Abstract
The genes involved in organic hydroperoxide protection in Agrobacterium tumefaciens were functionally evaluated. Gene inactivation studies and functional analyses have identified ohr, encoding a thiol peroxidase, as the gene primarily responsible for organic hydroperoxide protection in A. tumefaciens. An ohr mutant was sensitive to organic hydroperoxide killing and had a reduced capacity to metabolize organic hydroperoxides. ohr is located next to, and is divergently transcribed from, ohrR, encoding a sensor and transcription regulator of organic hydroperoxide stress. Transcription of both ohr and ohrR was induced by exposure to organic hydroperoxides but not by exposure to other oxidants. This induction required functional ohrR. The results of gel mobility shift and DNase I footprinting assays with purified OhrR, combined with in vivo promoter deletion analyses, confirmed that OhrR regulated both ohrR and ohr by binding to a single OhrR binding box that overlapped the ohrR and ohr promoters. ohrR and ohr are both required for the establishment of a novel cumene hydroperoxide-induced adaptive response. Inactivation or overexpression of other Prx family genes (prx1, prx2, prx3, bcp1, and bcp2) did not affect either the resistance to, or the ability to degrade, organic hydroperoxide. Taken together, the results of biochemical, gene regulation and physiological studies support the role of ohrR and ohr as the primary system in sensing and protecting A. tumefaciens from organic hydroperoxide stress.
Agrobacterium tumefaciens is a soil bacterium that causes crown gall disease in a wide range of plants by transferring some of its DNA (T-DNA) into the plant host. The T-DNA is then stably integrated into the plant genome, where its expression leads to the synthesis of plant hormones that stimulate tumor growth (5). In general, soil bacteria are exposed to hydroperoxides from various sources, such as their own aerobic metabolism and exposure to other soil bacteria and fungi that produce hydroperoxides. In addition, during plant microbe interactions, bacterial phytopathogens are exposed to reactive oxygen species (ROS), including H2O2, superoxide anions, and lipid hydroperoxides, that are generated as part of active plant defense responses. Although the levels of plant lipoxygenases that catalyze the formation of fatty acid hydroperoxides from fatty acid precursors have been shown to increase in response to microbial invasion (12), the role of ROS during Agrobacterium-plant interactions is not clear.
In order to grow and proliferate, bacterial phytopathogens and soil bacteria must overcome these ROS. In regard to the protection against organic hydroperoxide toxicity, there are two major families of enzymes, peroxiredoxins (Prx) and Ohr, that have been shown to be important in many bacteria (3, 16, 26). AhpC (alkyl hydroperoxide reductase), an enzyme of the Prx family that catalyzes the reduction of organic hydroperoxides to their corresponding alcohols, has been well characterized biochemically and genetically (26). The enzyme not only detoxifies organic hydroperoxides but is also involved in the degradation of low concentrations of intracellular H2O2 (29). The physiological functions and biochemical properties of other members of the Prx family, such as Tpx (thiol peroxidase), bactoferritin comigratory protein (BCP), 1-Cys Prx, and 2-Cys Prx, are less clear partly due to their limited distribution in only a few bacterial species (4, 13, 26, 34). Nonetheless, they have been shown to be capable of metabolizing organic hydroperoxide. Ohr (organic hydroperoxide resistance protein), a thiol peroxidase, was initially discovered in Xanthomonas campestris due to its ability to complement organic hydroperoxide-sensitive phenotypes in an Escherichia coli ahpC mutant (21). Ohr is uniquely regulated, and its expression is highly induced only by organic hydroperoxides. Purified Ohr has hydroperoxide peroxidase activity and catalyzes the reduction of organic hydroperoxides to their corresponding alcohols (6, 16). Both ahpC and ohr are found in diverse species of bacteria (3, 10, 16, 21, 23, 30). They have similar biochemical properties but differ in both their physiological function and pattern of gene expression in response to stresses. In many bacteria, the expression of ahpC is regulated by OxyR, a peroxide sensor and transcription regulator (17, 31); however, in a number of bacteria ahpC is regulated by the peroxide sensing repressor, PerR (19). ohr is controlled by OhrR, an organic hydroperoxide-inducible transcription repressor (3, 19, 20, 32).
The aim of the present study was to functionally evaluate the roles of genes predicted, based on sequence homology, to be involved in organic hydroperoxide resistance. The analysis of the biochemical properties of ohrR and ohr mutants and the expression patterns of ohrR and ohr indicate that this system plays a primary role in sensing and protecting A. tumefaciens from organic hydroperoxides.
MATERIALS AND METHODS
Bacterial growth conditions.
A. tumefaciens NTL4 (18) and mutant strains were grown aerobically in Luria-Bertani (LB) medium at 30°C with continuous shaking at 150 rpm. To ensure synchronous growth, overnight cultures were inoculated into fresh LB medium to give an optical density at 600 nm (OD600) of ca. 0.1. Exponential-phase (OD600 of ∼0.6, after 4 h of growth) and stationary-phase (OD600 of ∼5.0, after 30 h of growth) cells were used in all experiments. The peroxide induction experiments were executed with exponential treated with various concentrations of peroxides for 15 and 30 min for Northern analysis and enzymatic assays, respectively. The organic hydroperoxides, tert-butyl hydroperoxide and cumene hydroperoxide were obtained from Fluka (Buchs, Switzerland) and Merck (Darmstadt, Germany), respectively. Linoleic acid hydroperoxide was prepared from linoleic acid (Sigma, St. Louis, MO) as described by Evans et al. (9).
Molecular biology techniques.
General molecular genetics techniques, including genomic DNA preparation, plasmid preparation, restriction endonuclease digestions, ligation, transformation in E. coli, agarose gel electrophoresis, and Southern and Northern blot analyses were performed according to standard protocols (28). Plasmid purification for DNA sequencing was performed by using the QIAGEN Miniprep kit. DNA was sequenced by using a BigDye terminator cycle sequencing kit (PE Biosystems) and run on an ABI 310 automated DNA sequencer. Routinely, A. tumefaciens was transformed by electroporation as previously described (18).
Purification of OhrR.
A 472-bp PCR fragment containing ohrR, in which an NcoI site overlapping the start codon had been introduced, was generated by using pOhrR as a template and the specific oligonucleotide primers BT992 and BT486. The NcoI-digested fragment was cloned into NcoI-HincII-digested pETBlue-2 (Novagen), yielding pETohrR.
E. coli harboring pETohrR was grown to mid-log phase before 1 mM IPTG was added, followed by incubation for 3 h. The cultures were harvested by centrifugation, and cell pellets were resuspended in 50 mM PB, sonicated, and then spun at 10,000 × g for 15 min. The cleared lysate was then loaded onto an Affi-Gel heparin column (Bio-Rad), followed by extensive washing with column buffer (25 mM Tris-HCl [pH 8], 25 mM NaCl, 2 mM EDTA). The protein was eluted by the addition of elution buffer (25 mM Tris-HCl [pH 8], 500 mM NaCl, 2 mM EDTA, 1 mM dithiothreitol). The eluted fraction was dialyzed against 25 mM Tris-HCl [pH 8]-100 mM NaCl-2 mM EDTA-1 mM dithiothreitol. The purity of the protein was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Gel mobility shift and DNase I protection assays.
32P-labeled DNA fragments were prepared by PCR with the oligonucleotide primers BT536 and BT537 (see Table 1) and A. tumefaciens NTL4 genomic DNA as the template to generate a 363-bp fragment spanning the ohr and ohrR promoter region. Gel mobility shift assays were performed as previously described (20). Gel mobility shift reactions contained 3 fmol of labeled probe in 25 μl of reaction buffer (20 mM Tris [pH 7.0], 50 mM KCl, 1 mM EDTA, 5% glycerol, 50 μg of bovine serum albumin ml−1, 5 μg of calf thymus DNA ml−1, 0.5 mg of poly(dI-dC) ml−1, 400 ng of purified OhrR). DNase I footprinting assays with the 336-bp PCR-generated DNA fragment spanning the ohr-ohrR intergenic region and purified OhrR were performed as described previously (20).
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain plasmid, or primer | Description or sequence (5′ to 3′) | Source or reference |
|---|---|---|
| A. tumefaciens strains | ||
| NTL4 | pTiC58-cured derivative of strain C58 ΔtetC58 | 18 |
| bcp1 | bcp1::pKNOCK-Gm | This study |
| bcp2 | bcp2::pKNOCK-Gm | This study |
| prx1 | prx1::pKNOCK-Gm | This study |
| prx2 | prx2::pKNOCK-Gm | This study |
| prx3 | prx3::pKNOCK-Gm | This study |
| ohr | ohr::pKNOCK-Km | This study |
| ohrR | ohrR::pKNOCK-Gm | This study |
| ohr bcp1 | ohr::pKNOCK-Km, bcp1::pKNOCK-Gm | This study |
| ohr prx1 | ohr::pKNOCK-Km, prx1::pKNOCK-Gm | This study |
| Plasmids | ||
| pKNOCK | Broad-host-range suicide vector; RP4 oriT, R6K γ-ori | 1 |
| pBBR1MCS-4 | Broad-host-range cloning vector; rep, mob, lacZα, ApR | 15 |
| pBcp1 | pBBR1MCS-4 containing A. tumefaciens bcp1 | This study |
| pBcp2 | pBBR1MCS-4 containing A. tumefaciens bcp2 | This study |
| pPrx1 | pBBR1MCS-4 containing A. tumefaciens prx1 | This study |
| pPrx2 | pBBR1MCS-4 containing A. tumefaciens prx2 | This study |
| pPrx3 | pBBR1MCS-4 containing A. tumefaciens prx3 | This study |
| pOhr | pBBR1MCS-4 containing A. tumefaciens ohr | This study |
| pOhrR | pBBR1MCS-4 containing A. tumefaciens ohrR | This study |
| pPohr | pUFR027 containing ohr::lacZ | This study |
| pPohrR | pUFR027 containing ohrR::lacZ | This study |
| Primers | ||
| BT485 | TGGAGCAGGAAAATGGAC | |
| BT486 | GCCTATGGCCGGGAGAGA | |
| BT487 | GAAGGAGTAAATGCCATG | |
| BT488 | TAAGCCCGCTTTATCAGG | |
| BT532 | ATCGGGTAAGTGAGGACC | |
| BT533 | ATTCGCACCGCGACCGGC | |
| BT536 | CTGCGCGTAAAGGGCAA | |
| BT537 | GAGCGTGACGTCGAGAAC | |
| BT538 | CGGAACAGCTTTTCGCGG | |
| BT539 | TTCTTCGGCTTTCTCACG | |
| BT546 | TAGGCGTCAGCGCCACCA | |
| BT547 | TCGACCCGATCGGGGCTCA | |
| BT574 | GGAGAAAGCACACTATGA | |
| BT575 | TTCGTTAGCAGCTTAGCC | |
| BT907 | ATCGACTTCAGCGCGCTC | |
| BT908 | TTCGGCGATCTTGCCATC | |
| BT909 | GCGAAAGAAAGGTAGAAT | |
| BT910 | GGCTGGATGGCCCCTCAG | |
| BT911 | CACCTCAATCTTGCGCTT | |
| BT912 | CACCCGCTTCAAACATTG | |
| BT913 | TGCCAGGGATGGAAATGC | |
| BT914 | ATTGGCCAAGACACTCAT | |
| BT1046 | TAAGAATGGGCTTGTTAAA | |
| BT1047 | GTCTCCTCGTGCCTGTACT | |
| BT1173 | CGCTTAGAGCGCACACCAA | |
| BT1174 | TCTCGAAAATCGCGACGCC | |
| BT1236 | TAATTGTACGCTATAAGG | |
| BT1317 | AGAACAGGAGACAAGACATC | |
| BT1318 | AACCGAGATCAGGCCGCAGC | |
| BT1319 | CTATTGCCTTTCGGTCAACG | |
| BT1320 | ACTGCTCCACCAGACCGTCA |
Construction of A. tumefaciens bcp, prx, ohr, and ohrR mutants.
The specific primers used for PCR amplification of gene internal fragments of A. tumefaciens bcp1, bcp2, prx1, prx2, prx3, ohr, and ohrR were designed based on the nucleotide sequences corresponding to putative open reading frames (ORFs) Atu1830 (BT907 and BT908), Atu3655 (BT911 and BT912), Atu1480 (BT532 and BT533), Atu0779 (BT1173 and BT1174)), Atu2399 (BT1319 and BT1320), Atu0847 (BT538 and BT539), and Atu0846 (BT546 and BT547), respectively, in the A. tumefaciens genome sequence (35) (Table 1). The PCR products were ligated into pDrive prior to the subcloning of the EcoRI fragments into pKNOCK-Gm or pKNOCK-Km and insertion mutants were constructed by using a protocol previously described (1). Mutants were confirmed by PCR with two primers flanking the insertion site and by Southern blot analysis.
Construction of pBcp1, pBcp2, pPrx1, pPrx2, pPrx3, pOhr, and pOhrR.
The full-length genes were PCR amplified from A. tumefaciens genomic DNA by using specific pairs of primers (BT909 and BT910 for bcp1; BT913 and BT914 for bcp2; BT574 and BT575 for prx1;BT1046 and BT1047 for prx2; BT1317 and BT1318 for prx3; BT487 and BT488 for ohr and BT485 and BT486 for ohrR) (Table 1) and Pfu polymerase. The PCR products were cloned into pCR-Blunt (Invitrogen), sequenced, and subcloned into the broad-host-range plasmid pBBR1MSC-4 (15) to generate the high-expression plasmids pBcp1, pBcp2, pPrx1, pPrx2, pPrx3, pOhr, and pOhrR.
Organic hydroperoxide degradation assay.
The degradation of organic hydroperoxides was measured as previously described (23, 30) with some modifications. Overnight cultures of various A. tumefaciens strains were inoculated into 20 ml of LB medium at a final OD600 of 0.1. Exponential-phase cultures (after 4 h of growth) were adjusted to an OD600 of 0.5 with fresh medium prior to addition of tert-butyl hydroperoxide (tBOOH), cumene hydroperoxide (CuOOH), or linoleic acid hydroperoxide (LOOH) at a concentration of 200 μM. Residual organic hydroperoxide concentrations were determined at 10-min intervals using a xylenol orange-iron reaction. At various time intervals, 1 ml of the culture was removed, and the cells were pelleted. A total of 100 μl of the cleared supernatant was then added to 400 μl of 25 mM sulfuric acid in a 1-ml cuvette. A total of 500 μl of freshly prepared reaction buffer (200 μM ammonium ferrous sulfate, 200 μM xylenol orange, and 25 mM sulfuric acid) was then added to the mixture. After 10 min of incubation at room temperature, the absorbance at 540 nm was determined. The concentration of residual organic hydroperoxide in the culture was calculated from a standard curve generated by using LB medium containing known organic hydroperoxide concentrations.
Determination of oxidant resistance by inhibition zone and plate sensitivity assays.
The resistance levels of A. tumefaciens strains to oxidants were determined by using either growth inhibition zone (21) or a plate sensitivity assay as previously described (27). Briefly, 1 ml of exponential-phase cells were mixed with 10 ml of molten top agar (LB containing 0.7% agar) prewarmed at 50°C and overlaid onto LB plates (14-cm-diameter petri dishes containing 40 ml of LB agar). The plates were left at room temperature for 15 min to let the top agar solidify. Sterile 6-mm-diameter disks (prepared from Whatman filter paper no. 3) soaked with either 5 μl of 1.0 M H2O2, 1.0 M tBOOH, or 0.5 M CuOOH were placed on the cell lawn, and zones of growth inhibition were measured after 24 h of incubation at 30°C. For plate sensitivity assay, serial dilutions of exponential phase cells were made in LB medium and 10 μl of each dilution was spotted onto a LB agar plate containing either 200 μM CuOOH or 800 μM tBOOH. The plates were incubated at 30°C for 24 h before bacterial colonies were scored.
Determination of adaptive protection to CuOOH.
Induced adaptive resistance to CuOOH killing was measured by adding 50 μM CuOOH to exponential-phase cultures of A. tumefaciens strains prior to treatment with lethal concentrations of CuOOH (1, 5, and 10 mM) for 30 min. After treatment, the cells were washed with fresh LB medium, and the number of viable cells was determined as described previously (33). The surviving fraction was defined as the number of CFU recovered after treatment divided by the CFU prior to treatment. Three independent experiments were performed in each case.
β-Galactosidase assay.
Crude bacterial lysates were prepared, and protein assays were performed as previously described (21). In brief, 20 ml of exponential-phase cultures were harvested and washed once with 50 mM sodium phosphate buffer (pH 7.0; PB). Bacterial suspensions in 0.5 ml of PB containing 1 mM phenylmethylsulfonyl fluoride, a protease inhibitor, were lysed by intermittent sonication, followed by centrifugation at 10,000 × g for 20 min. The total protein concentration was determined for each of the cleared lysates prior to their use in enzyme assays. β-Galactosidase was assayed as described earlier (25).
RESULTS AND DISCUSSION
Physiological analysis of ohr and ohrR mutants.
As a first step in investigating the role of the ohrR-ohr system in oxidative stress defense, Agrobacterium NTL4 strains carrying insertions in either ohr or ohrR were constructed by using the pKNOCK system (1), and their ability to resist exposure to oxidants was evaluated. Inactivation of either ohrR or ohr had no effect on aerobic growth rate or colony formation on a complex medium and an ohr mutant was less resistant, as shown by zones of growth inhibition of 26.5 ± 1.0 mm and 25.5 ± 1.2 mm to the organic hydroperoxides, CuOOH and tBOOH, respectively, compared to the wild-type strain NTL4 (20.0 ± 0.7 mm and 21.5 ± 1.0 mm). However, the sensitivity of the ohr mutant to CuOOH was more pronounced, suggesting that the A. tumefaciens ohrR-ohr system has evolved to be more efficient at sensing and protecting the bacteria from moderately complex organic hydroperoxides such as CuOOH than to the simple organic hydroperoxide, tBOOH, or to linoleic acid hydroperoxide (LOOH), a more complex organic hydroperoxide. The increased sensitivity to CuOOH, in the ohr mutant, could be complemented by the introduction of plasmid-borne ohr. The ohr mutant harboring pOhr gave 18.5 ± 0.5 mm zone of inhibition with 0.5 M CuOOH compared to 26.5 ± 1.0 mm for the mutant and 20.0 ± 0.7 mm for wild-type NTL4.
In a few bacteria, ohr has been implicated in H2O2 protection and metabolism (6, 16). In A. tumefaciens, it is unlikely that ohr plays any protective role against H2O2 since ohr mutant or high-level expression of ohr on an expression vector in NTL4 had no effect on resistance to either H2O2 or the superoxide generator, menadione (data not shown). Inactivation of ohrR led to a small increase in the resistance level to CuOOH as shown by zone of growth inhibition of 18.5 ± 0.8 mm for the mutant compared to 20.0 ± 0.7 mm in NTL4. This was probably due to increased expression of ohr. Furthermore, no changes in the resistance levels to inorganic oxidants were detected in an ohrR mutant (data not shown).
The ohr insertion mutant was further evaluated for its ability to degrade CuOOH in the culture media. The ohr mutant, along with the wild-type strain NTL4 and the ohr mutant strain carrying the Ohr expression plasmid, pOhr, were incubated with CuOOH and the rate of hydroperoxide degradation was determined. The results, shown in Fig. 1, indicate that the wild-type strain NTL4 rapidly metabolized CuOOH, whereas only the ohr mutant showed a significant reduction in the ability to metabolize CuOOH. After 15 min of incubation in medium containing CuOOH, the amount of CuOOH remaining was 60% for the ohr mutant and 40% for wild-type strain NTL4 (Fig. 1). The reduced capacity to metabolize CuOOH in the ohr mutant could be complemented by the introduction of plasmid-borne ohr in pOhr, resulting in a rate of CuOOH degradation that was similar to that in NTL4 (Fig. 1). These observations indicate that ohr is the major detoxification enzyme involved in organic hydroperoxide degradation in A. tumefaciens.
FIG. 1.
Degradation of CuOOH by various A. tumefaciens strains. The rates of CuOOH degradation in culture medium containing 200 μM CuOOH by A. tumefaciens parental NTL4 (▪), ohr mutant (□), ohr prx1 mutant (▴), ohr bcp1 mutant (•), and ohr mutant harboring pOhr (▵) are indicated. The levels of CuOOH remaining in the culture medium at the various time points are reported, along with those of a medium control without bacteria (○).
ohrR- and ohr-mediated adaptive response.
The ability to adapt to stress is crucial for bacterial survival under stressful conditions. It has often been observed that low-level exposure to a particular stress can elicit an adaptive response that results in an increased resistance to a subsequent high-level exposure to the same stress. An adaptive response to H2O2 exposure has been observed in many bacteria, including A. tumefaciens (33). Although adaptive responses to organic hydroperoxide are rare, an adaptive response to lipid hydroperoxide involving the ohrR/ohr system has been reported in X. campestris pv. phaseoli (14). We previously reported the lack of an adaptive response in A. tumefaciens to the organic hydroperoxide, tBOOH (33). In light of the physiological data concerning the role played by the ohrR/ohr system in organic hydroperoxide defense, the adaptive response of A. tumefaciens to CuOOH was investigated. The results indicate that preexposure to a low concentration (50 μM) of CuOOH conferred a 10-fold increase in resistance to subsequent exposure to killing concentrations (1, 5, and 10 mM) of CuOOH relative to uninduced cells (Fig. 2). Moreover, inactivation of either ohr or ohrR resulted in a complete loss of the CuOOH adaptive response (Fig. 2), indicating that the establishment of an adaptive response to CuOOH in A. tumefaciens requires the ohr/ohrR system.
FIG. 2.
Induced adaptive protection to CuOOH in A. tumefaciens requires functional ohr and ohrR. CuOOH-induced adaptive response experiments were performed by incubating exponential-phase cultures of A. tumefaciens (A), ohr mutant (B), and ohrR mutant (C) in 50 μM CuOOH for 30 min before treatment with the indicated concentrations of CuOOH for 30 min. Cells that survived various treatments were scored after 48 h of incubation. The CuOOH survival curves against CuOOH concentration are plotted. Symbols: ▪, CuOOH-induced; □, uninduced cultures. Values presented are the means and the standard deviations (SD) of four replicate experiments.
Regulation of ohrR and ohr expression in response to stresses.
Given the primary role played by Ohr in organic oxidant defense, studies were conducted to investigate the regulatory mechanism of the ohrR/ohr system in Agrobacterium. The expression patterns of genes involved in stress protection should correlate with their physiological roles. Thus, regulators of these genes must have mechanisms to sense and respond to changes in the levels of the appropriate stresses. In general, genes involved in stress protection are tightly regulated, and their expression is highly induced by stresses. In order to determine the regulatory pattern of ohr transcription, the levels of ohr mRNA were determined under uninduced and oxidant-induced growth conditions by Northern analysis. Compared to uninduced cultures, the levels of ohr mRNA in strain NTL4 markedly increased during growth in the presence of 200 μM tBOOH, 50 μM CuOOH, or 50 μM LOOH by 20-, 30-, or 15-fold, respectively, as determined by densitometry (Fig. 3A). The observed pattern of oxidant-induced ohr expression was similar to the pattern observed in several other microorganisms, where ohr expression was highly induced only by organic hydroperoxides (21-23) and is consistent with ohr's proposed physiological role as the major protective system against organic hydroperoxide toxicity.
FIG. 3.
Organic hydroperoxide-induced gene expression of ohr and ohrR. Northern blots of total RNA extracted from exponential-phase cultures of A. tumefaciens parental strain NTL4 and an ohrR mutant under uninduced conditions (UN) and after exposure to 200 μM tBOOH (tB), 50 μM CuOOH (C), 50 μM LOOH (L), 200 μM H2O2 (H), and 200 μM menadione (M) and then hybridized with a radioactively labeled ohr (A)- and ohrR (B)-specific probe.
An increase in ohr expression upon exposure to an organic hydroperoxide (an Ohr substrate) would certainly contribute to bacterial survival under this stress condition. It should be noted that treatment of A. tumefaciens cultures with inorganic oxidants, including a superoxide generator (200 μM menadione), and 200 μM H2O2 failed to induce ohr expression (Fig. 3A). The lack of H2O2-induced expression of ohr is at odds with results in Pseudomonas and other bacteria showing that treatment with high concentrations of H2O2 resulted in low-level induction of ohr expression (22, 24), leading to the suggestion that ohr may also play some role in H2O2 protection (16). In these cases it is unclear whether the inducer is H2O2 or some by-product of H2O2 treatment such as organic hydroperoxides that are produced during exposure to high concentrations of H2O2. If ohr induction is due to the accumulation of by-products resulting from H2O2 exposure, then lack of induction in A. tumefaciens could be a reflection of the organism's ability to rapidly detoxify H2O2.
In other organisms, such as Bacillus subtilis, OhrR has been shown to be an organic hydroperoxide responsive repressor of ohr and ohrR transcription (10). Under reducing conditions the repressor is active and binds to the ohr and ohrR promoters. Exposure to organic oxidants renders the repressor incapable of DNA binding through the reversible oxidation of conserved cysteine residues (11). In order to assess the role of the peroxide-sensing repressor ohrR in regulating ohr expression, an ohrR insertion mutant was constructed and the mutation's effects on ohr transcription during uninduced and oxidant induced conditions were investigated. The results in Fig. 3A clearly demonstrate that A. tumefaciens ohrR is a repressor of ohr expression since its inactivation resulted in constitutively high expression of ohr that was unaffected by oxidant exposure. The expression analysis was extended to determine the pattern of oxidative stress induced expression of ohrR. ohrR expression was highly induced (10- to 15-fold) by treatments with the organic hydroperoxides, tBOOH and CuOOH, but not the superoxide generator menadione or H2O2 (Fig. 3B). Thus, ohrR shares a similar organic hydroperoxide inducible expression profile with ohr.
Further analysis of ohr regulation was done by using strains carrying ohr promoter-lacZ fusion constructs. A 363-bp fragment (PCR with BT536 and BT537 primers) containing the ohr promoter was transcriptionally fused to a promoterless lacZ in the promoter probe vector pUFR027lacZ, a derivative of pUFR027 (7) to yield pPohr. pPohr was then used to monitor ohr promoter activity in response to inducing concentrations of hydroperoxides and the superoxide generator, menadione in wild-type strain NTL4 and an ohrR mutant. The results shown in Fig. 4A mirror those of the Northern analyses and indicate that the organic hydroperoxides CuOOH, tBOOH, and LOOH were potent inducers of ohr promoter activity, with magnitudes of induction ranging from 2.5- to 3-fold. The increases in promoter activity appeared to be dose dependent in the low-dosage range (i.e., 200 μM and below) for tBOOH and LOOH. However, as the inducing concentrations of the various organic hydroperoxides increased, significant reductions in ohr promoter activity were observed (Fig. 4A). This was most likely due to organic hydroperoxide toxicity resulting in growth arrest and cell death.
FIG. 4.
In vivo characterization of the ohr and ohrR promoters. The β-galactosidase activity of exponential-phase cultures of A. tumefaciens strains, containing either an ohr or an ohrR promoter-lacZ transcriptional fusion plasmid, exposed to CuOOH, tBOOH, LOOH, H2O2, or menadione at various concentrations was determined. (A) A. tumefaciens harboring pPohr; (B) A. tumefaciens (NTL4), A. tumefaciens ohr mutant (ohr), and A. tumefaciens ohr mutant containing pOhrR (ohr/pOhrR) harboring pPohr exposed to tBOOH (░⃞), CuOOH (▪), or unexposed (□). (C) Experiments were performed as described in panel B but with A. tumefaciens strains containing the ohrR promoter-lacZ fusion plasmid, pPohrR. Values are the means and the SD of four replicate experiments.
It has recently been reported that expression of the X. campestris ohrR/ohr system is more responsive toward low concentrations of the complex organic hydroperoxide, LOOH, compared to the simple organic hydroperoxide, tBOOH (14). In A. tumefaciens, the ohr promoter was more responsive to the moderately complex hydroperoxide, CuOOH, than to either LOOH or tBOOH, suggesting that the relative sensitivity of regulatory system to respond to different organic hydroperoxides is organism specific. The differences in the sensitivity to the various organic hydroperoxide inducers between the two bacteria are probably due to differences in the structure of the OhrRs. A. tumefaciens and X. campestris pv. phaseoli OhrRs each contain a cysteine (Cys) residue at position 21 that is absolutely conserved among all OhrRs and has been shown to be required for sensing organic hydroperoxide (11, 25). X. campestris ohrR also contains additional Cys residues at positions 127 and 131, and there is evidence that Cys-127 interacts with Cys-21 during peroxide sensing (25a). A. tumefaciens OhrR lacks Cys-127, suggesting that the hydroperoxide sensing mechanisms of the A. tumefaciens and X. campestris proteins may be different. Minor differences in the efficiency of different types of organic hydroperoxides in inducing ohr could be advantageous to A. tumefaciens when it encounters mixtures of organic hydroperoxides of various toxicity. In any case, organic hydroperoxides were still much more efficient inducers of the system than either the inorganic oxidant H2O2 or the superoxide generator, menadione, regardless of concentration (Fig. 4A).
As expected, inactivation of ohrR resulted in ohr promoter activity that was constitutively high and unaffected by organic hydroperoxide treatments (Fig. 4B). Furthermore, ohr promoter activity in the ohrR mutant was twofold higher than the fully induced level observed in the wild-type strain (Fig. 4B), suggesting that, even under fully induced conditions, some OhrR probably still bound to the ohr promoter. This could provide additional fine-tuning of the expression of OhrR regulated genes. Finally, high-level expression of ohrR from an expression vector led to the repression of ohr promoter activity, and this effect could be negated by CuOOH treatment (Fig. 4B). This observation is consistent with the idea that OhrR acts as the transcription repressor of the ohr promoter.
The in vivo promoter analyses were extended to the ohrR promoter. The ohrR promoter activity was induced by organic hydroperoxide treatments, but was unaffected by either H2O2 or menadione treatment (data not shown). The pattern of sensitivity of the ohrR promoter to induction by organic hydroperoxides was similar to the pattern for the ohr promoter. CuOOH was the most potent inducer, followed by LOOH and tBOOH. The organic hydroperoxide inducibility of the ohrR promoter was lost in an ohrR mutant background with absolute levels of ohrR promoter activity that were higher than those in wild-type strain NTL4 (Fig. 4C). Moreover, complementation with plasmid-borne ohrR in pOhrR restored the normal pattern of hydroperoxide inducibility (Fig. 4C). These observations indicate that OhrR negatively autoregulated its own expression. Consistent with the results of the Northern blotting experiments (Fig. 3B), comparative analyses of induced ohr and ohrR promoter activities showed that the ohr promoter was the stronger of the two, with up to ninefold higher promoter activity under a given condition.
Mapping of regulatory elements within the ohr and ohrR promoters.
As a first step in the characterization of both ohrR and ohr promoters, primer extension experiments were performed to determine the transcription start sites of both genes. The results in Fig. 5A show that ohr transcription initiates at a C residue, 21 bases upstream from the translation initiation codon. Immediately upstream of the ohr transcription start site were found E. coli RNA polymerase δ70-like −10 (TATAAG) and −35 (TTGCGT) sequence elements that were separated by 17 bases (Fig. 5A). The transcriptional start site of ohrR was mapped to a G residue 81 bases upstream of the ATG codon. Examination of the region upstream of the transcription start also revealed the presence of E. coli RNA polymerase δ70-like −10 and −35 sequence motifs TTGAAT and GATAAT, respectively, separated by 17 bases (Fig. 5A). Quantitative analysis of ohr and ohrR primer extension products indicated that transcription initiation from these promoters was highly induced by CuOOH (Fig. 5A). Thus, the increase in ohr and ohrR transcripts in response to CuOOH treatment detected in Northern experiments was due to increases in transcription initiation.
FIG. 5.
Localization of ohr and ohrR promoters and alignment of OhrR binding box. (A) Primer extension of RNA extracted from uninduced (UN) and CuOOH-induced cultures. The experiment was performed with a 32P-labeled oligonucleotide primer as described in Materials and Methods. The C, T, A, and G lanes of a dideoxy sequencing ladder using the same primer as that used for the primer extension are shown. The ohr and ohrR transcription start sites are marked by arrowheads in the primer extension autoradiographs and as “+1” in the accompanying sequence. Putative −35 and −10 regions are shown in boldface italics. The translation initiation codons (ATG) are in boldface. The putative OhrR box is underlined. (B) Alignment of putative OhrR binding sites from X. campestris (32), B. subtilis (10), and A. tumefaciens. The numbers indicate the number of intervening nucleotides.
Genetic evidence indicates that ohrR regulates its own expression in addition to that of ohr. This fact, combined with the close proximity of the divergently transcribed ohr and ohrR promoters suggested that they might share regulatory sites. Examination of the ohr-ohrR intergenic region revealed the presence of the AT-rich inverted repeat sequence, 5′-gcgTACAATTnAATTGTAcgc-3′ (uppercase letters indicate part of the conserved OhrR box), that was similar to the putative OhrR box sequence thought to be involved in the binding of OhrR to target promoters in B. subtilis and X. campestris (10, 32) (Fig. 5B). The inverted repeat was situated 19 bp upstream of the ohrR −35 promoter element and overlapped the region between the ohr −35 and −10 promoter elements (Fig. 5A), suggesting that a single OhrR box could be involved in the regulation of both the ohr and ohrR promoters.
In order to probe the function of the putative OhrR binding site, a number of promoter-lacZ transcriptional fusion plasmids were constructed that contained various amounts of sequence upstream of the ohr and ohrR promoters (Fig. 6A). The ability of each fusion to be induced by organic hydroperoxide treatments was tested in vivo. The results shown in Fig. 6B and C indicate that the OhrR box is necessary for normal organic hydroperoxide inducible regulation of both promoters. Deletion of the sequence upstream of position −55 (p921) in the ohr promoter had no appreciable effect on promoter function relative to the full-length control promoter (pPohr) (Fig. 6B). Deletion of the sequence upstream of −22, in p1236, that removed the upstream half of the putative OhrR binding box along with the −35 promoter element resulted in inactivation of the promoter (Fig. 6B). Thus, the ohr promoter resides in the region within 55 bp of the ohr transcription start containing the OhrR box and the −10 and −35 promoter elements.
FIG. 6.
ohr and ohrR promoter deletion analyses. (A) Map of the ohr-ohrR intergenic region showing the upstream end points of promoter fragments used to construct the various promoter-lacZ fusion plasmids; (B) β-galactosidase activity of A. tumefaciens harboring the ohr promoter-lacZ fusion pPohr or its deletions p921 and p1236; (C) β-galactosidase activity of A. tumefaciens harboring pPohrR or its deletions. p975/pOhrR represents A. tumefaciens containing p975 and carrying pOhrR for the expression of ohrR. Cells were cultured to exponential phase before induction with tBOOH (░⃞), CuOOH (▪), or uninduced (□). Values are the means and SD from four replicate experiments.
A similar analysis of the ohrR promoter showed that a fusion (p974) containing 80 bp upstream of the ohrR transcription start and spanning the OhrR-box, as well as the −10 and −35 promoter elements, was regulated normally. Deletion of all or part of the Ohr box, in fusion plasmids p920 (deleted to position −36), and p975 (deleted to position −61), respectively, yielded expression patterns that were similar to that of the full-length ohrR promoter in an ohrR mutant, i.e., high-level constitutive expression that was unaffected by organic hydroperoxide (Fig. 4C and 6C). Taken together, the data indicate that the ohrR promoter was located within 80 bp of the ohrR transcription start and that the OhrR box was required for organic hydroperoxide dependent regulation. One interesting finding was the fact that overexpression of OhrR from plasmid pOhrR restored organic hydroperoxide-dependent regulation to the lacZ fusion plasmid p975 (Fig. 6C). Since this fusion contained only the proximal half of the OhrR box, the result suggested that OhrR could still bind to this site, albeit with a lower affinity than to the full OhrR box.
Binding of OhrR to the ohr-ohrR intergenic region.
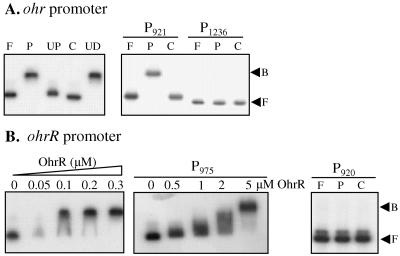
The direct interaction of OhrR with the ohr and ohrR promoters was tested by using purified A. tumefaciens OhrR and a 363-bp DNA fragment spanning the ohr-ohrR intergenic region, that contained the putative OhrR binding box, using gel mobility shift assays. OhrR specifically bound to the intergenic region since binding was blocked by the addition of excess unlabeled probe fragment (UP) but not by nonspecific competitor DNA, pBBR1MSC-4 (UD) (Fig. 7A). The genetic and physiological analyses reported in the present study indicate that the likely role of OhrR is as a sensor of organic hydroperoxide. More direct evidence of this was obtained when the organic hydroperoxide CuOOH was added to the gel mobility shift reactions containing purified OhrR and the 363-bp intergenic region probe (Fig. 7A). The addition of CuOOH to the binding reaction leads to the loss of OhrR binding to its target site (Fig. 7A). This is consistent with the proposed mechanism of OhrR sensing of organic hydroperoxide in which oxidation of a sensing Cys residue(s) leads to inactivation of the repressor that, in turn, allows RNA polymerase to bind to the promoter and activate transcription (11, 19, 25). In light of both the in vivo and in vitro data, it is clear that A. tumefaciens OhrR has evolved to sense and respond to organic hydroperoxide.
FIG. 7.
OhrR binds the ohr and ohrR promoters. The results of DNA mobility shift assays with 32P-labeled ohr (A) and ohrR (B) promoter fragments and purified OhrR. F, free probe; P, a reaction containing purified OhrR and labeled probe. UD and UP indicate reactions containing 2 μg of unrelated DNA (pBBR1MCS-4 plasmid) and 1 μg of unlabeled promoter, respectively. C, reactions in which CuOOH (1.0 mM) was added to the binding reaction. If not indicated, the amount of purified OhrR in the binding reaction was 0.3 μM. B, bound probe.
Similar gel mobility shift experiments were performed with deleted OhrR promoter fragments spanning either all (p920) or part (p975) of the OhrR box. Consistent with the lacZ fusion results, no binding of OhrR to fragment p920 was detected. However, fragment p975, containing half of the OhrR box, was still bound by OhrR (Fig. 7B). This was in good agreement with the lacZ-fusion results with p975, where hydroperoxide inducibility of this promoter deletion was restored when OhrR was expressed at high levels from plasmid pOhrR (Fig. 6C). This implies that OhrR binds to the target half-site in the proper configuration and retains its function.
Finally, precise localization of the OhrR binding site within the ohr-ohrR intergenic region was accomplished by DNase I footprinting (Fig. 8). OhrR protected a region a 49-bp region from positions −6 to −54 relative to the ohr transcription start. The extent of protection was typical of previously mapped OhrR binding sites in B. subtilis and X. campestris (10, 20) and indicates that OhrR binding represses expression of both genes by covering the −10 and −35 elements of the ohr promoter, as well as the −35 region of the ohrR promoter. Given the data presented here, it seems reasonable to assume that maximal repression requires the binding of multiple OhrRs within this region. The binding of a single OhrR dimer to the high-affinity consensus ohr-box could function as a nucleation site for the cooperative binding of additional dimmers that would further stabilize the complex. Such a scenario might allow for the fine-tuning of ohr expression under conditions in which organic hydroperoxide levels are low and full derepression of ohr is not required.
FIG. 8.
DNase I protection assay of OhrR binding to the ohr-ohrR intergenic region. The results of a DNase I footprinting assay using purified OhrR and a 32P-labeled probe spanning the ohr-ohrR intergenic region are presented. The minus sign (−) represents the probe fragment treated with DNase I in the absence of OhrR. The plus sign (+) represents the probe fragment treated with DNase I in the presence of OhrR. Arrowheads and numbers indicate the limits of the protected sites and their corresponding position relative to the ohr transcription start (+1). The sequence of the ohr-ohrR intergenic region is also shown in which the OhrR protected region is shaded. Divergent arrows indicate the putative OhrR box. The −10 and −35 regions of ohr and ohrR promoters are shown in boldface.
Evaluation of the physiological and biochemical role of putative genes encoding organic hydroperoxide scavenging enzymes other than ohr.
The objective of the investigation was to evaluate the roles of various genes encoding putative organic hydroperoxide-metabolizing enzymes in protecting A. tumefaciens from organic hydroperoxide exposure. The physiological analyses of the ohrR/ohr system mutants clearly indicate that this system is the major organic hydroperoxide defense system in Agrobacterium; however, it should be noted that the ohr mutant still retained a significant capacity to degrade CuOOH, suggesting that other enzymes are also involved in the process. BLAST algorithm (2) searches of the annotated genome of A. tumefaciens (35) identified at least six predicted ORFs that had a high degree of sequence similarity to enzymes that have been shown to be involved in organic hydroperoxide metabolism in other organisms. These ORFs could be grouped into either the peroxiredoxin (TSA/AhpC) or Ohr families (3, 13, 16, 21). The ORFs belonging to the peroxiredoxin family were prx1 (peroxiredoxin, Atu1480), prx2 (Atu0779), prx3 (Atu2399), bcp1 (bacterioferritin comigratory protein, Atu1830), and bcp2 (Atu3655). The deduced amino acid sequence of prx1 is 20% identical to that of E. coli ahpC, whereas prx2 and prx3 are more similar to the atypical 2-cysteine peroxiredoxin, prxS, of Rhizobium etli (8) with sequence identities of 32 and 75%, respectively. This is in contrast to the 9% sequence identity between prx1 and prxS. A. tumefaciens bcp1 and bcp2 are 40 and 26% identical, respectively, to E. coli bcp, while the deduced amino acid sequence of A. tumefaciens ohr (Atu0847), a member of the Ohr family of thiol peroxidases, is 51% identical to that of X. campestris ohr (21).
As an initial step toward understanding the physiological function of these genes in protecting A. tumefaciens from lethal doses of organic hydroperoxides, mutants lacking a functional copy of either; prx1, prx2, prx3, bcp1, or bcp2 were constructed by insertional inactivation using the pKNOCK system (1). The resistance levels of these mutants to organic hydroperoxides were determined by using both growth inhibition zone and a more sensitive plate sensitivity assays and compared to those of the wild-type strain NTL4 and the ohr mutant. Only the ohr mutant showed increased sensitivity toward CuOOH and tBOOH, whereas none of the single peroxiredoxin mutants showed any change in resistance relative to wild-type NTL4 (data not shown). It was possible that prx1, prx2, prx3, bcp1, and bcp2 played minor roles in organic hydroperoxide resistance such that expression of the Ohr system masked the effects of mutations in these genes. Thus, double mutants were constructed in which ohr was inactivated along with either prx1, prx2, prx3, bcp1, or bcp2. Each of the double mutants showed resistance levels to tBOOH and CuOOH that were similar to those of the ohr mutant (data not shown).
Another approach used to evaluate the in vivo function of the putative organic hydroperoxide protective genes was to test whether high-level expression of plasmid-borne prx1, prx2, prx3, bcp1, or bcp2 affected the organic hydroperoxide-sensitive phenotype of an ohr mutant background. Each of the genes was cloned into pBBR1MSC-4 to create pPrx1, pPrx2, pPrx3, pBcp1 pBcp2, and pOhr (see Materials and Methods). Each plasmid was introduced into an A. tumefaciens ohr mutant, and the organic hydroperoxide resistance levels were determined. As expected, pOhr restored the CuOOH resistance level of an ohr mutant to that of wild type (data not shown). In contrast, expression of plasmid-borne prx1, prx2, prx3, bcp1, or bcp2 did not alter the CuOOH resistance level of the ohr mutant strain (data not shown). Hence, prx1, prx2, prx3, bcp1, or bcp2, individually, are unlikely to play important roles in the protection of A. tumefaciens from organic hydroperoxide toxicity under the conditions tested.
It is possible that some of these gene products could contribute to organic hydroperoxide degradation; however, their contributions might not be sufficient to confer significant resistance to the lethal concentrations of organic hydroperoxide used in the study. In order to detect more subtle changes in the capacity to detoxify organic hydroperoxides, the effects of either gene inactivation or overexpression, on a strain's ability to degrade organic hydroperoxide, were determined. The ohr single and ohr prx1 and ohr bcp1 double mutants were incubated with CuOOH and the rate of hydroperoxide degradation was determined. These genes were initially chosen for further analysis due to the fact that homologs of both prx1 and bcp1 had been shown to be involved in organic hydroperoxide resistance in other bacteria (13, 36). As previously stated, wild-type strain NTL4 rapidly metabolized CuOOH, whereas only the ohr mutant showed a significant reduction in the ability to metabolize CuOOH that could be complemented by the introduction of plasmid-borne ohr in pOhr (Fig. 1). The double mutants, i.e., the ohr prx1 and ohr bcp1 mutants, had rates of CuOOH degradation that were similar to that of the ohr single mutant (Fig. 1).
The CuOOH degradation assay was also used to assess the effects of overexpression of genes, putatively involved in organic hydroperoxide metabolism, on an ohr mutant's ability to degrade CuOOH. The expression plasmids were transformed into ohr mutant, and the transformant's ability to degrade CuOOH was determined. Overexpression of prx1, prx2, prx3, bcp1, or bcp2 in an ohr mutant did not significantly alter the rate of CuOOH degradation (data not shown).
It should be noted that the ohr mutant still retained a significant capacity to degrade CuOOH, suggesting that other enzymes are also involved in the process. Obvious candidates for this role were the peroxiredoxin homologs encoded by prx1, prx2, prx3, bcp1, and bcp2. However, inactivation of each of these genes had no effect on the ability of the bacterium to either resist lethal exposure to CuOOH or to degrade CuOOH present in the culture medium. Although participation of these genes in organic hydroperoxide metabolism cannot be ruled out, it is likely that other, as yet unidentified, enzymes are responsible for the residual CuOOH degradation observed in the ohr mutant
The genetic and physiological data clearly indicate that ohr is the major protective system against organic hydroperoxide stress. The finding that prx1, prx2, prx3, bcp1, and bcp2 did not participate in organic hydroperoxide resistance was surprising. This was especially true for A. tumefaciens prx1 that encodes an AhpC (alkyl hydroperoxide reductase) homologue. AhpC is a structurally and functionally conserved hydroperoxide-metabolizing enzyme that has been shown to be involved in organic hydroperoxide resistance in other bacteria (26). It remains to be seen whether some of these genes might have functions under specific conditions. Alternatively, it is possible that the genes may have overlapping functions such that phenotypic effects would only be seen in multiple mutants.
Acknowledgments
We thank A. Phagakhayai and P. Saenkham for technical assistance.
This research was supported by a Research Team Strengthening Grant from the National Center for Genetic Engineering and Biotechnology and a Senior Research Scholar Grant RTA4580010 from the Thailand Research Fund to S.M. and by a grant from the ESTM under the Higher Education Development Project of the Ministry of Education. T.C. was supported by the Royal Golden Jubilee Scholarship (PHD/0160/2544) from the TRF, and parts of this study are from her dissertation submitted for a Ph.D. degree from Mahidol University.
REFERENCES
- 1.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26:824-826, 828. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atichartpongkul, S., S. Loprasert, P. Vattanaviboon, W. Whangsuk, J. D. Helmann, and S. Mongkolsuk. 2001. Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology 147:1775-1782. [DOI] [PubMed] [Google Scholar]
- 4.Cha, M. K., S. K. Hong, D. S. Lee, and I. H. Kim. 2004. Vibrio cholerae thiol peroxidase-glutaredoxin fusion is a 2-Cys TSA/AhpC subfamily acting as a lipid hydroperoxide reductase. J. Biol. Chem. 279:11035-11041. [DOI] [PubMed] [Google Scholar]
- 5.Chilton, M. D., M. H. Drummond, D. J. Merio, D. Sciaky, A. L. Montoya, M. P. Gordon, and E. W. Nester. 1977. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell 11:263-271. [DOI] [PubMed] [Google Scholar]
- 6.Cussiol, J. R., S. V. Alves, M. A. de Oliveira, and L. E. Netto. 2003. Organic hydroperoxide resistance gene encodes a thiol-dependent peroxidase. J. Biol. Chem. 278:11570-11578. [DOI] [PubMed] [Google Scholar]
- 7.DeFeyter, R., C. I. Kado, and D. W. Gabriel. 1990. Small, stable shuttle vectors for use in Xanthomonas. Gene 88:65-72. [DOI] [PubMed] [Google Scholar]
- 8.Dombrecht, B., C. Heusdens, S. Beullens, C. Verreth, E. Mulkers, P. Proost, J. Vanderleyden, and J. Michiels. 2005. Defence of Rhizobium etli bacteroids against oxidative stress involves a complexly regulated atypical 2-Cys peroxiredoxin. Mol. Microbiol. 55:1207-1221. [DOI] [PubMed] [Google Scholar]
- 9.Evans, M. V., H. E. Turton, C. M. Grant, and I. W. Dawes. 1998. Toxicity of linoleic acid hydroperoxide to Saccharomyces cerevisiae: involvement of a respiration-related process for maximal sensitivity and adaptive response. J. Bacteriol. 180:483-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuangthong, M., S. Atichartpongkul, S. Mongkolsuk, and J. D. Helmann. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183:4134-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuangthong, M., and J. D. Helmann. 2002. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci. USA 99:6690-6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jalloul, A., J. L. Montillet, K. Assigbetse, J. P. Agnel, E. Delannoy, C. Triantaphylides, J. F. Daniel, P. Marmey, J. P. Geiger, and M. Nicole. 2002. Lipid peroxidation in cotton: Xanthomonas interactions and the role of lipoxygenases during the hypersensitive reaction. Plant J. 32:1-12. [DOI] [PubMed] [Google Scholar]
- 13.Jeong, W., M. K. Cha, and I. H. Kim. 2000. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J. Biol. Chem. 275:2924-2930. [DOI] [PubMed] [Google Scholar]
- 14.Klomsiri, C., W. Panmanee, S. Dharmsthiti, P. Vattanaviboon, and S. Mongkolsuk. 2005. Novel roles of ohrR-ohr in Xanthomonas sensing, metabolism, and physiological adaptive response to lipid hydroperoxide. J. Bacteriol. 187:3277-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, 2nd, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 16.Lesniak, J., W. A. Barton, and D. B. Nikolov. 2002. Structural and functional characterization of the Pseudomonas hydroperoxide resistance protein Ohr. EMBO J. 21:6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loprasert, S., M. Fuangthong, W. Whangsuk, S. Atichartpongkul, and S. Mongkolsuk. 2000. Molecular and physiological analysis of an OxyR-regulated ahpC promoter in Xanthomonas campestris pv. phaseoli. Mol. Microbiol. 37:1504-1514. [DOI] [PubMed] [Google Scholar]
- 18.Luo, Z. Q., T. E. Clemente, and S. K. Farrand. 2001. Construction of a derivative of Agrobacterium tumefaciens C58 that does not mutate to tetracycline resistance. Mol. Plant-Microbe Interact. 14:98-103. [DOI] [PubMed] [Google Scholar]
- 19.Mongkolsuk, S., and J. D. Helmann. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45:9-15. [DOI] [PubMed] [Google Scholar]
- 20.Mongkolsuk, S., W. Panmanee, S. Atichartpongkul, P. Vattanaviboon, W. Whangsuk, M. Fuangthong, W. Eiamphungporn, R. Sukchawalit, and S. Utamapongchai. 2002. The repressor for an organic peroxide-inducible operon is uniquely regulated at multiple levels. Mol. Microbiol. 44:793-802. [DOI] [PubMed] [Google Scholar]
- 21.Mongkolsuk, S., W. Praituan, S. Loprasert, M. Fuangthong, and S. Chamnongpol. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J. Bacteriol. 180:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 23.Ochsner, U. A., D. J. Hassett, and M. L. Vasil. 2001. Genetic and physiological characterization of ohr, encoding a protein involved in organic hydroperoxide resistance in Pseudomonas aeruginosa. J. Bacteriol. 183:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palma, M., D. DeLuca, S. Worgall, and L. E. Quadri. 2004. Transcriptome analysis of the response of Pseudomonas aeruginosa to hydrogen peroxide. J. Bacteriol. 186:248-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panmanee, W., P. Vattanaviboon, W. Eiamphungporn, W. Whangsuk, R. Sallabhan, and S. Mongkolsuk. 2002. OhrR, a transcription repressor that senses and responds to changes in organic peroxide levels in Xanthomonas campestris pv. phaseoli. Mol. Microbiol. 45:1647-1654. [DOI] [PubMed] [Google Scholar]
- 25a.Panmanee, W., P. Vattanaviboon, L. B. Poole, and S. Mongkolsuk. Novel organic hydroperoxide sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 26.Poole, L. B. 2005. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch. Biochem. Biophys. 433:240-254. [DOI] [PubMed] [Google Scholar]
- 27.Prapagdee, B., P. Vattanaviboon, and S. Mongkolsuk. 2004. The role of a bifunctional catalase-peroxidase KatA in protection of Agrobacterium tumefaciens from menadione toxicity. FEMS Microbiol. Lett. 232:217-223. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Seaver, L. C., and J. A. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shea, R. J., and M. H. Mulks. 2002. ohr, Encoding an organic hydroperoxide reductase, is an in vivo-induced gene in Actinobacillus pleuropneumoniae. Infect. Immun. 70:794-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 32.Sukchawalit, R., S. Loprasert, S. Atichartpongkul, and S. Mongkolsuk. 2001. Complex regulation of the organic hydroperoxide resistance gene (ohr) from Xanthomonas involves OhrR, a novel organic peroxide-inducible negative regulator, and posttranscriptional modifications. J. Bacteriol. 183:4405-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vattanaviboon, P., W. Eiamphungporn, and S. Mongkolsuk. 2003. Atypical adaptive and cross-protective responses against peroxide killing in a bacterial plant pathogen, Agrobacterium tumefaciens. Curr. Microbiol. 47:323-326. [DOI] [PubMed] [Google Scholar]
- 34.Wang, G., A. A. Olczak, J. P. Walton, and R. J. Maier. 2005. Contribution of the Helicobacter pylori thiol peroxidase bacterioferritin comigratory protein to oxidative stress resistance and host colonization. Infect. Immun. 73:378-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 36.Wood, Z. A., E. Schroder, J. Robin Harris, and L. B. Poole. 2003. Structure, mechanism, and regulation of peroxiredoxins. Trends Biochem. Sci. 28:32-40. [DOI] [PubMed] [Google Scholar]