53 | 57 | 523 | 23382-24953 | IS66-like transposase, E. coli (0.0) | CAD33772.1 | pfam03050 | Transposase |
| aec-54 | 55 | 268 | C 25214-26020 | (ORF truncated) Putative arabinose exporter (MFS superfamily), Acinetobacter sp. ADP1 (3e-29) | YP_047119.1 | COG2814 | Carbohydrate transport and metabolism |
| aec-55 | 54 | 129 | C 26375-26764 | (ORF truncated) Oxidoreductase, aldo/ ketoreductase family, Silicibacter pomeroyi DSS-3 (3e-31) | YP_165172.1 | pfam00248 | Oxidoreductase |
| aec-56 | 52 | 247 | 26921-27664 | (Truncated ORF) IS100 transposase, Yersinia pestis (e-122) | YP_094025.1 | COG4584, partial cd00093 | Transposase |
| aec-57 | 55 | 132 | 27544-27942 | (Truncated ORF) Transposase for insertion sequence IS100, Yersinia pestis biovar Medievalis str. 91001 (e-71) | AAS60948.1 | Partial COG4584 | Transposase |
| aec-58 | 50 | 260 | 27939-28721 | Insertion sequence IS100, ATP-binding protein, Yersinia pestis biovar Medievalis str. 91001 (e-132) | NP_993067.1 | smart00382 | Transposase |
| aec-59 | 53 | 141 | C 28827-29252 | Hypothetical protein SF2980, Shigella flexneri 2a str. 301 (1e-69) | NP_708754.1 | None | Unknown |
| aec-60 | 55 | 129 | C 29249-29638 | Hypothetical protein SF2981, Shigella flexneri 2a str. 301 (1e-59) | NP_708755.1 | None | Unknown |
| aec-61 | 59 | 189 | C 29799-30368 | ORF31, E. coli RW1374 (3e-91) | CAI43834.1 | None | Unknown |
| aec-62 | 51 | 169 | 32482-32991 | ORF33, E. coli RW1374 (2e-96) | CAI43836.1 | None | Unknown |
| aec-63 | 45 | 500 | C 33313-34815 | Hypothetical protein YfjI, E. coli CFT073 (0.0) | NP_755527.1 | None | Unknown |
| aec-64 | 58 | 303 | 35989-36900 | YeeP protein, E. coli (e-162) | CAE85196.1 | COG3596 | GTP binding protein |
| aec-65 | 52 | 225 | C 37213-37890 | Putative transposase, E. coli RW1374 (e-129) | CAI43898.1 | pfam00665 | Transposase |
| aec-66 | 50 | 115 | C 38079-38426 | Unknown protein encoded by IS911 within prophage CP-933L, E. coli EDL933 (2e-46) | AAG58804.1 | pfam01527 | Transposase |
| aec-67 | 58 | 727 | 38411-40594 | (Truncated ORF) Adhesin AIDA-I precursor, E. coli (0.0) | BAA15832.1 | cd01344, pfam03797 | Autotransporter |
| aec-68 | 52 | 840 | 40709-43231 | ORF36, E. coli RW1374 (0.0) | CAI43839.1 | None | Unknown |
| aec-69 | 48 | 151 | 43307-43762 | Hypothetical protein c3665, E. coli CFT073 (2e-83) | NP_755540.1 | None | Unknown |
| aec-70 | 55 | 77 | 43841-44074 | ORF39, E. coli RW1374 (8e-37) | CAI43842.1 | None | Unknown |
| aec-71 | 57 | 272 | 44174-44992 | ORF40, E. coli RW1374 (e-146) | CAI43843.1 | pfam06067 | Unknown |
| aec-72 | 54 | 161 | 45047-45532 | ORF41, E. coli RW1374 (4e-84) | CAI43844.1 | pfam03230 | Antirestriction |
| aec-73 | 58 | 163 | 45533-46024 | YeeS protein, E. coli 536 (1e-74) | CAE85201.1 | pfam04002 | DNA repair protein |
| aec-74 | 53 | 73 | 46094-46315 | ORF44 (YeeT), E. coli RW1374 (5e-34) | CAI43847.1 | None | Unknown |
| aec-75 | 57 | 122 | 46478-46846 | Putative structural protein, E. coli EDL933 (4e-64) | AAG55773.1 | None | Unknown |
| aec-76 | 53 | 124 | 46936-47310 | ORF45, E. coli RW1374 (4e-66) | CAI43848.1 | None | Unknown |
| aec-77 | 51 | 162 | 47307-47795 | YeeW, E. coli RW1374 (5e-87) | CAI43849.1 | None | Unknown |
| aec-78 | 50 | 80 | 47762-48004 | ORF104, E. coli RW1374 (1e-29) | CAI43905.1 | None | Unknown |
| aec-79 | 48 | 281 | 48089-48934 | Z1226 protein, E. coli 536 (e-151) | CAD33792.1 | None | Unknown |
| aec-80 | 39 | 135 | 48980-49387 | (Truncated ORF) Hypothetical protein c4580, E. coli CFT073 (5e-34) | NP_756440.1 | Partial COG3943 | Unknown |
| aec-81 | 34 | 122 | 49530-49898 | Hypothetical protein c4581, E. coli CFT073 (3e-36) | NP_756441.1 | None | Unknown |
C indicates ORFs transcribed on the complementary strand.
str., strain; MFS, major facilitator superfamily.
Putative mobile genetic elements present on AGI-3.
AGI-3 contains genes and putative mobile elements homologous to bacteriophage and insertion sequence (IS) elements that could promote horizontal gene transfer. The aec-33 gene potentially encodes a polypeptide of 394 amino acids that is 86% similar to the integrase of the retronphage φR73, belonging to the tyrosine recombinase family (61). Aec-33 is identical to putative phage integrases identified in pathogenicity islands located at the selC locus of both E. coli strains CFT073 (ORF c4491) and 4797/97 (ORF L01), and almost identical (more than 90% of similar amino acids) to those found in PAIs of Shigella flexneri 2a strain 301 (ORF SF3698), E. coli EDL933 (ORF Z5087), Yersinia pseudotuberculosis IP 32953 (ORF YPTB3886), and Photorhabdus luminescens subsp. laumondii TTO1 (ORF plu0125) (11, 22, 33, 51, 57, 65). Fourteen ORFs of AGI-3 are homologous to IS elements or transposons of the IS1, IS1222, IS2, ISEc8, IS66, IS100, or IS911 type (Fig. 1 and Table 3). Only the IS1 element (aec-38 to aec-40) seems to be intact and to possess its left and right terminal inverted repeats (47). All the other elements are disrupted either by frameshift or by integration of another IS element. These data strongly suggest that AGI-3 has evolved by repeated recombination events.
Genetic features of AGI-3.
Among the 49 ORFs identified, 35 are not related to mobility. These 35 ORFs can be divided into five modules bound by either repeats, insertion elements, or bacteriophage sequences (Fig. 1, Table 3).
Module 1, which is composed of gene aec-34 and of the aec-35-aec-37 gene cluster, was also found to be present at the same location and with the same organization in the uropathogenic E. coli strain CFT073 (99% identical nucleotide) (Table 4) (65). The G+C content (average: 43%) of the genes in module 1 is lower than the average for the E. coli genome (51%) (Table 3). The aec-34 gene encodes a protein highly homologous to ShiA of Shigella flexneri, involved in the attenuation of inflammation (31). The shiA gene is located downstream of the integrase gene of the SHI-2 pathogenicity island present at the selC locus (44).
TABLE 4.
Orthologs of Aec-35, Aec-36, and Aec-37 found in the genomes of sequenced pathogenic bacteria
| Species and strain | Homolog (% amino acid similarity)
|
||
|---|---|---|---|
| Aec-35 | Aec-36 | Aec-37 | |
| Escherichia coli CFT073 | C4494 (99) | C4494 (100) | C4496 and C4495 (99) |
| Shigella sonnei Ss046 | SSO_0802 (87) | SSO_0801 (94) | SSO_0800 (91) |
| Erwinia carotovora subsp. atroseptica SCRI1043 | ECA1966 (75) | ECA1967 (89) | ECA1968 (84) |
| Yersinia pseudotuberculosis IP 32953 | YPTB3091 (74) | YPTB3092 (85) | YPTB3093 (83) |
| Yersinia pestis biovar Orientalis CO92 | YP00846 (74) | YP00847 (85) | YP00848 (83) |
| Yersinia pestis biovar Mediaevalis KIM | Y3231 (74) | Y3232 (85) | Y3233 (83) |
| Yersinia pestis biovar Mediaevalis 9100 | YP3543 (74) | YP3544 (85) | YP3545 (83) |
| Burkholderia cepacia R18194 | Not present | Bcepa03004976 (62) | Bcepa03004977 (59) |
| Ralstonia solanacearum GMI1000 | Not present | RSc1080 (62) | RSc1081 (61) |
The aec-35 to aec-37 gene cluster is potentially involved in carbohydrate transport and metabolism. The aec-35 product is homologous to transcriptional regulators of the LacI family. Aec-35 has a helix-turn-helix DNA-binding motif (position 5 to 26) and a sugar binding domain of the LacI family (position 73 to 313). The aec-36 product shares similarity (62% similar amino acids) with ExuT of E. coli K-12 MG1655, a major facilitator superfamily transporter of the anion:cation symporter family 14 (48). ExuT is involved in galacturonate uptake in E. coli and in other bacteria such as Erwinia chrysanthemi and Ralstonia solanacearum (26, 45, 63). Aec-36, like ExuT, is predicted to be an integral membrane protein.
The aec-37 gene encodes a putative glycosylhydrolase of family 31. This family is very diverse and contains alpha-glucosidases, glucoamylases, sucrase-isomaltases, alpha-xylosidases, alpha-glucan lyases, and isomaltosyltransferases (13). The aec-35-aec-37 region is present in the sequenced genome of other pathogenic bacterial species (Table 4). The cluster is present in Shigella sonnei, Yersinia pestis biovar Orientalis CO92, Yersinia pseudotuberculosis IP32953, Yersinia pestis biovar Medievalis KIM and 91001, and Erwinia carotovora subsp. atroseptica SCRI1043 (3, 11, 15, 49, 60). In these species, the organization of the aec-35-aec-37 gene cluster is the same as in E. coli BEN2908 and CFT073 but the cluster is not inserted at the same genomic location. The genes aec-36 and aec-37 but not aec-35 are also present in Burkholderia cepacia and in Ralstonia solanacearum GMI1000 (55). In these two species, the aec-36 and aec-37 orthologs are linked to a transcriptional regulator that is divergently transcribed but that does not show any similarity with Aec-35.
Module 2 is composed of the aec-41 to aec-47 genes. The gene products show homologies with proteins putatively involved in carbohydrate transport and metabolism (putative fructokinase, glycosylhydrolases, and sugar transporter of the major facilitator oligosaccharide:H+ symporter family 5). Such an arrangement of genes has not been described before.
Module 3 includes ORFs aec-54 and aec-55. Due to the insertion of the IS elements aec-53 and aec-56, the predicted products of aec-54 and aec-55 are truncated compared to their homologs (a putative arabinose exporter of Acinetobacter sp. strain ADP1 and an oxidoreductase of the aldo/ketoreductase family of Silicibacter pomeroyi strain DSS-3) (Table 3). Thus, module 3 is probably nonfunctional.
Module 4, comprising ORFs aec-59 to aec-64, is very similar to the region containing ORF29 to ORF34 in the enterohemorrhagic E. coli strain RW1374, located in a 111-kb pathogenicity island inserted at the pheV locus (34). The products of the ORFs of module 4 are of unknown function.
Module 5 is composed of aec-67 to aec-81. A nearly identical region is present in the 111-kb pathogenicity island of E. coli strain RW1374 (ORF35, ORF36, and ORF39 to ORF47) (34). This module is also present in the she pathogenicity island of Shigella flexneri 2a strain 301 inserted at the pheV locus, in genomic island I of E. coli strain Nissle 1917 inserted at the serX locus, and in two different genomic islands of E. coli strain CFT073, inserted at the serX and pheV loci (1, 27, 65). Various ORFs of module 5 show homology with the CP4-44 prophage of E. coli K-12 MG1655 (6). Gene aec-67 product is highly related to the AIDA-I precursor encoded by the flu gene. However, aec-67 is truncated at its 5′ end by the insertion of the IS911 transposase gene, leading to a deletion of 380 amino acids compared to the native AIDA-I precursor of E. coli MG1655. AIDA belongs to the autotransporter protein family, which comprises many virulence factors (29).
Cluster aec-35 to aec-37 is implicated in carbohydrate metabolism and virulence.
As the products of aec-35 to aec-37 are homologous to proteins implicated in carbohydrate transport and metabolism, we tested the role of this gene cluster in the metabolism of strain BEN2908. For that aim, the aec-35-aec-37 region (5,124 bp, Fig. 1) was replaced by a kanamycin resistance cassette by the method of Datsenko and Wanner, leading to mutant strain BEN2929 (BEN2908Δaec-35-37::Kanr) (14).
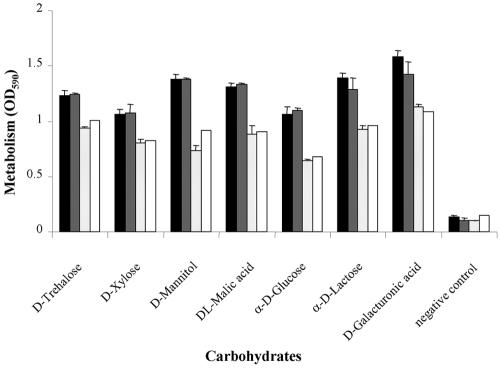
The capacity of wild-type strain BEN2908 and mutant BEN2929 to metabolize 190 different carbon sources was then tested using PM1 and PM2 phenotype microarrays from Biolog. A statistically significant reduction in the metabolism of d-mannitol (P = 0.0073), α-d-glucose (P = 0.0081), α-d-lactose (P = 0.0074), d-xylose (P = 0.0309), d,l-malic acid (P = 0.0035), d-galacturonic acid (P = 0.0085), and d-threhalose (P= 0.0084) was observed for the mutant strain BEN2929 (Fig. 2). The wild phenotype was restored by complementation of the deleted mutant BEN2929 with the entire aec-35-aec-37 region previously cloned into the high-copy-number plasmid pCR2.1-Topo (strain BEN2929/pBEN182). When tested on PM1 and PM2 phenotype microarray plates, the complemented strain metabolized the above-listed carbohydrates as well as the wild-type strain, confirming that at least one of the aec-35-aec-37 genes is involved in the metabolism of these carbohydrates (Fig. 2). Furthermore, the generation times of the wild-type and the mutant strains were measured in minimum medium containing Na-d-galacturonate or trehalose. In both conditions, the generation time of the mutant strain was twofold less than those of the wild-type strain (1,500 min versus 750 min and 750 min versus 375 min in the presence of Na-d-galacturonate or trehalose, respectively).
FIG. 2.
Role of the aec-35-aec-37 gene cluster in carbohydrate metabolism. Data for the seven carbohydrates that were differently metabolized by strain BEN2908 and its derivatives are reported as well as data obtained for the negative control (a well without carbohydrate). Strains BEN2908, BEN2929/pBEN182, BEN2929, and BEN2929/pCR2.1 (from left to right within each group) were seeded into Biolog PM1 and PM2 microplates containing a panel of 190 carbohydrates. The microplates were incubated aerobically at 37°C for 24 h and the optical density at 590 nm (OD590) was measured. Experiments were done in triplicate (except for strain BEN2929/pCR2.1); means and standard deviations are indicated.
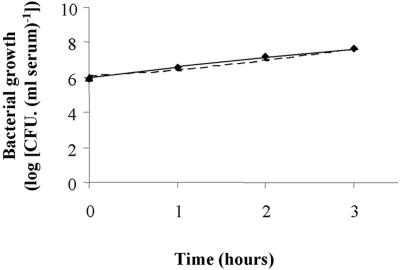
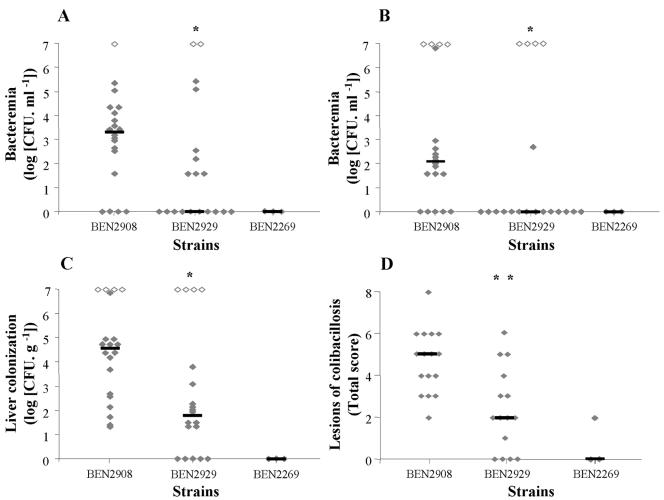
As the aec-35-aec-37 gene cluster is well conserved in several pathogenic bacteria of the family Enterobacteriaceae (Erwinia carotovora, Shigella sonnei, and Yersinia spp., Table 4), we hypothesized that the ability to efficiently metabolize the seven carbohydrates identified above would give an adaptive advantage to E. coli for the colonization of several tissues and body fluids. This was first tested by comparing the growth of the wild-type strain BEN2908 and its mutant derivative BEN2929 in chicken serum. Both strains were able to multiply equally well in serum (Fig. 3). We then compared the virulence of both strains in experimental colibacillosis of chicken. Compared to the wild-type strain, the mutant derivative was significantly (P = 0.0016) less able to induce specific lesions of colibacillosis such as aerosacculitis, perihepatitis, and pericarditis (Fig. 4). The mutant was also less bacteremic at 24 h (P = 0.0457) and 48 h postinoculation (P = 0.0328), and less able to colonize the liver (P = 0.0175) than the wild-type strain (Fig. 4). These results indicate that cluster aec-35-aec-37 is involved in the virulence mechanism of E. coli BEN2908 for chicken.
FIG. 3.
Bacterial growth in chicken serum. Strain BEN2908 (⧫), its derivative BEN2929 (dotted line) were grown in 90% normal chicken serum for 3 h. Viable cells were estimated by plating serial dilutions of the cultures at 1-hour intervals.
FIG. 4.
Role of the aec-35-aec-37 gene cluster in the colonization of liver and blood and in the development of colibacillosis lesions in chickens. Twenty and 19 25-day-old specific-pathogen-free White Leghorn chickens were inoculated in the right air sac with 5 × 106 CFU of strains BEN2908 or BEN2929, respectively. Three chickens were inoculated with the nonpathogenic avian strain BEN2269. Each animal is represented by a diamond symbol (⧫, ⋄). Bacteremia was determined 24 h (A) and 48 h (B) postinoculation. Animals were euthanized 48 h postinoculation by injection of Nesdonal and then necropsied. Liver bacterial colonization (C) and colibacillosis lesions (D) were recorded. The lesion score was not determined for dead animals. In each group infected by either BEN2908 or BEN2929, four chickens died and were given the highest rank. They are represented by an open diamond symbol (⋄). Bars indicate the median for each group of chickens. A Wilcoxon-Mann-Whitney test was used to analyze the differences between groups of chickens inoculated with the wild-type and the mutant strains (*, P < 0.05; **, P < 0.01).
Prevalence of aec-35, aec-36, and aec-37 in a collection of pathogenic and nonpathogenic E. coli strains.
The prevalence of the aec-35-aec-37 gene cluster was examined among 36 nonpathogenic strains of avian origin and 249 ExPEC strains: 205 strains of avian origin, 7 strains isolated from animals other than poultry, and 37 strains of human origin. For that aim, PCRs were performed on each strain to amplify a 948-bp internal fragment of aec-35 (primers IC79 and IC36, Table 2), a 1,225-bp DNA fragment of aec-36 (primers IC39 and IC64, Table 2), and a 1,574-bp DNA fragment of aec-37 (primers IC45 and IC50, Table 2). These three ORFs were amplified in 33 (30 strains of avian origin and 3 strains of human origin) of the strains analyzed (11.6%), whereas in two strains (one of avian origin and one of an animal other than avian origin) only the aec-35 ORF could be amplified (Table 5).
TABLE 5.
Distribution and genetic link to selC of genes aec-35, aec-36, and aec-37 in a collection of pathogenic and nonpathogenic E. coli strainsa
| DNA region amplified | No. of strains positive
|
||
|---|---|---|---|
| Avian pathogenic strains (n = 205) | Avian nonpathogenic strains (n = 44) | Human and animal pathogenic strains (n = 36) | |
| aec-35 | 27 | 4 | 4 |
| aec-36 | 26 | 4 | 3 |
| aec-37 | 26 | 4 | 3 |
| aec-35 to aec-37 | 26 | 4 | 3 |
| selC to aec-35 | 25 | 4 | 3 |
Pathogenic strains of avian origin were isolated from chickens or ducks with signs of colibacillosis and were able to kill at least four of five day-old chicks inoculated subcutaneously with 108 CFU. Nonpathogenic strains of avian origin were isolated from the gut of healthy birds and were unable to kill any chick in the day-old chick lethality model (16). Other pathogenic strains were isolated from pathological samples.
The locations of the three ORFs (aec-35 to aec-37) were determined in the 33 positive strains by amplification of a fragment overlapping aec-35 and aec-36 (primers IC37 and IC60, Table 2) and a fragment overlapping aec-36 and aec-37 (primers IC43 and IC44, Table 2). In all 33 of these strains the aec-35, aec-36, and aec-37 genes were linked in one cluster and in the same order as in BEN2908. A PCR amplifying a fragment of 4,143 bp covering the region between selC and aec-35 (primers SelC1 and IC80, Table 2) was then performed to investigate if the aec-35-aec-37 cluster was linked to selC. In 32 strains of the 33 strains carrying the aec-35-aec-37 cluster, it was linked to selC (Table 5). Among the 33 strains carrying aec-35-aec-37 genes, 29 belonged to the pathogenic group and 4 to the nonpathogenic group (Table 5). The prevalence of the region was therefore not significantly different (around 12%) between the pathogenic and the nonpathogenic strains tested. It is interesting that the aec-35-aec-37 cluster was present in all 15 avian pathogenic strains tested belonging to the O5 serogroup.
DISCUSSION
Genomic islands have been described as horizontally acquired DNA regions that are frequently chromosomally inserted in the vicinity of tRNA genes (28). They encode various functions which are related to virulence, symbiosis, metabolism, resistance to antibiotics or degradation of xenobiotic compounds. A typical genomic island is flanked by direct repeat structures and carries several genes encoding traits that may increase bacterial adaptability or fitness under certain growth conditions. Typically, GEIs carry multiple functional and fragmented insertion sequence elements and other mobility-related genes, as well as a functional integrase (int) gene, the product of which is involved in insertion and deletion of the DNA region that is flanked by direct repeat structures (19).
We have characterized a novel genomic island (AGI-3) which fulfills most of the criteria described above. AGI-3 is inserted at the selC tRNA gene (a hot spot insertion site for PAIs) in the avian ExPEC strain BEN2908. The presence of a putative active integrase gene (aec-33) the product of which is highly similar (86%) to the integrase of the retronphage φR73, the vicinity of the selC tRNA locus, integration site of φR73 phage, and the presence of direct repeats of 16 bp corresponding to the half site of the attP site of φR73 strongly suggest that AGI-3 or a primitive core of AGI-3 was acquired by horizontal gene transfer via a bacteriophage of the φR73 family. These elements are also in favor of putative mobility of the AGI-3 island as it was observed for PAI-I of the uropathogenic E. coli strain 536 (18).
AGI-3 shows a modular structure composed of five modules bound by IS elements. Parts of modules 4 and 5 (aec-61 to aec-78, with the exception of aec-65 and aec-66, which encode transposases) are also found with the same organization in the 111-kb PAI of the enterohemorrhagic E. coli strain RW1374 inserted at the pheV locus (34). Moreover, from aec-64 (located at the right end of module 4) to aec-81, very similar regions exist in the she pathogenicity island of Shigella flexneri 2a strain 301 inserted at the pheV locus, in genomic island I of E. coli strain Nissle 1917 inserted at the serX locus, and in two different islands of E. coli strain CFT073, inserted at the serX and pheV loci (1, 27, 65). The aec-65 and aec-66 ORFs could not be found in these PAIs. Taken together, these observations indicate that module 4 and module 5 initially consisted of one module that was later disrupted by the aec-65-aec-66 transposase genes in strain BEN2908 or modified by the insertion of transposase-encoding genes at various locations in the other cited PAIs.
Various ORFs of modules 4 and 5 (aec-64, -67, -68, -73, -74, -75, -76, and -77) are also similar to the putative prophage CP4-44, suggesting that they could have a phage origin (6, 10). Modules 1 and 3 are also found in other pathogenic E. coli strains and in other species of pathogenic bacteria, whereas the organization of module 2 has never been described before. The presence of modules with a similar organization in other bacteria as well as the presence of IS elements and transposase genes strongly argue in favor of stepwise acquisition of the modules from other strains into an AGI-3 primitive core, resulting in a mosaic structure.
The aec-35-aec-37 gene cluster of module 1 was shown to be implicated in the uptake of seven carbohydrates and faster growth of the strain in minimal medium containing galacturonate or trehalose. The aec-35 gene was predicted to encode a regulator of the LacI family. As the bacterial LacI family has been described to repress transcription of genes involved in carbohydrate transport and utilization, deletion of the aec-35 gene would lead to abolition of the repression of such genes and to an increase in the transport of carbohydrates (64). Such results have not been observed in our phenotype microarray assays since we only demonstrated the lesser utilization of seven carbohydrates by the mutant strain BEN2929. We can thus rule out the hypothesis that the observed decrease in the assimilation of seven carbohydrates could be due to a pleiotropic effect resulting from the deletion of aec-35.
The aec-36 gene was predicted to encode a major facilitator superfamily sugar permease similar to the hexuronate transporter ExuT of E. coli (52). Among the seven carbohydrates identified only the d-galacturonic acid is a hexuronate, the others are pentose (d-xylose), tetrose (d,l-malic acid), hexoses (α-d-glucose, d-xylose and d-mannitol) or disaccharides (d-lactose and d-trehalose). We can thus hypothesize that the Aec-36 sugar permease encoded by strain BEN2908 could be able to transport different kind of carbohydrates. However all these carbohydrates can be also transported by other permeases as we do not have a complete abolition of their assimilation in the mutant strain (Fig. 2). Furthermore, as only 190 different carbohydrates were tested with phenotype microarrays, we cannot completely exclude that the main substrate of the system is another carbohydrate that has not been identified yet.
The aec-37 gene product, a putative glycosylhydrolase, has been assigned to family 31 in Henrissat's classification based on amino acid sequence (30). Enzymes in this family cleave only α-glucosidic linkages (1-1, 1-2, 1-3, 1-4, and 1-6) from various di- and/or oligosaccharides. Among the seven identified carbohydrates, only d-lactose and d-trehalose are disaccharides but only d-trehalose is formed by two glucose molecules bound in αα-1,1 linkage and might be a substrate for Aec-37. To better understand the activity of the enzyme encoded by aec-37, it would be interesting to clearly determine its substrate.
The mutant strain deleted of the aec-35-aec-37 gene cluster was also found to be less able to induce specific lesions of colibacillosis, bacteremia, and colonization of the liver of chickens compared to the wild-type strain. These results strongly suggest that the aec-35-aec-37 gene cluster is involved in the virulence of strain BEN2908 for chickens. Avian colibacillosis generally starts as a respiratory infection that evolves to a generalized infection resulting in fibrinopurulent lesions of internal organs. Passage into the bloodstream is an important step for the dissemination of the bacteria in avian colibacillosis and allows further multiplication in blood and colonization of internal organs.
Pourbakhsh et al. have shown that the invasion of the vascular system via the lung and the damaged air sac interstitium may be an important portal of entry for bacteria (53). We have observed that the mutant was less bacteremic than the wild-type strain in experimental colibacillosis. However, in in vitro tests, the mutant strain showed the same capacity to multiply in chicken serum as the wild-type strain. We thus hypothesize that the deletion of the aec-35-aec-37 gene cluster affects the early steps of colibacillosis, such as colonization of the air sacs and/or the lung, due to reduced fitness, leading to a reduced ability to invade the bloodstream and to disseminate in internal organs.
Indeed, the importance of hexuronate and other sugar catabolism in the fitness of E. coli has recently been stressed. It has been shown that hexuronate catabolism plays a role in the maintenance of E. coli strain MG1655 in the mouse intestine. Furthermore, several genes, such as exuT, that are involved in catabolism of hexuronate are significantly induced by mucus (12). Analysis of the transcriptome of the uropathogenic E. coli strain CFT073 during urinary tract infection also revealed that genes involved in hexuronate metabolism were upregulated. The authors suggested that in the urinary tract metabolism shifted so that hexuronates could be used to support growth (59).
Recently, in the APEC strain IMT5155, which harbors the same serotype (O2:K1:H5) as BEN2908, it was shown that metabolic functions play a role in host infectivity (38). Using signature-tagged mutagenesis, Li et al. (38) identified several genes, mutation of which results in reduced septicemia, that are also involved in metabolism and nutrient uptake. Of these gene products, one is the transketolase TktA, involved in the catabolism of pentose sugars, the formation of d-ribose 5-phosphate and in the provision of d-erythrose 4-phosphate, a precursor of aromatic amino acids, aromatic vitamins and of pyridoxine. Another is a β-cystathionase involved in methionine biosynthesis, and the latter is involved in pyrimidine pathway regulation.
Taken together, all of these observations indicate that metabolism in ExPEC is probably closely linked to virulence. In addition to the aec-35-aec-37 gene cluster of module 1, we have also identified module 2 of AGI-3, which is potentially involved in carbohydrate uptake and assimilation and could also be linked to virulence.
In conclusion, we reported here the description of a new APEC genomic island associated with the selC locus and we demonstrated its role in carbohydrate assimilation and virulence. As three of the five modules described in AGI-3 of the APEC strain BEN2908 are also present in other pathogenic E. coli and in other pathogenic bacterial species, it is tempting to hypothesize that these modules also play a similar role in other pathogenic bacteria. As the AGI-3 genomic island possesses all the potential to be mobile, future work will be directed towards evaluation of its ability to be transferred between E. coli strains and between various bacterial species.
Acknowledgments
Some of the avian E. coli strains used for this study are part of the European collection of APEC strains (European Project FAIR6-CT-98-4093). We thank J. Blanco (Faculdad de Veterinaria, Universidad de Santiago de Compostella, Lugo, Spain) for the serotyping of most avian E. coli strains and R. Quentin (EA 3854, UFR Medecine, Tours, France) for providing some of the ExPEC strains of human origin. We are grateful to E. Guédon and S. D. Ehrlich (Laboratoire de Génétique Microbienne, INRA, Jouy en Josas, France) for the use of the Bioscreen C apparatus. We are also grateful to Patrice Cousin and Nelly Rouet for their assistance in the in vivo virulence assays, and we thank M. Répérent for technical assistance.
This work was supported by the European Community under FP5 (COLIRISK-QLK2-CT-2002-00944) and by the INRA under the AIP séquençage.
REFERENCES
- 1.Al-Hasani, K., K. Rajakumar, D. Bulach, R. Robins-Browne, B. Adler, and H. Sakellaris. 2001. Genetic organization of the she pathogenicity island in Shigella flexneri 2a. Microb. Pathog. 30:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, H. J., J.-P. Vaillancourt, and W. B. Gross. 2003. Colibacillosis, p. 631-652. In H. J. B. Y. M. Saif, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), Diseases of poultry, 11th ed. Iowa State University Press, Ames, Iowa.
- 3.Bell, K. S., M. Sebaihia, L. Pritchard, M. T. Holden, L. J. Hyman, M. C. Holeva, N. R. Thomson, S. D. Bentley, L. J. Churcher, K. Mungall, R. Atkin, N. Bason, K. Brooks, T. Chillingworth, K. Clark, J. Doggett, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, H. Norbertczak, D. Ormond, C. Price, M. A. Quail, M. Sanders, D. Walker, S. Whitehead, G. P. Salmond, P. R. Birch, J. Parkhill, and I. K. Toth. 2004. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. USA 101:11105-11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco, J. E., M. Blanco, A. Mora, W. H. Jansen, V. Garcia, M. L. Vazquez, and J. Blanco. 1998. Serotypes of Escherichia coli isolated from septicaemic chickens in Galicia (northwest Spain). Vet. Microbiol. 61:229-235. [DOI] [PubMed] [Google Scholar]
- 5.Blanc-Potard, A. B., and E. A. Groisman. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 16:5376-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschape, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochner, B. R., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, P. K., and R. Curtiss 3rd. 1996. Unique chromosomal regions associated with virulence of an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 93:11149-11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277-300. [DOI] [PubMed] [Google Scholar]
- 11.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, D. E., D. J. Smalley, D. L. Tucker, M. P. Leatham, W. E. Norris, S. J. Stevenson, A. B. Anderson, J. E. Grissom, D. C. Laux, P. S. Cohen, and T. Conway. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 101:7427-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328:307-317. [DOI] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, W., V. Burland, G. Plunkett 3rd, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dho, M., and J. P. Lafont. 1982. Escherichia coli colonization of the trachea in poultry: comparison of virulent and avirulent strains in gnotoxenic chickens. Avian Dis. 26:787-797. [PubMed] [Google Scholar]
- 17.Dho-Moulin, M., and J. M. Fairbrother. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30:299-316. [PubMed] [Google Scholar]
- 18.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 20.Dozois, C. M., F. Daigle, and R. Curtiss 3rd. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 100:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dozois, C. M., M. Dho-Moulin, A. Bree, J. M. Fairbrother, C. Desautels, and R. Curtiss 3rd. 2000. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect. Immun. 68:4145-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J. F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Medigue, A. Lanois, K. Powell, P. Siguier, R. Vincent, V. Wingate, M. Zouine, P. Glaser, N. Boemare, A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 23.Elliott, S. J., V. Sperandio, J. A. Giron, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleckenstein, J. M., L. E. Lindler, E. A. Elsinghorst, and J. B. Dale. 2000. Identification of a gene within a pathogenicity island of enterotoxigenic Escherichia coli H10407 required for maximal secretion of the heat-labile enterotoxin. Infect. Immun. 68:2766-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Germon, P., Y. H. Chen, L. He, J. E. Blanco, A. Bree, C. Schouler, S. H. Huang, and M. Moulin-Schouleur. 2005. ibeA, a virulence factor of avian pathogenic Escherichia coli. Microbiology 151:1179-1186. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez, E. T., and C. Allen. 2003. Characterization of a Ralstonia solanacearum operon required for polygalacturonate degradation and uptake of galacturonic acid. Mol. Plant-Microbe Interact. 16:536-544. [DOI] [PubMed] [Google Scholar]
- 27.Grozdanov, L., C. Raasch, J. Schulze, U. Sonnenborn, G. Gottschalk, J. Hacker, and U. Dobrindt. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 186:5432-5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 29.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingersoll, M. A., J. E. Moss, Y. Weinrauch, P. E. Fisher, E. A. Groisman, and A. Zychlinsky. 2003. The ShiA protein encoded by the Shigella flexneri SHI-2 pathogenicity island attenuates inflammation. Cell. Microbiol. 5:797-807. [DOI] [PubMed] [Google Scholar]
- 32.Inouye, S., M. G. Sunshine, E. W. Six, and M. Inouye. 1991. Retronphage φR73: an E. coli phage that contains a retroelement and integrates into a tRNA gene. Science 252:969-971. [DOI] [PubMed] [Google Scholar]
- 33.Jin, Q., Z. Yuan, J. Xu, Y. Wang, Y. Shen, W. Lu, J. Wang, H. Liu, J. Yang, F. Yang, X. Zhang, J. Zhang, G. Yang, H. Wu, D. Qu, J. Dong, L. Sun, Y. Xue, A. Zhao, Y. Gao, J. Zhu, B. Kan, K. Ding, S. Chen, H. Cheng, Z. Yao, B. He, R. Chen, D. Ma, B. Qiang, Y. Wen, Y. Hou, and J. Yu. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jores, J., S. Wagner, L. Rumer, J. Eichberg, C. Laturnus, P. Kirsch, P. Schierack, H. Tschape, and L. H. Wieler. 2005. Description of a 111-kb pathogenicity island (PAI) encoding various virulence features in the enterohemorrhagic E. coli (EHEC) strain RW1374 (O103:H2) and detection of a similar PAI in other EHEC strains of serotype 0103:H2. Int. J. Med. Microbiol. 294:417-425. [DOI] [PubMed] [Google Scholar]
- 35.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 36.Lafont, J. P., M. Dho, H. M. D'Hauteville, A. Bree, and P. J. Sansonetti. 1987. Presence and expression of aerobactin genes in virulent avian strains of Escherichia coli. Infect. Immun. 55:193-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.La Ragione, R. M., A. R. Sayers, and M. J. Woodward. 2000. The role of fimbriae and flagella in the colonization, invasion and persistence of Escherichia coli O78:K80 in the day-old-chick model. Epidemiol. Infect. 124:351-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, G., C. Laturnus, C. Ewers, and L. H. Wieler. 2005. Identification of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect. Immun. 73:2818-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marc, D., and M. Dho-Moulin. 1996. Analysis of the fim cluster of an avian O2 strain of Escherichia coli: serogroup-specific sites within fimA and nucleotide sequence of fimI. J. Med. Microbiol. 44:444-452. [DOI] [PubMed] [Google Scholar]
- 41.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mellata, M., M. Dho-Moulin, C. M. Dozois, R. Curtiss 3rd, P. K. Brown, P. Arne, A. Bree, C. Desautels, and J. M. Fairbrother. 2003. Role of virulence factors in resistance of avian pathogenic Escherichia coli to serum and in pathogenicity. Infect. Immun. 71:536-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, J. H. 1972. Experiments in molecular genetics, p. 31-36. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 44.Moss, J. E., T. J. Cardozo, A. Zychlinsky, and E. A. Groisman. 1999. The selC-associated SHI-2 pathogenicity island of Shigella flexneri. Mol. Microbiol. 33:74-83. [DOI] [PubMed] [Google Scholar]
- 45.Nemoz, G., J. Robert-Baudouy, and F. Stoeber. 1976. Physiological and genetic regulation of the aldohexuronate transport system in Escherichia coli. J. Bacteriol. 127:706-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nolan, L. K., S. M. Horne, C. W. Giddings, S. L. Foley, T. J. Johnson, A. M. Lynne, and J. Skyberg. 2003. Resistance to serum complement, iss, and virulence of avian Escherichia coli. Vet. Res. Commun. 27:101-110. [DOI] [PubMed] [Google Scholar]
- 47.Ohta, S., K. Tsuchida, S. Choi, Y. Sekine, Y. Shiga, and E. Ohtsubo. 2002. Presence of a characteristic D-D-E motif in IS1 transposase. J. Bacteriol. 184:6146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 50.Parreira, V. R., and C. L. Gyles. 2003. A novel pathogenicity island integrated adjacent to the thrW tRNA gene of avian pathogenic Escherichia coli encodes a vacuolating autotransporter toxin. Infect. Immun. 71:5087-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 52.Portalier, R., J. Robert-Baudouy, and F. Stoeber. 1980. Regulation of Escherichia coli K-12 hexuronate system genes: exu regulon. J. Bacteriol. 143:1095-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pourbakhsh, S. A., M. Boulianne, B. Martineau-Doize, C. M. Dozois, C. Desautels, and J. M. Fairbrother. 1997. Dynamics of Escherichia coil infection in experimentally inoculated chickens. Avian Dis. 41:221-233. [PubMed] [Google Scholar]
- 54.Russo, T. A., and J. R. Johnson. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181:1753-1754. [DOI] [PubMed] [Google Scholar]
- 55.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 57.Schmidt, H., W. L. Zhang, U. Hemmrich, S. Jelacic, W. Brunder, P. I. Tarr, U. Dobrindt, J. Hacker, and H. Karch. 2001. Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect. Immun. 69:6863-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schouler, C., F. Koffmann, C. Amory, S. Leroy-Setrin, and M. Moulin-Schouleur. 2004. Genomic subtraction for the identification of putative new virulence factors of an avian pathogenic Escherichia coli strain of O2 serogroup. Microbiology 150:2973-2984. [DOI] [PubMed] [Google Scholar]
- 59.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song, Y., Z. Tong, J. Wang, L. Wang, Z. Guo, Y. Han, J. Zhang, D. Pei, D. Zhou, H. Qin, X. Pang, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, F. Chen, S. Li, C. Ye, Z. Du, W. Lin, J. Yu, H. Yang, P. Huang, and R. Yang. 2004. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 11:179-197. [DOI] [PubMed] [Google Scholar]
- 61.Sun, J., M. Inouye, and S. Inouye. 1991. Association of a retroelement with a P4-like cryptic prophage (retronphage φR73) integrated into the selenocystyl tRNA gene of Escherichia coli. J. Bacteriol. 173:4171-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tung, W. L., and K. C. Chow. 1995. A modified medium for efficient electrotransformation of E. coli. Trends Genet. 11:128-129. [DOI] [PubMed] [Google Scholar]
- 63.Valmeekam, V., Y. L. Loh, and M. J. San Francisco. 2001. Control of exuT activity for galacturonate transport by the negative regulator ExuR in Erwinia chrysanthemi EC16. Mol. Plant-Microbe Interact. 14:816-820. [DOI] [PubMed] [Google Scholar]
- 64.Weickert, M. J., and S. Adhya. 1992. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 267:15869-15874. [PubMed] [Google Scholar]
- 65.Welch, R. A., V. Burland, G. Plunkett 3rd, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou, L., X. H. Lei, B. R. Bochner, and B. L. Wanner. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 185:4956-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]