Abstract
Helicobacter pylori is one of the most diverse bacterial species known. A rational basis for this genetic variation may be provided by its natural competence for genetic transformation and high-frequency recombination. Many bacterial competence systems have homology with proteins that are involved in the assembly of type IV pili and type II secretion systems. In H. pylori, DNA uptake relies on a transport system related to type IV secretion systems (T4SS) designated the comB system. The prototype of a T4SS in Agrobacterium tumefaciens consists of 11 VirB proteins and VirD4, which form the core unit necessary for the delivery of single proteins or large nucleoprotein complexes into target cells. In the past we identified proteins ComB4 and ComB7 through ComB10 as being involved in the process of DNA uptake in H. pylori. In this study we identified and functionally characterized further (T4SS-homologous) components of the comB transformation competence system. By combining computer prediction modeling, experimental topology determination, generation of knockout strains, and genetic complementation studies we identified ComB2, ComB3, and ComB6 as essential components of the transformation apparatus, structurally and functionally homologous to VirB2, VirB3, and VirB6, respectively. comB2, comB3, and comB4 are organized as a separate operon. Thus, for the H. pylori comB system, all T4SS core components have been identified except for homologues to VirB1, VirD4, VirB5, and VirB11.
Helicobacter pylori, a highly motile gram-negative bacterial pathogen, is the principal cause of chronic active gastritis and peptic ulcer disease in humans and has been implicated in the development of gastric mucosa-associated lymphoid tissue lymphoma and adenocarcinoma (38, 49). H. pylori colonizes a very special habitat at the surface of gastric epithelial cells or in the mucus layer covering the epithelium, which suggests that this bacterium has evolved specialized features for gastric adaptation. A comparison of the two published genome sequences shows considerable diversity in gene content, with about 7% of all putative genes being strain specific (2). The panmictic population structure of H. pylori is believed to result from frequent recombination during mixed colonization by unrelated strains (20, 50).
H. pylori is naturally competent for genetic transformation (36). Natural transformation competence in bacteria is a complex process, involving DNA binding, uptake/translocation, and recombination. In general, bacterial competence systems have homology with proteins that are involved in the assembly of type IV pili and type II secretion systems and form a structure that partially spans the cell envelope (see reference 13 for a recent review). In H. pylori, DNA uptake is not mediated by a type IV pilus-based apparatus but relies on a transport system related to type IV secretion systems (T4SS) designated the comB system (26).
The Vir system of Agrobacterium tumefaciens, necessary for the transfer of its transferred DNA (T-DNA) into plant cells, is considered the prototype for type IV secretion (10). It is assembled from the products of the virB operon (the VirB1 through VirB11 proteins) and the virD4 gene, which form a cell envelope-spanning channel for the translocation of the substrate. Three putative ATPases, VirD4, VirB4, and VirB11, are involved in this process. The transmembrane channel itself, connecting the inner and outer membranes, is thought to be composed of the proteins VirB7 through VirB10; the properties and interactions between these proteins and the substrate have been studied in detail (11, 43). The polytopic inner-membrane protein VirB6 is considered to form a pore or channel at the cytoplasmic membrane, and the pilin subunit, VirB2, assembles as the T pilus in association with VirB5 and the VirB7 lipoprotein. In addition to the virB system, A. tumefaciens also carries two other T4SS, the trb (32) and the avhB (14) systems. The trb system encodes functions required for the conjugal transfer of the Ti plasmid between Agrobacterium spp., whereas the avhB T4SS mediates the conjugal transfer of the pAtC58 cryptic plasmid between cells of A. tumefaciens.
The H. pylori ComB proteins are named after their orthologues from the A. tumefaciens VirB system. As known so far, they consist of five open reading frames (ORFs): comB4, encoding a putative ATPase, and comB7 through comB10, supposed to be structural components of the membrane-spanning secretion pore (24, 25). ComB8 through ComB10 are tightly associated with the membrane, probably forming a bridge between the inner and outer membranes (24). ComB8, ComB9, and ComB10 are essential for transformation competence, whereas ComB7 is not but seems to stabilize the ComB protein complex (25). In addition to the comB system, which seems to be universally present in all H. pylori strains, a subgroup of H. pylori strains is equipped with a second T4SS, the cag system, which is dedicated to the translocation of the effector protein CagA into eukaryotic cells. The cag and the com systems are functionally independent (25, 27). Certain H. pylori strains carry a third T4SS, named tfs3 (30).
A number of additional proteins not classified as ComB proteins seem to be involved at different steps in the transformation competence process, such as the cytoplasmic proteins RecA (42) and DprA (3, 46), the inner-membrane protein ComE3 (53), and the secreted protein ComH (45). The goal of this study was to identify additional components of the T4SS-based ComB DNA uptake machinery. We therefore started a computer-based in silico search for typical type IV-homologous proteins missing in the comB system. By this approach we identified putative homologues of VirB2, VirB3, and VirB6, which we named ComB2, ComB3, and ComB6, respectively, and we verified their function in the process of natural transformation competence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
For routine culture, H. pylori strains (Table 1) were grown on GC agar plates (Difco) supplemented with vitamin mix (1%), horse serum (8%), vancomycin (10 mg/liter), trimethoprim (5 mg/liter), and nystatin (1 mg/liter) (serum plates). Inoculated plates were incubated for 24 to 48 h under microaerobic conditions (85% N2, 10% CO2, 5% O2) at 37°C. Escherichia coli strains HB101 (8) and DH5α (Bethesda Research Laboratories) were grown on Luria-Bertani (LB) agar plates or in LB liquid medium (40) supplemented with ampicillin (100 mg/liter), chloramphenicol (30 mg/liter), or kanamycin (40 mg/liter), as appropriate. For conjugation experiments from E. coli into H. pylori, strain β2155 (17) was used grown on the same media supplemented with diaminopimelic acid (0.2 mM). For quantification of phoA activity, E. coli CC118 (35) was used.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristicsa | Reference or source |
|---|---|---|
| H. pylori strains | ||
| P1 | H. pylori wild type, naturally competent | 26 |
| P12 | H. pylori wild type, naturally competent | 25 |
| J99 | Wild-type, genome-sequenced strain | 2 |
| 26695 | Wild-type, genome-sequenced strain | 52 |
| P149 | Wild-type, Mongolian gerbil-adapted strain | 29 |
| P224 | P12; deletion of hp0015 (comB2), cat insertion | This study |
| P225 | P12; deletion of hp0016 (comB3), cat insertion | This study |
| P226 | P1; deletion of hp0015-hp0017, aphA-3 insertion | This study |
| P227 | P1; deletion of hp0037 (comB6), cat insertion | This study |
| P228 | P1; deletion of region spanning hp0037 (comB6)-hp0042 (comB10) | This study |
| P229 | P1Δhp0015-hp0017 complemented with pHel2 plasmid carrying comB2-comB4 (pAK20) | This study |
| P230 | P1Δhp0015-hp0017 complemented with pHel2 plasmid carrying comB3-comB4 (pAK21) | This study |
| P231 | P1Δhp0015-hp0017 complemented with pHel2 plasmid carrying comB4 (pAK22) | This study |
| P232 | P1Δhp0037-hp0042 complemented with pHel2 plasmid carrying comB6-comB10 (pAK24) | This study |
| P233 | P1Δhp0037-hp0042 complemented with pHel2 plasmid carrying comB7-comB10 (pDHO46) | This study |
| P234 | P149[hp1421::TnHK9] Tn insertion at codon 20 | 29 |
| E. coli strains | ||
| CC118 | Δ(ara-leu)7697 ΔlacX74 ΔphoA20 galE galK thi rpsE rpoB argE(Am) recA1 | 31 |
| β2155 | thrB1004 pro thi strA hsdS lacZΔM15 (F′ lacZΔM15 lacIqtraD36 proA+ proB+) ΔdapA::erm (Ermr) pir::RP4 [::kan (Kmr) from SM10] | 17 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15] Tn10 (Tetr) | Stratagene |
| Plasmids | ||
| pRB67 | pMin2 plasmid carrying phoA without start codon | This study |
| pAK03 | pBA with region hp0014-hp0018, primers AK04-AK05 | This study |
| pAK04 | pBA with hp0015::cat and flanking regions | This study |
| pAK05 | pBA with hp0016::cat and flanking regions | This study |
| pAK07 | pBA with region hp0036-hp0038, AK02-AK03 | This study |
| pAK08 | pBA with hp0037::cat and flanking regions | This study |
| pAK09a | pRB67 with hp0037 fragment AK12-AK13 (34 aa) | This study |
| pAK09b | pRB67 with hp0037 fragment AK12-AK14 (62 aa) | This study |
| pAK09c | pRB67 with hp0037 fragment AK12-AK15 (115 aa) | This study |
| pAK09d | pRB67 with hp0037 fragment AK12-AK16 (198 aa) | This study |
| pAK09e | pRB67 with hp0037 fragment AK12-AK23 (236 aa) | This study |
| pAK09f | pRB67 with hp0037 fragment AK12-AK18 (296 aa) | This study |
| pAK19 | pBluescriptIISK comB2-comB4 deletion plasmid, aphA-3 | This study |
| pAK20 | pDH80, comB2-comB4 | This study |
| pAK21 | pDH80, comB3-comB4 | This study |
| pAK22 | pDH80, comB4 | This study |
| pAK23 | pMin1, comB6-comB10 deletion plasmid, aphA-3 | This study |
| pAK24 | pDH80, comB6-comB10 | This study |
| pDHO36 | pMin1, comB7-comB10 deletion plasmid, aphA-3 | 17 |
| pDHO46 | pDH80, comB7-comB10 | 25 |
aa, amino acids.
DNA manipulations and plasmid and strain constructions.
Cloning and DNA analysis procedures were performed according to Sambrook et al. (40). Chromosomal DNA from H. pylori was isolated with the QIAamp tissue kit (QIAGEN, Hilden, Germany). Plasmid DNA was purified from E. coli either by the boiling procedure or with the QIAprep Spin Miniprep kit (QIAGEN, Hilden, Germany).
The ΔcomB2-comB4 (hp0015-hp0017) mutant P226 was generated by transformation of plasmid pAK19 into H. pylori strain P1. To establish plasmid pAK19, two gene fragments, one upstream of hp0015 and another downstream of hp0017, were amplified with primers AK37-AK38 and AK39-AK40 and cloned into the vector pBluescriptIISK. Next, an aphA-3 resistance gene cassette was inserted between those two gene fragments. The plasmid was introduced into H. pylori strain P1 by natural transformation and integrated into the chromosome via homologous recombination, which was verified by PCR. For genetic complementation of mutant P226, plasmids pAK20, pAK21, and pAK22 were introduced by electroporation. The plasmids contained different parts of the deleted genes hp0015 to hp0017, which were cloned into pDH80 (23) with primers AK49, AK50, AK51, and AK52. pAK20 carried the whole deleted region hp0015 to hp0017, pAK21 contained genes hp0016 and hp0017, and pAK22 harbored hp0017 only.
For construction of the single-gene-deletion mutants P224 (Δhp0015) and P225 (Δhp0016), a gene fragment containing hp0015-hp0016 and about 500 bp upstream and downstream was amplified with primers AK04 and AK05 and cloned into vector pBA, creating plasmid pAK03. Using primers AK06, AK07, AK08, and AK09, the vector backbone except hp0015 and hp0016 was PCR amplified. A cat cassette without a terminator was introduced to replace gene hp0015 or hp0016, resulting in plasmids pAK04 and pAK05, respectively. The plasmids were transferred into strain P12 by natural transformation. Correct chromosomal integration was verified by PCR.
As described for the deletion mutants P224 and P225, the single-gene mutant P227 (Δhp0037) was also created by cloning a PCR fragment with primers AK02 and AK03 containing hp0037 together with flanking regions in vector pBA (pAK07). After amplifying the vector with flanking regions of hp0037 with primers AK19 and AK20, a cat cassette without a terminator was introduced instead of hp0037, resulting in plasmid pAK08, which was subsequently transformed in H. pylori P12.
The creation of the ΔcomB6-comB10 mutant P228 was achieved by PCR amplification of a gene fragment upstream of hp0037 with primers AK59 and AK65. This fragment was introduced by using EcoRI restriction sites into EcoRI-digested pDHO36 (25), resulting in pAK23. Successful integration in the H. pylori chromosome by homologous recombination resulted in the replacement of genes hp0037 to hp0042 by an aphA-3 resistance cassette. Genetic complementation of deletion mutant P228 was performed starting from shuttle plasmid pDHO46. Using an internal NdeI restriction site in hp0037, the amplified N-terminal part of this gene was integrated with primers AK57 and AK58, resulting in plasmid pAK24. Shuttle plasmid pAK24 contained genes hp0037 to hp0042. The plasmid was introduced into mutant strain P228 via electroporation. The deletion mutant P228 was also complemented with unmodified pDHO46 in an analogous way to obtain P233. The mutant gene hp1421 used in this study was an insertion mutant obtained by using TnHK9, as previously described (29).
Electroporation, natural transformation competence, and conjugation of H. pylori.
Transformation of H. pylori strains was performed with plasmid or chromosomal DNA according to a procedure described previously (22). Bacteria were harvested from serum plates and suspended to an optical density at 550 nm (OD550) of 0.1 in brucella broth containing 10% fetal calf serum. DNA was added (1 μg), and incubation was extended for 5 h under microaerophilic conditions before the suspension was plated on selective serum plates.
Conjugation of the shuttle vector pHel2 carrying the comB genes from E. coli to H. pylori was performed essentially as described previously (23). Growth of the dapA mutant β2155 is strictly dependent on exogenously supplied diaminopimelic acid; hence, the removal of diaminopimelic acid provides an efficient counterselection against this donor.
For electroporation of H. pylori, bacterial cells were harvested from serum plates and suspended in 1 ml of cold phosphate-buffered saline (PBS) solution. For each electroporation, bacteria were diluted to an OD550 of 1 and 1 ml bacterial suspension was washed twice with PBS and suspended first in 500 μl and then in 40 μl electroporation buffer (44). Forty microliters of H. pylori competent cells was mixed with 1 to 2 μl DNA in prechilled 0.2-cm electroporation cuvettes. Electroporation was performed at 2.5 kV, 200 Ω, and 25 μF by a Gene Pulser (Bio-Rad, Munich, Germany). After electroporation, 1 ml of brucella broth-10% fetal calf serum was added to each sample. The aliquots were incubated for 4 h in a CO2 incubator before being plated on selective agar plates.
Quantification of PhoA and green fluorescent protein (GFP) fluorescence.
Alkaline phosphatase activities of fusion proteins were determined by partial gene sequences of comB6 amplified with primers AK12, AK13, AK14, AK15, AK16, AK18, and AK23, which were cloned in front of a phoA gene without a start codon in the modified pMin2 vector pRB67 (R. Buhrdorf et al., submitted). The plasmids (pAK09a to pAK09f [Table 1]) were transformed into E. coli strain CC118, and the alkaline phosphatase activities were measured as described previously (34).
For analysis of the GFP activities of the ComB6 fusion proteins, the same PCR products that were described for the alkaline phosphatase assay were cloned into the pBluescriptIIKS vector in front of a gfp gene without a start codon. The fusion constructs were under the control of the lac promoter. Constitutively expressed gfp fusions were found to be toxic in E. coli DH5α; therefore, the plasmids were transferred into E. coli XL1-Blue, and the lac promoter was repressed by the addition of 2% glucose in the medium. For the activity assay, the cells were grown in 2 ml LB medium supplemented with 2% glucose with ampicillin, up to an OD550 of about 0.8. The cells were then washed with PBS and resuspended in fresh LB medium with ampicillin and 100 μM isopropyl-β-d-thiogalactoside instead of glucose. After growth for another 2 to 4 h, the cells were harvested. One hundred microliters of a suspension in PBS with an OD550 of 1.0 together with dilutions was applied to a 96-well plate, and the GFP activities were measured with Fuji FLA 3000 (Raytest, Straubenhardt, Germany).
For both activity assays, at least three different measurements were performed. To exclude negative results due to sequencing errors, the gene constructs of low-activity fusion proteins were sequenced.
RNA extraction, cDNA synthesis, and RT-PCR.
Total RNA was extracted from H. pylori (wild-type [wt] and mutant strains) using the RNeasy mini kit (QIAGEN) according to the manufacturer's instructions. The RNA yield, purity, and integrity were determined by measuring the absorbance ratio at 260/280 nm (ratio, >1.8) and checked with a 1% agarose-formaldehyde gel. The RNA samples were digested with DNase I (Roche Diagnostics, Mannheim, Germany). For cDNA synthesis, 800 ng of each RNA sample was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) and gene-specific primers (Table 2). The absence of genomic DNA was verified by PCR using untreated RNA as a template. Reverse transcription (RT)-PCR was carried out using PowerScript polymerase (PAN Biotech GmbH) and the oligonucleotide primers listed in Table 2.
TABLE 2.
Oligonucleotide sequences used in this study
| Use | Primer | Sequence |
|---|---|---|
| cDNA synthesis and PCR | CH80 | 5′-TCAAAAGTGATAGAACTAGCG-3′ |
| CH82 | 5′-GATTAGCGTGTTCTTTGG-3′ | |
| PCR | AK02 | 5′-CGGGCTCGAGATCCACTCATTAGCGGAG-3′ |
| AK03 | 5′-GAAGATCTCGCCCCAATGAGCGAACG-3′ | |
| AK04 | 5′-GCCGCTCGAGGGGGTGTGTAACAATTTC-3′ | |
| AK05 | 5′-GGGGATCCCCCATTAATGTATTCCGC-3′ | |
| AK06 | 5′-GCCGCTCGAGCAAAACCCTTCTGTTTAA-3′ | |
| AK07 | 5′-GCGGATCCAGCGAATTGGTTTATGGG-3′ | |
| AK08 | 5′-GCCGCTCGAGTAAAAAATTCCCATAAAC-3′ | |
| AK09 | 5′-GCGGATCCGTTATAGAGCGTAGAATG-3′ | |
| AK12 | 5′-ACCGCTCGAGCTTTAAGAAGGAGATATACATATGAAAAATGACGCTTATG-3′ | |
| AK13 | 5′-GCGGATCCCGCATGCAAAGTGTAGAG-3′ | |
| AK14 | 5′-GCGGATCCAGCGCTGAAGAAATTCTG-3′ | |
| AK15 | 5′-GCGGATCCAGAAAAGTTACTCAAGCT-3′ | |
| AK16 | 5′-GCGGATCCTAGGCATATAACCACAAC-3′ | |
| AK18 | 5′-GCGGATCCTGGGGTCGTGATGTATTG-3′ | |
| AK19 | 5′-GCCGCTCGAGCAATACTTCAACGGACTT-3′ | |
| AK20 | 5′-GCGGATCCAGGAATTTAATGAGAATT-3′ | |
| AK23 | 5′-GCGGATCCTAAGTCTTGTTTTTCTTG-3′ | |
| AK37 | 5′-CCGCTCGAGTATCGCTTTAGGCTATGCTA-3′ | |
| AK38 | 5′-GCATCGATCAAAACCCTTCTGTTTAATT-3′ | |
| AK39 | 5′-GGAATTCGCACTCCTATTCAGATGGCT-3′ | |
| AK40 | 5′-ACGAGCTCCGGCTCTTTACCATCTTCAA-3′ | |
| AK49 | 5′-ATGCCATATGTCCGCTCATTTTTTAAA-3′ | |
| AK50 | 5′-ATGCCTCGAGAGCCATCTGAATAGGAGTGC-3′ | |
| AK51 | 5′-ATGCCATATGATTATCCTGTCAGCGAG-3′ | |
| AK52 | 5′-CGGCCATATGTTAGAAAAGCTTTTAAG-3′ | |
| AK57 | 5′-CAGCATATGTAAAGTCCGTTGAAGTATTG-3′ | |
| AK58 | 5′-TCTTGCATATGAGCGTCTA-3′ | |
| AK59 | 5′-GGAATTCGAAGAACATCATAAGCGTTT-3′ | |
| AK65 | 5′-CAGAATTCCAATACTTCAACGGACTTT-3′ | |
| CH77 | 5′-CCAAAGAACACGCTAATC-3′ | |
| CH78 | 5′-ATGGAGAAAAAAATCACTGG-3′ | |
| CH79 | 5′-TTCTTCACGCTCGCTG-3′ | |
| CH81 | 5′-TAGTAGGCATGTGCGTTTC-3′ | |
| CH83 | 5′-GCAAGAGTTTTTAGGTTTTG-3′ |
RESULTS
In silico identification of novel components of the ComB transformation competence system.
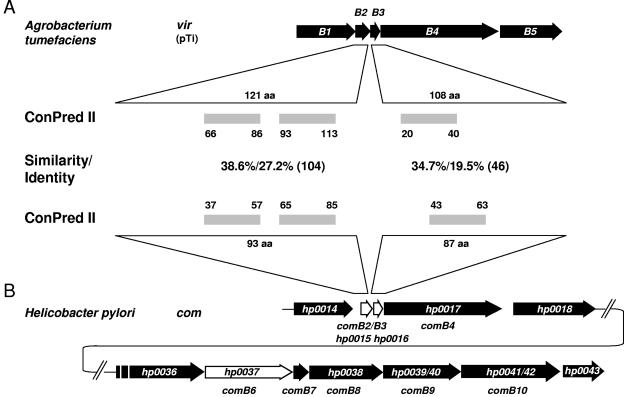
Increasing amounts of published sequence data about T4SS during the last few years have revealed that the organization of these operons could differ significantly, but it seems that the two gene clusters consisting of virB2 to virB4 and virB6 to virB10 often maintain a unit. The comB transformation competence system consists of the genes comB4 and comB7 to comB10. To identify further components of the comB DNA uptake machinery, we analyzed the regions upstream and downstream of the comB operon by intensive database comparison of individual ORFs. The criteria used for this analysis were (i) their position relative to the already known basic T4SS genes comB4 and comB7 to comB10, (ii) a similar secondary structure prediction of the putative ORFs, and (iii) sequence identity of the predicted ORFs to their corresponding VirB/Trb/AvhB counterparts of A. tumefaciens. This search resulted in the identification of three putative virB-related ORFs encoding proteins homologous to VirB2 (HP0015), VirB3 (HP0016), and VirB6 (HP0037). The putative comB2 (hp0015) and comB3 (hp0016) genes are located directly upstream of comB4 (hp0017) (Fig. 1), whereas the putative comB6 gene (hp0037) is positioned directly upstream of the comB7-to-comB10 operon (hp0038- hp0042) (Fig. 1B). Using ConPred II, which is a prediction server of transmembrane (TM) topology based on a consensus approach by combining the results of several proposed methods (4), three TM regions were predicted for HP0015, with the first TM domain having the property of a Sec-dependent signal peptide. Therefore, HP0015 has a secondary structure similar to VirB2. ConPred II predicted one central TM domain for HP0016 (Fig. 1), a characteristic shared with VirB3. For HP0037, several TM domains, including a Sec-dependent signal peptide, are predicted, similar to VirB6, suggesting that it is as well a polytopic inner-membrane protein. Direct amino acid sequence comparison revealed limited primary sequence conservation between AvhB2/ComB2 (38.6% similarity and 27.2% identity over 104 amino acids) and VirB3/ComB3 (34.7% similarity and 19.5% identity over 46 amino acids) (Fig. 1). The best primary amino acid sequence homology of HP0037 was found with A. tumefaciens TrbL (34.1% similarity and 21.2% identity over 108 amino acids), which is in the range of similarity and identity usually observed between T4SS-homologous proteins in different bacterial species. In summary, with this bioinformatics approach we were able to identify three additional putative components of the comB DNA uptake machinery.
FIG. 1.
Identification of novel type IV-related genes of the comB gene cluster. (A) Schematic representation of the A. tumefaciens virB locus with the number and location of putative TM domains in the VirB2 and VirB3 proteins, as predicted by ConPred II. (B) Overview of the complete H. pylori comB gene cluster and depiction of the comB region hp0014 to hp0018, with a comparison of the ComB2 (HP0015) and ComB3 (HP0016) TM domains, as predicted by ConPred II. Amino acid (aa) similarities and identities of the VirB and ComB homologues are shown between the corresponding ORFs.
Identification and functional characterization of hp0015 and hp0016 as essential genes for H. pylori natural transformation competence.
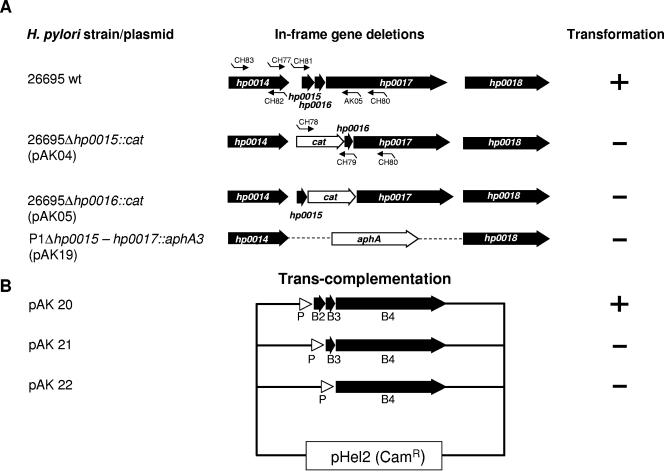
Next, we were interested in verifying whether or not HP0015 or HP0016 is essential for transformation competence in H. pylori. Therefore, in H. pylori strain P12 precise chromosomal deletions of hp0015 and hp0016 were generated separately by homologous recombination with plasmids pAK04 and pAK05, respectively (Fig. 2A; Table 1). The ORFs were replaced by a promoterless and terminatorless chloramphenicol acetyltransferase gene (cat), which usually does not cause a polar effect on downstream genes. The correct deletions were verified by PCR (data not shown). To test the individual P12Δhp0015 (P224) and P12Δhp0016 (P225) mutant strains for their natural transformation competence, they were transformed with (i) plasmid pDH29 carrying an H. pylori recA gene interrupted by an erythromycin resistance gene cassette (erm) or (ii) chromosomal DNA carrying a streptomycin resistance mutation (str) (21). Each of the deletions resulted in complete abrogation of natural transformation competence for plasmid as well as chromosomal DNA in the corresponding mutant strains (transformation frequency, <10−9) (Table 3; Fig. 2A).
FIG. 2.
Construction of precise deletions of comB2 (hp0015) or comB3 (hp0016) and genetic complementation. (A) Precise deletions of the hp0015, hp0016, or hp0015 to hp0017 genes were constructed by replacing the corresponding gene sequences with a chloramphenicol acetyltransferase (cat) or kanamycin (aphA-3) resistance gene cassette. (B) Complementation of the corresponding genes was obtained by cloning the comB4, comB3-comB4, or comB2 to comB4 gene(s) in the shuttle vector pHel2 behind the H. pylori flaA promoter (P) of plasmid pDH80 (pAK20 to pAK22) and transfer of the shuttle plasmid into P1Δhp0015-hp0017::aphA-3. The competence for genetic transformation of the corresponding strains is indicated (+ or −). For quantitative data of transformation experiments, see Table 3.
TABLE 3.
Transformation frequencies of H. pylori wt and mutant strains
| H. pylori strain and mutation | Transformation frequency with:
|
|
|---|---|---|
| Chromosomal H. pylori DNA (str)a | Plasmid DNA (pDH29) (erm)b | |
| P1 | (1.3 ± 0.5) × 10−5 | (1.1 ± 0.3) × 10−5 |
| P12 | (1.3 ± 0.6) × 10−6 | (2.0 ± 0.9) × 10−5 |
| P12ΔcomB2 | <10−9 | <10−9 |
| P12ΔcomB3 | <10−9 | <10−9 |
| P1ΔcomB2-comB4 | <10−9 | <10−9 |
| P1ΔcomB2-comB4[pAK20] | (5.2 ± 1.1) × 10−6 | (3.4 ± 1.2) × 10−6 |
| P1ΔcomB2-comB4[pAK21] | <10−9 | <10−9 |
| P1ΔcomB2-comB4[pAK22] | <10−9 | <10−9 |
| P1ΔcomB6 | <10−9 | <10−9 |
| P1ΔcomB6-comB10 | <10−9 | <10−9 |
| P1ΔcomB6-comB10[pAK24] | (0.4 ± 0.1) × 10−7 | (1.9 ± 0.7) × 10−7 |
| P1ΔcomB6-comB10[pDHO46] | <10−9 | <10−9 |
| P149 | ND | (5.4 ± 1.9) × 10−4 |
| P149[hp1421::TnHK9] | ND | (3.8 ± 1.4) × 10−4 |
Chromosomal DNA from strain P12 (str) or P1 (str) was used for transformation. str, streptomycin resistance gene cassette. ND, not done.
Plasmid pDH29 was used for transformation. It consists of pBluescript plasmid with the H. pylori recA gene interrupted by an erm (erythromycin resistance) cassette (25).
Genes hp0015, hp0016, and hp0017 are organized as an operon.
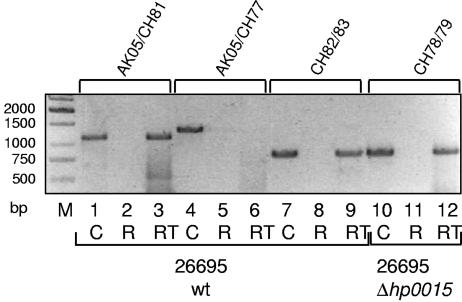
The genetic organizations of hp0015-hp0017 and jhp0013-jhp0015 in H. pylori strains 26695 and J99, respectively, are very similar. The stop and start codons of hp0015-hp0016 (jhp0013-jhp0014) partially overlap, and the stop codon of hp0016 (jhp0014) and start codon of hp0017 (jhp0015) are separated by one nucleotide only. This indicated that the corresponding region might be organized as an operon. To test this hypothesis experimentally, we used an approach to verify a common transcript of hp0015 to hp0017 by RT-PCR. Therefore, RNAs of the wild-type H. pylori strain 26695 and 26695Δhp0015 were prepared, and cDNA was generated with primers CH80 and CH82 (see Materials and Methods and Fig. 2). CH80-generated cDNA, 26695 chromosomal DNA, and RNA without the RT step were used as templates for RT-PCR using primer pair AK05-CH81 (Fig. 3). For chromosomal DNA and cDNA, but not for RNA templates, PCR fragments of the expected size were obtained (Fig. 3, lanes 1 through 3). To verify whether or not hp0014 is also cotranscribed with the comB operon, the same templates were used with primer pair AK05-CH77 (Fig. 2; Table 2), which did not result in a PCR product (Fig. 3, lanes 4 through 6). In contrast, when a cDNA was generated from the sequence upstream of hp0015 (primer pair CH82-CH83), an RT-PCR product comprising the hp0014 gene was obtained (Fig. 3, lanes 7 through 9), demonstrating the presence of a separate transcriptional unit upstream of hp0015. Finally, we also demonstrated that insertion of the cat resistance gene into hp0015 (in strain 26695Δhp0015::cat) did not cause a polar effect (primer pair CH78-CH79), since a common transcript comprising the cat and the hp0016 genes was obtained (Fig. 3, lanes 9 though 12). Taken together, these data show that hp0015-hp0017 is organized as an operon.
FIG. 3.
RT-PCR analysis of comB mRNA expression in H. pylori 26695 wt and ΔcomB mutant strains. Lanes 1 through 9, H. pylori 26695 wt; lanes 10 through 12, 26695Δhp0015. RT-PCR was performed with H. pylori 26695 wt cDNA (RT) (see Materials and Methods) using primer pairs AK05/CH81 (lane 3), AK05/CH77 (lane 6), and CH82/CH83 (lane 9) and with H. pylori 26695Δhp0015 cDNA using primer pair CH78/CH79 (lane 12). As positive controls, analogous PCRs with chromosomal DNA (C) of H. pylori 26695 wt (lanes 1, 4, and 7) and H. pylori 26695Δhp0015 (lane 10) were performed. No amplification products were detected in the negative controls (lanes 2, 5, 8, and 11), where RNA only, without cDNA synthesis (R), served as a template for PCR. Lane M, DNA size marker.
Genetic complementation of the Δhp0015 and Δhp0016 mutant strains in H. pylori 26695.
The deletion mutants showed a very drastic effect on transformation competence. The RT-PCR data for the wt and mutant strains already ruled out a polar effect of the cat gene replacement mutations on the expression of comB4, which is well described to be essential for transformation competence (25). To verify these data independently and to rule out any secondary mutation in another locus essential for natural competence, we sought to complement strain P1Δhp0015-hp0017 (strain P226) in trans, using the pHel2 shuttle vector (23). For complementation, hp0015 to hp0017, hp0016-hp0017, or hp0017 alone was used (strains P229, P230, and P231, respectively [Table 1]) (Fig. 2B). The complementing genes were cloned into the shuttle vector pHel2 under the control of the H. pylori flaA promoter. Since none of the constructs resulted in a detectable transformation frequency except for complementation with the complete set of genes (hp0015-hp0016-hp0017), we conclude that both comB2 (hp0015) and comB3 (hp0016) are essential for transformation competence.
HP0037 is an essential component of the ComB type IV transformation competence system.
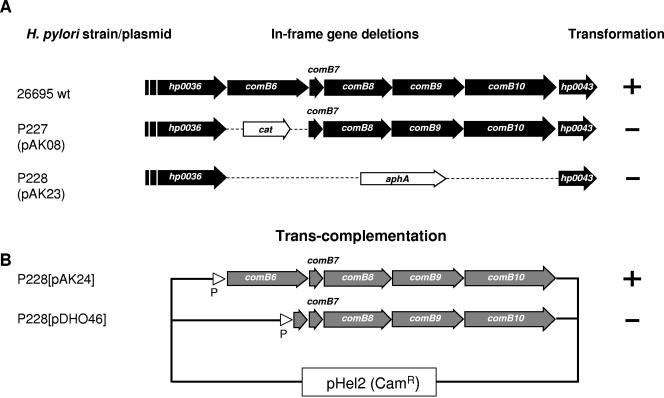
Our computer predictions identified the putative protein encoded by hp0037, which is located directly upstream of comB7, as a polytopic membrane protein like VirB6 from A. tumefaciens (Fig. 4A). Depending on the prediction program used, HP0037 possesses five to seven putative TM regions, very similar in their distribution to those of VirB6. To test whether HP0037 is an essential component of the comB transformation competence system, hp0037 was precisely deleted and replaced by the terminatorless cat resistance gene. Plasmid pAK08 was used to generate a gene knockout for hp0037 in H. pylori P1 by transformation and homologous recombination, resulting in P1ΔcomB6 (P227) (see Materials and Methods). The mutant strain completely lost its natural transformation competence for plasmid or chromosomal DNA (Table 3). Interestingly, transfer of plasmid or chromosomal DNA into the H. pylori mutant strain by electroporation was still possible, supporting our assumption that ComB6 is involved only in the process of DNA binding or uptake in a bacterial membrane compartment, rather than having an additional function via its cytoplasmic domain in a later step of transformation competence in the cytoplasm.
FIG. 4.
Essential role of comB6 for transformation competence as revealed by deletion and complementation studies. (A) Precise deletions of comB6 (hp0037) or the region comprising comB6 to comB10 (hp0037 to hp0042) were constructed by replacing the corresponding gene sequences with a cat or a kanamycin (aphA-3) resistance gene cassette, respectively. (B) Complementation of the corresponding genes was obtained by cloning the comB6 to comB10 or the comB7 to comB10 genes in the shuttle vector pHel2 behind the H. pylori flaA promoter (plasmid pDH80) to result in pAK24 or pDHO46 (25), respectively. The shuttle plasmids were transferred into strain P228 (P1ΔcomB6-comB10::aphA-3). The competence for genetic transformation of the corresponding strains is indicated at right (+ or −). For quantitative data of transformation experiments, see Table 3.
Production of the ComB8 to ComB10 proteins was reduced in the P1Δhp0037 mutant compared to the P1 wt strain (data not shown). To confirm that the DNA uptake defect was dependent on comB6, rather than being a polar effect of the mutation on expression of comB7 to comB10, we performed a genetic complementation experiment. First, a mutant strain was generated in which the chromosomal region spanning comB6 to comB10 was deleted and replaced by a kanamycin (aphA-3) resistance cassette (P1ΔcomB6-comB10) (data not shown). The corresponding region was then inserted into H. pylori P228 by using the shuttle vector pHel2, either including (pAK24, strain P232) or excluding (pDHO46, strain P233) hp0037 (Fig. 4B). The complemented strain, but not the deletion mutant strain, produced the ComB8, ComB9, and ComB10 proteins, as detected by immunoblot analysis (data not shown). Though lower than that of the wt, competence for genetic transformation was restored in the complemented strain carrying hp0037 but not in the strain without comB6. These results clearly identified comB6 as an essential gene for the process of natural transformation competence.
Topology mapping of HP0037 identifies the protein as a structural VirB6 homologue.
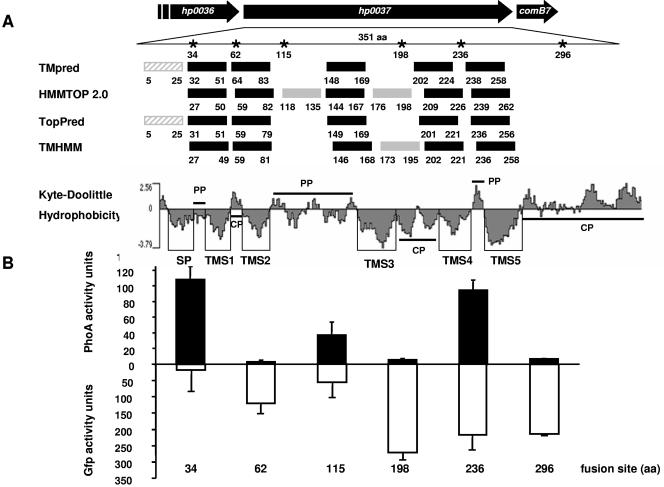
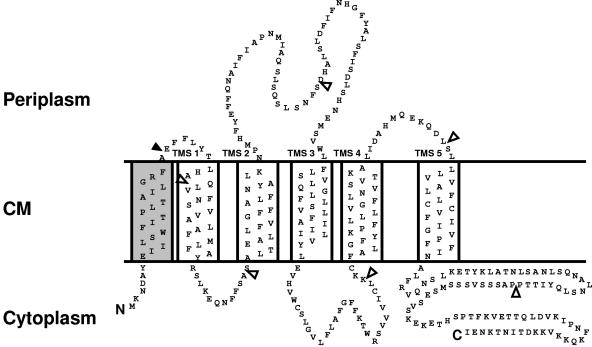
The HP0037 protein was predicted by several computer algorithms to be a typical transmembrane protein (Fig. 5A). Recent studies indicate that the topology of a membrane protein can be predicted reliably when several topology prediction methods show a consensus (18). We used the topology prediction programs TMpred, HMMTOP 2.0, TopPred, and TMHMM. In order to verify these predictions experimentally, we chose a reporter technology by fusing HP0037 N-terminal fragments to alkaline phosphatase (PhoA), which is active only in the periplasm, or to GFP, which is active only in the cytoplasm (18, 34). Recombinant E. coli strains producing the fusion proteins displayed a fairly consistent pattern of reporter protein activities. These proteins could be demonstrated to be stable fusion proteins with corresponding immunoblots (data not shown). E. coli strains producing fusion proteins with junction sites at positions 34, 115, and 236 of HP0037 exhibited intermediate to high levels of PhoA but low levels of GFP fluorescence activity, suggestive of a periplasmic location of the corresponding junction sites (Fig. 5B). Conversely, E. coli strains producing fusion proteins with junction sites at positions 62, 198, and 296 each displayed relatively high levels of GFP fluorescence but low levels of PhoA activity, suggestive of a cytoplasmic location (Fig. 5). One critical position was the GFP fusion at position 236, which was found always with rather high levels of activity. Since the predictions at this position were identical among all prediction programs and the hydrophobicity plot also indicated a clear distribution of TM sequences (Fig. 5A), we assume that the GFP fusion becomes mislocated (in the cytoplasm) in this case. The C terminus of the protein is located in the cytoplasm, as suggested by the low-level PhoA activity and high-level GFP fluorescence of the corresponding fusions (Fig. 5B). The topology prediction for the N terminus of HP0037 was less definite: not all programs predicted a TM domain for the first 25 amino acids, and in addition the SIGCLEAVE program but not SIGNALP predicted a Sec-dependent signal peptide. As we were able to generate an active PhoA fusion protein at position 34, we conclude that this is strong evidence for an N-terminal Sec-dependent secretion signal. Therefore, our topology model, which is supported by the computer predictions and the reporter gene assays, confirms five TM regions for ComB6, with the N terminus located in the periplasm and an extended periplasmic loop between TM domains 2 and 3, as well as a long C-terminal region in the cytoplasm (Fig. 6). Thus, the overall topology of HP0037 is reminiscent of the structure of VirB6 of A. tumefaciens (28), which, together with its essential function and its conserved location, justifies the designation ComB6 for this protein.
FIG. 5.
Topology analyses of the ComB6 subunit of the H. pylori transformation competence-mediating type IV transport system. (A) The computer programs listed at left predicted the transmembrane segments shown as solid rectangles with residue numbers relative to the N terminus. Solid rectangles identify transmembrane segments supported by results of reporter protein fusion studies; shaded rectangles are not supported by experimental data. *, amino acid (aa) positions of the PhoA or GFP fusions represented in panel B. Kyte-Doolittle hydrophobicity blot of HP0037: SP, signal peptide; TMS1 to TMS5, transmembrane segments 1 to 5; CP, cytoplasmic location; PP, periplasmic location. (B) Reporter protein activity levels of E. coli CC118 (phoA) or XL1-Blue (gfp) expressing six defined fragments of comB6 translationally fused to phoA (above the x axis) or gfp (below). Numbers at the bottom refer to the fusion site of the corresponding ComB fusion protein tested. Reporter protein activities represent the average of at least three experiments, with standard deviations indicated.
FIG. 6.
Proposed membrane topology model of ComB6. The model is derived from predictions by four widely used computer algorithms and the results of PhoA and GFP reporter protein fusion studies. The experimental data provide support for the five transmembrane segments (TMS) shown. The N-terminal signal sequence is shown as an additional TMS (shaded). A long C-terminal segment is located in the cytoplasm. The filled arrowhead represents a postulated signal sequence cleavage site; open arrowheads mark GFP or PhoA fusion sites. CM, cytoplasmic membrane.
HP1421, a putative VirB11-homologous ATPase, is not essential for the function of the ComB T4SS.
Bacterial T4SS involved in DNA transfer (conjugation systems, T-DNA transfer system) or protein translocation use several inner-membrane-associated ATPases for substrate transfer, such as VirB4, VirB11, and VirD4 homologues. For the ComB system, only one such ATPase homologue has been identified, ComB4, a functional homologue of VirB4, which is essential for the function of the T4SS (25). In A. tumefaciens, VirB11 interacts with VirB9 and VirB10 and is involved in the assembly of the transport system. Two genes in the H. pylori 26695 genome, hp0525 and hp1421, encode putative VirB11 homologues. The product of hp0525 has been identified as the VirB11-homologous ATPase of the cag pathogenicity island-encoded T4SS, and our earlier results showed that HP0525 is not functionally involved in DNA transformation competence (25). The H. pylori virB11 homologue might also be part of the comB system. We therefore analyzed hp1421, the second putative virB11-homologous gene in H. pylori, which might be involved in natural transformation competence, as suggested earlier (47). Therefore, a set of independent transposon insertion mutants was generated with the in vitro transposon pTnHK9 (29), a derivative of EZ::Tn (Epicenter, Madison, WI). A functional inactivation of the hp1421 gene was generated in H. pylori strain P149 and verified by PCR and DNA sequencing of the transposon insertion site (P234 [Table 1]). The transformation efficiency of the mutant strain was not significantly lower than that of the corresponding wt strain (Table 3), suggesting that the putative HP1421 ATPase is not functionally involved in the natural transformation competence of H. pylori.
DISCUSSION
Gram-positive and gram-negative bacteria use related proteins and molecular machines to import DNA. Components of these competence systems have homology with proteins that are involved in the assembly of type IV pili and type II secretion systems and form a structure that partially spans the cell envelope. The only exceptions known so far are the comB systems of H. pylori (26) and Campylobacter jejuni (5), which both use components of a T4SS for natural transformation competence. The comB apparatus as described previously hitherto consisted of the proteins ComB4 and ComB7 to ComB10 (24, 25). In this paper we report the identification and functional characterization of three additional basic components of T4SS for the comB system, ComB2 (HP0015), ComB3 (HP0016), and ComB6 (HP0037).
The genes comB2-comB3 and comB6 were first identified by their location in respect to the known T4SS components comB4 and comB7 to comB10, respectively. The involvement of the genes hp0015 and hp0037 in natural transformation competence was identified previously with a transposon mutagenesis screen (12), but polar effects on comB4 or the comB7-to-comB10 operon had not been ruled out. In our study, the generation of knockout mutant strains in the corresponding genes and subsequent complementation studies verified their essential function for the natural transformation competence of H. pylori. In addition, we were able to demonstrate a topology of the corresponding proteins similar to the homologous proteins of the prototype VirB T4SS in A. tumefaciens. The basis for these topological studies was a solid secondary structure prediction, performed either by a set of independent secondary structure prediction programs (TMPred, HMMTOP 2.0, TopPred, and TMHMM) or by ConPred II, a software package that combines a number of individual programs in a single package (4). Interestingly, despite rather low sequence identities between VirB2-ComB2 (27.2%) and VirB3-ComB3 (19.5%), the total length of the putative proteins, the elongated signal sequence (for VirB2-ComB2), and the secondary structure prediction were well conserved between these putative paralogous pairs of proteins.
The VirB2 protein is a subunit of a surface-exposed pilus structure of the VirB system. Whether the ComB transformation competence system also uses a surface-located pilus and what the function of such a pilus might be during DNA uptake and transformation are completely unknown. For H. pylori, flagella (48) and cag pathogenicity island-encoded surface appendages (39, 51), but no typical pili, have been described. Therefore the ComB2 protein might be part of a “stump structure” in the cell envelope (43) that might facilitate DNA uptake but does not form an extended extracellular pilus. Such a scenario is also known from type IV pili in gram-positive as well as gram-negative bacteria. In Neisseria gonorrhoeae, the PilE protein is able to form long, extended surface pili; although such extended pilus structures are neither necessary nor sufficient for transformation competence, the pilin protein and other pilE homologues are essential (1). VirB2 is expressed as a propilin, which is cleaved at the N terminus by the signal peptidase. In certain systems VirB2 is processed further at the C terminus, which results in the cyclization of the protein (19). We did not find in ComB2 a conserved AEIA motif, characteristic of certain TrbC-like pilins, indicating that ComB2 does not belong to the group of cyclic pilins.
The VirB3 homologues are essential components of T4SS, the precise function of which remains largely unknown. In A. tumefaciens, ConPred II predicts one hydrophobic region for VirB3. Originally the protein was suggested to localize to the periplasmic face of the outer membrane. More recent analyses suggest that VirB3 subunits assemble as a functional complex with the VirB4 ATPase at the inner membrane (16). For ComB3 of H. pylori we found a similar arrangement of the hydrophobic sequence (Fig. 1), but the localization of the protein and its function for DNA transformation competence are unknown.
The identification of a VirB6-homologous protein (HP0037, ComB6) in the H. pylori comB system was unexpected, since HP0037 was annotated as an NADH-ubiquinone oxidoreductase subunit in the H. pylori 26695 genome sequence (52). Furthermore, several years ago we reported on an H. pylori mutant strain, designated P87, in which a TnMax5 insertion in comB6 interrupted the gene at its 3′ end, resulting in a truncation of the last 29 amino acids of the protein (26). The corresponding strain showed a reduction in its transformation rate by 10 or 20%, depending on the use of chromosomal or plasmid DNA for transformation, respectively. At that time the gene was considered nonessential for transformation competence (26). The secondary structure prediction of ComB6 reveals now a five-transmembrane-spanning topology with the C-terminal 92 amino acids in the cytoplasm. Thus, the truncated ComB6 protein in strain P87 might still be active, since all membrane-spanning segments and part of the cytoplasmic tail are still present. Now we show that ComB6 is absolutely essential for DNA transformation competence. A precise, complete deletion of the gene resulted in a complete loss of transformation competence of the corresponding mutant strain, and genetic complementation resulted in a restoration of the phenotype, at least in part. A complete restoration might depend on an exact stoichiometry between ComB6 and other components of the apparatus. Due to the lack of an antibody, we were unable to test the level of comB6 expression directly in our complementation assay, but we can exclude a defect in the production of ComB8, ComB9, and ComB10 from this plasmid in the corresponding H. pylori P1 genetic background (25). Thus, our data indicate that ComB6 is an essential component of the T4SS involved in DNA transformation competence.
Recently, Yeh and coworkers identified a protein (HP1361) with sequence homology to Bacillus subtilis ComEC (also known as ComE3), which was absolutely essential for DNA transformation competence in H. pylori (53). ComEC is a putative channel protein for DNA uptake in B. subtilis and other genetically transformable bacteria. Thus, ComEC might have a function in the classical competence systems similar to that of VirB6 in the T4SS. H. pylori apparently has both types of proteins, a ComEC homologue and a ComB6 protein, both of which are considered to be multispanning transmembrane proteins and absolutely essential for transformation competence in these bacteria. HP1361 has been reported to be involved in DNA binding as well as in DNA uptake (53), although a defect in DNA binding might be sufficient to eliminate DNA uptake. Whether both proteins are involved in DNA transfer through the cytoplasmic membrane is not known so far, but since they can apparently not complement each other, they seem to fulfill independent functions. Interestingly, an H. pylori comEC knockout mutant was resistant to transformation via electroporation, a method that should be independent of DNA binding or a DNA uptake channel in the bacterial membrane (53). Our H. pylori comB6 mutant was readily transformable via electroporation, as shown by the complementation experiment, indicating that the corresponding protein is involved in DNA binding or in the mechanism of uptake through the bacterial membrane.
It was suggested that T4SS-mediated T-DNA transfer of A. tumefaciens might be a two-step process (15, 37) and that conjugation-mediated uptake of plasmid DNA in A. tumefaciens is enhanced by components of the VirB T4SS (7, 33). As H. pylori produces neither a type IV prepilin peptidase nor (pseudo)pilin proteins, we speculate that the ComB-mediated apparatus bridging the space between the outer and inner membranes might facilitate DNA uptake via the outer membrane into the periplasm. The transfer of the DNA across the inner membrane into the cytoplasm could be mediated by components of the comB T4SS and/or the ComEC homologue HP1361.
With the data presented here, we demonstrate that the comB DNA transformation competence system is comprised of a nearly complete set of basic T4SS components. These consist of proteins ComB2 to ComB4 and the core complex ComB6 to ComB10 but lack VirB1 and VirB5 homologues, as well as a VirB11-like ATPase. We cannot exclude the possibility that functional homologues of the latter two VirB proteins with only slight primary sequence homology might be encoded by H. pylori. It was shown that the murein hydrolase VirB1 is not essential for substrate translocation by the T4SS in A. tumefaciens (6); therefore, it is possible that the ComB apparatus of H. pylori could be assembled without the VirB1-mediated local disruption of the peptidoglycan cell wall. VirB5 is a secreted protein that localizes mainly to the T pilus (41). In addition, an adhesionlike function was demonstrated for its homologue, TraC (54). The establishing of cell-to-cell contact is required for T-DNA transfer and bacterial conjugation but not for DNA uptake by natural transformation competence, which could explain the absence of a VirB5 homologue in H. pylori. Interestingly, the ptl T4SS of Bordetella pertussis, which is essential for the contact-independent secretion of the pertussis toxin, also lacks a virB5 homologue (9).
Although we have identified here a further set of basic components of the H. pylori DNA uptake system, further studies are necessary to completely understand the composition and function of the comB transformation competence system in detail.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG HA 2697/3-3 and 2697/3-4).
We thank Renate Buhrdorf for providing plasmid pRB67 and Susanne Erhard for technical assistance during a practical course.
REFERENCES
- 1.Aas, F. E., C. Lovold, and M. Koomey. 2002. An inhibitor of DNA binding and uptake events dictates the proficiency of genetic transformation in Neisseria gonorrhoeae: mechanism of action and links to type IV pilus expression. Mol. Microbiol. 46:1441-1450. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Ando, T., D. A. Israel, K. Kusugami, and M. J. Blaser. 1999. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J. Bacteriol. 181:5572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai, M., H. Mitsuke, M. Ikeda, J. X. Xia, T. Kikuchi, M. Satake, and T. Shimizu. 2004. ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 32:W390-W393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, B. R., and P. J. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176:3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohne, J., A. Yim, and A. N. Binns. 1998. The Ti plasmid increases the efficiency of Agrobacterium tumefaciens as a recipient in virB-mediated conjugal transfer of an IncQ plasmid. Proc. Natl. Acad. Sci. USA 95:7057-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 9.Burns, D. L. 2003. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6:29-34. [DOI] [PubMed] [Google Scholar]
- 10.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascales, E., and P. J. Christie. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, K. C., Y. C. Yeh, T. L. Lin, and J. T. Wang. 2001. Identification of genes associated with natural competence in Helicobacter pylori by transposon shuttle random mutagenesis. Biochem. Biophys. Res. Commun. 288:961-968. [DOI] [PubMed] [Google Scholar]
- 13.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241-249. [DOI] [PubMed] [Google Scholar]
- 14.Chen, L., Y. Chen, D. W. Wood, and E. W. Nester. 2002. A new type IV secretion system promotes conjugal transfer in Agrobacterium tumefaciens. J. Bacteriol. 184:4838-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, L., C. M. Li, and E. W. Nester. 2000. Transferred DNA (T-DNA)-associated proteins of Agrobacterium tumefaciens are exported independently of virB. Proc. Natl. Acad. Sci. USA 97:7545-7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dehio, C., and M. Meyer. 1997. Maintenance of broad-host-range incompatibility group-P and group-Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179:538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drew, D., D. Sjostrand, J. Nilsson, T. Urbig, C. N. Chin, J. W. De Gier, and G. von Heijne. 2002. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. USA 99:2690-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenbrandt, R., M. Kalkum, E. M. Lai, R. Lurz, C. I. Kado, and E. Lanka. 1999. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 274:22548-22555. [DOI] [PubMed] [Google Scholar]
- 20.Falush, D., C. Kraft, N. S. Taylor, P. Correa, J. G. Fox, M. Achtman, and S. Suerbaum. 2001. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl. Acad. Sci. USA 98:15056-15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer, W., R. Buhrdorf, E. Gerland, and R. Haas. 2001. Outer membrane targeting of passenger proteins by the vacuolating cytotoxin autotransporter of Helicobacter pylori. Infect. Immun. 69:6769-6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas, R., T. F. Meyer, and J. P. M. van Putten. 1993. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol. Microbiol. 8:753-760. [DOI] [PubMed] [Google Scholar]
- 23.Heuermann, D., and R. Haas. 1998. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol. Gen. Genet. 257:519-528. [DOI] [PubMed] [Google Scholar]
- 24.Hofreuter, D., A. Karnholz, and R. Haas. 2003. Topology and membrane interaction of Helicobacter pylori ComB proteins involved in natural transformation competence. Int. J. Med. Microbiol. 293:153-165. [DOI] [PubMed] [Google Scholar]
- 25.Hofreuter, D., S. Odenbreit, and R. Haas. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379-391. [DOI] [PubMed] [Google Scholar]
- 26.Hofreuter, D., S. Odenbreit, G. Henke, and R. Haas. 1998. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol. Microbiol. 28:1027-1038. [DOI] [PubMed] [Google Scholar]
- 27.Israel, D. A., A. S. Lou, and M. J. Blaser. 2000. Characteristics of Helicobacter pylori natural transformation. FEMS Microbiol. Lett. 186:275-280. [DOI] [PubMed] [Google Scholar]
- 28.Jakubowski, S. J., V. Krishnamoorthy, E. Cascales, and P. J. Christie. 2004. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion System. J. Mol. Biol. 341:961-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kavermann, H., B. P. Burns, K. Angermüller, S. Odenbreit, W. Fischer, K. Melchers, and R. Haas. 2003. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197:813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kersulyte, D., B. Velapatino, A. K. Mukhopadhyay, L. Cahuayme, A. Bussalleu, J. Combe, R. H. Gilman, and D. E. Berg. 2003. Cluster of type IV secretion genes in Helicobacter pylori's plasticity zone. J. Bacteriol. 185:3764-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, E., and C. Manoil. 1994. Mutations eliminating the protein export function of a membrane-spanning sequence. J. Biol. Chem. 269:28822-28828. [PubMed] [Google Scholar]
- 32.Li, P. L., D. M. Everhart, and S. K. Farrand. 1998. Genetic and sequence analysis of the pTiC58 trb locus, encoding a mating-pair formation system related to members of the type IV secretion family. J. Bacteriol. 180:6164-6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, Z., and A. N. Binns. 2003. Functional subsets of the VirB type IV transport complex proteins involved in the capacity of Agrobacterium tumefaciens to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010. J. Bacteriol. 185:3259-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manoil, C. 1991. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 34:61-75. [DOI] [PubMed] [Google Scholar]
- 35.Manoil, C., and J. Beckwith. 1986. A genetic approach to analyzing membrane protein topology. Science 233:1403-1408. [DOI] [PubMed] [Google Scholar]
- 36.Nedenskov-Sorensen, P., G. Bukholm, and K. Bovre. 1990. Natural competence for genetic transformation in Campylobacter pylori. J. Infect. Dis. 161:365-366. [DOI] [PubMed] [Google Scholar]
- 37.Pantoja, M., L. Chen, Y. Chen, and E. W. Nester. 2002. Agrobacterium type IV secretion is a two-step process in which export substrates associate with the virulence protein VirJ in the periplasm. Mol. Microbiol. 45:1325-1335. [DOI] [PubMed] [Google Scholar]
- 38.Peek, R. M., and M. J. Blaser. 2002. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2:28-37. [DOI] [PubMed] [Google Scholar]
- 39.Rohde, M., J. Püls, R. Buhrdorf, W. Fischer, and R. Haas. 2003. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 49:219-234. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Schmidt-Eisenlohr, H., N. Domke, C. Angerer, G. Wanner, P. C. Zambryski, and C. Baron. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181:7485-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt, W., S. Odenbreit, D. Heuermann, and R. Haas. 1995. Cloning of the Helicobacter pylori recA gene and functional characterization of its product. Mol. Gen. Genet. 248:563-572. [DOI] [PubMed] [Google Scholar]
- 43.Schröder, G., and E. Lanka. 2005. The mating pair formation system of conjugative plasmids—a versatile secretion machinery for transfer of proteins and DNA. Plasmid 54:1-25. [DOI] [PubMed] [Google Scholar]
- 44.Segal, E. D., and L. S. Tompkins. 1993. Transformation of Helicobacter pylori by electroporation. BioTechniques 14:225-226. [PubMed] [Google Scholar]
- 45.Smeets, L. C., J. J. Bijlsma, S. Y. Boomkens, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 182:3948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smeets, L. C., J. J. Bijlsma, E. J. Kuipers, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. The dprA gene is required for natural transformation of Helicobacter pylori. FEMS Immunobiol. Med. Microbiol. 27:99-102. [DOI] [PubMed] [Google Scholar]
- 47.Smeets, L. C., and J. G. Kusters. 2002. Natural transformation in Helicobacter pylori: DNA transport in an unexpected way. Trends Microbiol. 10:159-162. [DOI] [PubMed] [Google Scholar]
- 48.Suerbaum, S. 1995. The complex flagella of gastric Helicobacter species. Trends Microbiol. 3:168-170. [DOI] [PubMed] [Google Scholar]
- 49.Suerbaum, S., and P. Michetti. 2002. Helicobacter pylori infection. N. Engl. J. Med. 347:1175-1186. [DOI] [PubMed] [Google Scholar]
- 50.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, J. Maynard Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka, J., T. Suzuki, H. Mimuro, and C. Sasakawa. 2003. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell Microbiol. 5:395-404. [DOI] [PubMed] [Google Scholar]
- 52.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quakenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 53.Yeh, Y. C., T. L. Lin, K. C. Chang, and J. T. Wang. 2003. Characterization of a ComE3 homologue essential for DNA transformation in Helicobacter pylori. Infect. Immun. 71:5427-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeo, H. J., Q. Yuan, M. R. Beck, C. Baron, and G. Waksman. 2003. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc. Natl. Acad. Sci. USA 100:15947-15952. [DOI] [PMC free article] [PubMed] [Google Scholar]