Abstract
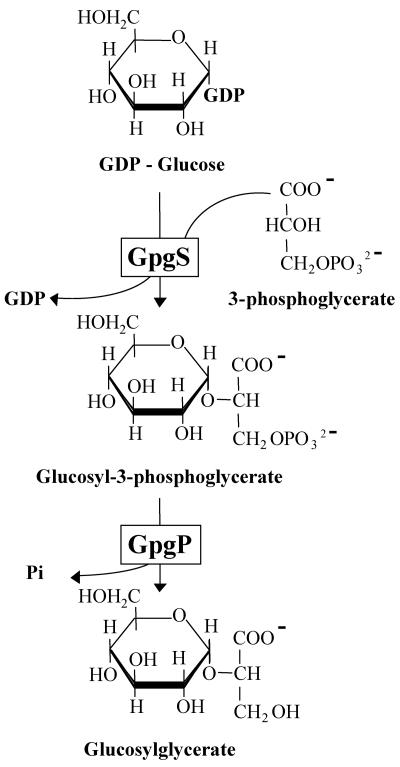
The pathway for the synthesis of the organic solute glucosylglycerate (GG) is proposed based on the activities of the recombinant glucosyl-3-phosphoglycerate synthase (GpgS) and glucosyl-3-phosphoglycerate phosphatase (GpgP) from Methanococcoides burtonii. A mannosyl-3-phosphoglycerate phosphatase gene homologue (mpgP) was found in the genome of M. burtonii (http://www.jgi.doe.gov), but an mpgS gene coding for mannosyl-3-phosphoglycerate synthase (MpgS) was absent. The gene upstream of the mpgP homologue encoded a putative glucosyltransferase that was expressed in Escherichia coli. The recombinant product had GpgS activity, catalyzing the synthesis of glucosyl-3-phosphoglycerate (GPG) from GDP-glucose and d-3-phosphoglycerate, with a high substrate specificity. The recombinant MpgP protein dephosphorylated GPG to GG and was also able to dephosphorylate mannosyl-3-phosphoglycerate (MPG) but no other substrate tested. Similar flexibilities in substrate specificity were confirmed in vitro for the MpgPs from Thermus thermophilus, Pyrococcus horikoshii, and “Dehalococcoides ethenogenes.” GpgS had maximal activity at 50°C. The maximal activity of GpgP was at 50°C with GPG as the substrate and at 60°C with MPG. Despite the similarity of the sugar donors GDP-glucose and GDP-mannose, the enzymes for the synthesis of GPG or MPG share no amino acid sequence identity, save for short motifs. However, the hydrolysis of GPG and MPG is carried out by phosphatases encoded by homologous genes and capable of using both substrates. To our knowledge, this is the first report of the elucidation of a biosynthetic pathway for glucosylglycerate.
Most microorganisms capable of osmotic adjustment accumulate low-molecular-weight organic compounds, commonly designated compatible solutes, which can be taken up from the environment or synthesized de novo (6). Although the uptake of organic solutes such as glycine betaine and trehalose, among others, by microorganisms under salt stress is preferred because it is energetically favorable (10), many organisms synthesize specific compatible solutes because these are not present in the environment or those that are present do not fulfill the specific requirements of the organisms.
Ectoine, glycine betaine, trehalose, and glutamate are among the most common compatible solutes of bacteria and archaea, where they normally accumulate in response to stress imposed by salt (10). Mannosylglycerate (MG), di-myo-inositol-phosphate, and diglycerol-phosphate have a more restricted distribution, being found primarily in (hyper)thermophilic prokaryotes. Mannosylglycerate is widespread in (hyper)thermophilic bacteria and archaea and is also found in some red algae (21, 35). Cyclic 2,3-bisphosphoglycerate has only been encountered in methanogens (20, 25, 36).
The rare solute glucosylglycerate (GG) was originally identified in the cyanobacterium Agmenellum quadruplicatum strain PCC7002 grown under nitrogen-limiting conditions (22). This solute was also detected in the archaeon Methanohalophilus portucalensis strain FDF-1 and in a salt-sensitive mutant of Halomonas elongata, but its accumulation did not appear to be related to salt stress in either organism (7, 33). However, GG was also recently found to behave as a compatible solute in the γ-proteobacterium Erwinia chrysanthemi strain 3937 under nitrogen-limiting conditions, replacing glutamate and glutamine which accumulate under salt stress in the medium when nitrogen is not limiting for growth (18). It is therefore possible that GG also accumulates in other microorganisms during salt stress, under conditions when nitrogen has to be mobilized for the synthesis of other nitrogenous cell components (22). Moreover, GG is chemically related to MG, which has been shown to serve as a compatible solute under salt stress in (hyper)thermophilic prokaryotes (1, 35, 37). It is interesting that GG has only been detected in mesophiles (7, 18, 22, 33).
Our knowledge of the biosynthetic pathways for compatible solutes in prokaryotes has increased significantly in recent years. The synthesis of sugar-derived compatible solutes, such as sucrose, trehalose, glucosylglycerol, galactosylglycerol, and MG, has been elucidated for several organisms (9, 12, 17, 19, 41). We have now identified the genes involved in the synthesis of GG from Methanococcoides burtonii, characterized the recombinant enzymes, and showed that the synthesis of this compatible solute proceeds via a two-step pathway involving glucosyl-3-phosphoglycerate synthase (GpgS) and glucosyl-3-phosphoglycerate phosphatase (GpgP).
MATERIALS AND METHODS
Strains, identification and cloning of gpgS and gpgP from Methanococcoides burtonii, and functional overexpression in Escherichia coli.
Sequences for mannosyl-3-phosphoglycerate phosphatase (MpgP) from Pyrococcus horikoshii (GenPept accession no. BAA30022), Aeropyrum pernix (BAA79870), Thermus thermophilus HB27 (AAO43098), “Dehalococcoides ethenogenes” (YP_182074), and Rhodothermus marinus (AAP74553) were used for BLAST searches in the M. burtonii genome database (DOE Joint Genome Institute [http://www.jgi.doe.gov]).
Methanococcoides burtonii (DSM 6242) and Methanohalophilus portucalensis strain FDF-1 (DSM 7471) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ; Braunschweig, Germany). M. burtonii was cultured in modified methanogenium medium (www.dsmz.de/media/med141.htm), with (2 g/liter) or without Trypticase, under anaerobic conditions with N2-CO2 (80:20 [vol/vol]) as the gas phase at 23°C, with the medium containing 1.8 or 3.0% NaCl. Trimethylamine (50 mM) or methanol (250 mM) was used as the energy source. The medium was supplemented with NH4Cl from 2.5 to 10 mM when growth was attempted on methanol. Cells were harvested during the late exponential phase by centrifugation and washed twice with NaCl at the same concentration as the growth medium. The biomass was freeze-dried and stored at −20°C until extracted with ethanol.
The chromosomal DNA of M. burtonii was isolated according to the method of Rainey et al. (31). The putative glucosyltransferase gene was amplified using 5′ and 3′ primers containing additional EcoRI and HindIII recognition sites, respectively. The mpgP homologue was amplified using 5′ and 3′ primers with NcoI and HindIII sites, respectively. PCR amplification mixtures containing 100 ng of M. burtonii DNA were made up with Pfu Turbo DNA polymerase (Stratagene) as described elsewhere (12). Amplification products were visualized on agarose gels and purified (Promega). After digestion with the suitable restriction enzymes, the fragments were ligated into the corresponding sites of the expression vector pTRC99A (Amersham Pharmacia) to obtain plasmids pMbGT and pMbP, respectively. DNA manipulations followed standard procedures (34). The constructs were then sequenced (AGOWA, Berlin, Germany).
E. coli DH5α was used as the host for expression. E. coli cells carrying pMbGT or pMbP were grown to an optical density at 600 nm of 0.8 in LB medium at 37°C, induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and grown further for 5 h. Ampicillin was added to a final concentration of 100 μg/ml. Cells were harvested, and pellets were suspended in Tris-HCl (20 mM, pH 7.6) containing a protease inhibitor cocktail (Roche) and disrupted by sonication followed by centrifugation. GpgS and GpgP activities were confirmed in cell extracts. An E. coli cell extract with empty pTRC99A was used as a negative control. The supernatants were filtered and used for purification of the enzymes. The protein contents were determined by the Bradford assay (5).
The glucosyltransferase gene was also cloned into pTRCGE, a plasmid carrying a glutathione S-transferase (GST) gene, to facilitate purification of the enzyme (38).
Substrate specificities of GpgS and GpgP.
The substrate specificity of GpgS was determined using ADP-ribose, UDP-acetylgalactosamine, UDP-glucuronic acid, UDP-galactose, GDP-fucose, GDP-mannose, UDP-mannose, ADP-mannose, GDP-glucose, UDP-glucose, TDP-glucose, and ADP-glucose as sugar donors and glycerol, d-3-phosphoglycerate, d-2-phosphoglycerate, l-glycerol-3-phosphate, 2,3-diphospho-d-glycerate, and phosphoenolpyruvate as the sugar acceptors (all from Sigma-Aldrich). The reaction mixtures (50 μl), containing 15 μl cell extract, a 2.5 mM concentration of each substrate, and 20 mM MgCl2 in 25 mM Tris-HCl (pH 7.6), were incubated at 30°C for 30 min, followed by incubation with 2 U of alkaline phosphatase (Sigma-Aldrich) for 30 min at 37°C. The products were visualized by thin-layer chromatography (TLC) (13). Cell extracts from E. coli carrying the empty vector were used as negative controls.
Several sugar phosphates, namely, mannosyl-3-phosphoglycerate (MPG), mannose-1-phosphate, mannose-6-phosphate, glucose-1-phosphate, glucose-6-phosphate, glucose-1,6-bisphosphate, fructose-1-phosphate, fructose-6-phosphate, and trehalose-6-phosphate, as well as GDP and GMP (all from Sigma-Aldrich), were examined as possible substrates for GpgP in a mixture containing a 2.5 mM concentration of each substrate, 25 mM morpholinoethanesulfonic acid (MES; pH 6.0), and 10 mM CoCl2. The mixtures were incubated at 50°C for 5 min and cooled on ice, followed by separation by TLC (13). Cell extracts from E. coli containing the plasmid without an insert were used as negative controls.
Substrate specificity and kinetic parameters of MpgP from T. thermophilus strain HB27.
The dephosphorylation of glucosyl-3-phosphoglycerate (GPG) by MpgP from T. thermophilus (13) was determined in a reaction mixture containing 2.0 mM GPG and 1.0 mM CoCl2 in 25 mM MES (pH 6.0) incubated at 50°C for 15 min and cooled on ice. The free phosphate was quantified, and the complete dephosphorylation of GPG was verified by TLC (13). The reaction to determine the kinetic parameters of MpgP from T. thermophilus was initiated by the addition of known amounts of MpgP (0.8 μg) to 25 mM bis-Tris propane (pH 6.0) solutions containing either MPG or GPG (0.25 to 2.0 mM). These parameters were determined at 30, 50, 70, and 90°C. Reactions were stopped at different times by an ice-ethanol bath. The phosphate released was quantified. All experiments were performed in duplicate.
Synthesis and purification of GPG.
GPG was synthesized using the partially purified recombinant GpgS protein in a mixture containing 10 mM GDP-glucose, 10 mM d-3-phosphoglycerate (3-PGA) in 25 mM bis-Tris propane buffer (pH 8.0) with 1 mM MnCl2. Synthesis proceeded for 30 min at 30°C. After the removal of denatured host proteins, the GPG-containing supernatant was used for the GpgP assays. The concentration of GPG was determined by dephosphorylation of 10 μl of the supernatant of the previous reaction mixtures with 1.0 μg of pure MpgP from T. thermophilus at 50°C for 15 min in a total volume of 25 μl containing 25 mM Tris-HCl (pH 6.0) and 10 mM MgCl2. The volume was adjusted to 300 μl with distilled water prior to phosphate quantification (2). The complete dephosphorylation of GPG was confirmed by TLC. The solvent system used separated GPG from residual GDP-glucose, and visualization of the compounds was achieved with an α-naphthol-sulfuric acid solution (13). The reaction mixture containing GPG was loaded onto a QAE-Sephadex column (Pharmacia, Uppsala, Sweden) equilibrated at pH 9.8 with 5 mM sodium carbonate buffer. The sample was eluted with a linear gradient (5 mM to 1 M NaHCO3 buffer). Fractions were analyzed by nuclear magnetic resonance (NMR) analysis. The buffer was removed in an activated Dowex 50W-X8 column. GG was synthesized from GPG; the specific dephosphorylation of GPG to GG was carried out in a reaction mixture containing GPG, 1 μg of pure MpgP from T. thermophilus HB27, 25 mM Tris-HCl (pH 6.0), and 10 mM MgCl2 in a total volume of 250 μl. The complete dephosphorylation of GPG was confirmed by TLC as described above.
Enzyme assays during purification of proteins.
The activity of GpgS in E. coli cell extracts and during purification was detected by using the reaction mixtures described above, with a 2.5 mM concentration of each substrate. Each mixture was incubated at 50°C for 15 min, followed by incubation with 2 U of alkaline phosphatase for 15 min at 37°C. The activity of GpgP was detected in reaction mixtures (50 μl) containing 15 μl of the eluted fraction, 1.0 mM GPG, 25 mM MES (pH 6.0), and 10 mM CoCl2 for 15 min at 50°C. All products were visualized by TLC (13).
Purification of recombinant GpgS.
GpgS was partially purified with two sequential Q-Sepharose fast-flow columns (Hi-Load 16/10) equilibrated with 20 mM Tris-HCl, pH 7.6. Elution was carried out with linear NaCl gradients (0.0 to 1.0 M), and activity was located as described above. The active fractions were equilibrated with 20 mM Tris-HCl, pH 7.6, loaded onto a Mono Q fast-flow column (Mono Q HR 5/5) equilibrated with the same buffer and eluted by a linear gradient of NaCl (0.0 to 1.0 M). The active fractions were loaded into a Resource-Q fast-flow column as described above. Elution was carried out with a linear NaCl gradient (0.0 to 1.0 M). Active fractions were concentrated and equilibrated by ultrafiltration and then loaded into a Superose 12 column equilibrated with 0.25 M NaCl in 50 mM Tris-HCl, pH 7.6. The purity of the fractions was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Extracts containing pTRGST-GpgS were loaded onto a GSTprep column (FF16/10) and eluted with 50 mM Tris-HCl (pH 7.6) containing 10 mM glutathione (Sigma). Fractions were tested for activity as described above. To separate GpgS from the GST tag, samples were treated with enterokinase according to the manufacturer's instructions (Novagen). The sample was loaded into the Mono-Q column and eluted as described above. The fractions were tested for activity, and purity was determined by SDS-PAGE.
Purification of recombinant GpgP.
The GpgP-containing supernatant was purified with two sequential Q-Sepharose fast-flow columns (Hi-Load 16/10) as described for recombinant GgpS. After elution, the active fractions were concentrated in a 30-kDa-cutoff Centricon column (Amicon), equilibrated with 20 mM Tris-HCl, pH 7.6, loaded onto a strong anionic Q-Sepharose fast-flow column (HiPrep 16/10 Q FF), and eluted. Active fractions were loaded onto a Mono Q fast-flow column (Mono Q 5/50 GL), eluted, concentrated, equilibrated with 50 mM MES (pH 6.0), and loaded into an SP-Sepharose fast-flow column (HiPrep 16/10 SP FF). All elution steps were carried out with linear NaCl gradients (0.0 to 1.0 M). Active fractions were concentrated by ultrafiltration, equilibrated with 50 mM MES (pH 6.0), and applied to a Superose 12 column equilibrated with 0.25 M NaCl in the same buffer. The purity of the samples was judged by SDS-PAGE.
Characterization of recombinant GpgS and GpgP.
The temperature profile, pH dependence, effect of cations, and thermal stability of GpgS were calculated from the synthesis of GPG. Reactions were initiated by adding exact amounts of GpgS and stopped at different times by cooling on ethanol-ice. Samples were then incubated for 2 min at 80°C with MpgP (2 μg) from T. thermophilus to ensure rapid and complete dephosphorylation of GPG, which was confirmed by TLC (13). The free phosphate was quantified.
The activity of GpgP was calculated based on the release of inorganic phosphate from GPG and MPG. Reactions were commenced by adding an exact amount of protein and stopped as described above. Free phosphate was quantified.
The temperature profiles for GpgS and GpgP were determined between 4 and 80°C in 25 mM bis-Tris propane buffer (pH 8.0) with 1 mM MnCl2 and in 25 mM MES buffer (pH 6.0) with 10 mM CoCl2, respectively. The effects of pH on the activities of GpgS and GpgP (with GPG as the substrate) were determined in 25 mM acetate buffer (pH 4.0 to 5.5), 25 mM MES (pH 6.0 to 6.5), bis-Tris propane (pH 6.5 to 9.5), 25 mM Tris-HCl (pH 7.0 to 9.0), and 25 mM CAPS (pH 9.0 to 11.0) at 50°C. The effect of pH on the activity of GpgP with MPG as the substrate was determined at 60°C. All pH values were measured at room temperature. The thermal stabilities of both enzymes were determined at 4, 23, and 50°C as follows. GpgS aliquots (10 μl of a solution of 1.0 mg/ml) were incubated in 25 mM bis-Tris propane (pH 8.0). GpgP (10 μl of a solution of 1.5 mg/ml) was incubated in 25 mM MES (pH 6.0). At appropriate times, samples were withdrawn and immediately examined for residual activities at 30°C with the assays described above.
The Km value for each substrate of GpgS was determined as follows. Reactions were initiated by the addition of known amounts of partially purified GpgS to 25 mM bis-Tris propane solutions (pH 8.0) containing either GDP-glucose (0.1 to 5.0 mM) plus 3-PGA (5 mM) or GDP-glucose (5 mM) plus 3-PGA (0.1 to 5.0 mM) and an excess of MpgP (2.0 μg) from T. thermophilus (23). The reactions were performed at 30 and 50°C and were stopped at 1-min intervals by cooling on ice-ethanol, and the amount of phosphate released was determined. The reactions used to determine the kinetic parameters of GpgP were initiated by the addition of known amounts of GpgP to 25 mM MES solutions (pH 6.0) containing either GPG or MPG (0.25 to 2.0 mM), incubated at 30 and 50°C, and stopped at 1-min intervals by cooling on ice-ethanol. The phosphate released was quantified. All experiments were performed in duplicate. Protein contents were determined by the Bradford assay (5).
Identification of gene encoding GpgS from Methanohalophilus portucalensis.
To amplify the putative gpgS gene from M. portucalensis, degenerate primers were designed on the basis of conserved regions of the GpgS homologues. A fragment of the gpgS gene was amplified with the 5′ primer TTL2 (5′-ACNACNATYCATGAYTTY-3′) and the 3′ primer designated FRY (5′-CNCCNGCNAGNGTRTAYCTRAA-3′). PCR was carried out as previously described, except for the annealing step, which was done at 45°C for 1.5 min. The amplification product was purified and sequenced (AGOWA, Berlin, Germany).
Extraction, quantification of intracellular solutes, and cell protein determination.
Organic solutes were extracted twice with boiling 80% ethanol as previously described (26). Freeze-dried ethanolic extracts were extracted with chloroform to remove lipids. The extracts were freeze-dried and suspended in D2O for NMR analysis. Analyses of organic compounds were performed by 1H-NMR. The protein contents of the cells were determined with a BCA protein assay kit (Pierce) after sonication (B. Braun, Biotech SA).
NMR spectroscopy.
All spectra were acquired on a Bruker DRX500 spectrometer. 31P-NMR and 13C-NMR spectra were recorded with broadband proton decoupling at 202.45 MHz and 125.77 MHz, respectively. Some 1H-NMR spectra were acquired with selective irradiation of the phosphorous signal at 4.4 ppm. Two-dimensional spectra, namely, homonuclear correlation spectroscopy (COSY), nuclear Overhauser effect spectroscopy (NOESY), total correlation spectroscopy (TOCSY), and heteronuclear multiple quantum correlation spectroscopy (1H-13C-HMQC), were performed using standard Bruker pulse programs. Mixing times of 600 ms and 100 ms were employed for the NOESY and TOCSY spectra, respectively. A delay of 3.5 ms was used for the evolution of 1JCH in the carbon correlation spectrum. The pH of the samples was 9.0.
Nucleotide sequence accession number.
A 700-bp sequence corresponding to the partial gpgS gene amplified from Methanohalophilus portucalensis strain FDF-1 (DSM 7471) has been deposited in GenBank under accession number DQ148457.
RESULTS
Identification of genes encoding GpgS and GpgP in M. burtonii.
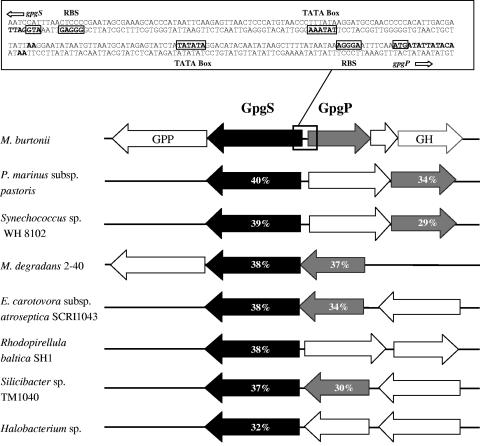
The M. burtonii genome sequence (http://www.jgi.doe.gov) was screened with the BLAST tool, using the amino acid sequences of several mannosyl-3-phosphoglycerate synthase (MpgS; EC 2.4.1.217) and mannosyl-3-phosphoglycerate phosphatase (MpgP; EC 3.1.3.70) proteins. An open reading frame encoding a protein (ZP_00561712) with high homology to MpgP has been assigned as a putative hydrolase of the haloacid dehalogenase superfamily (23). Conserved D-X-D-X-T/V-X and G-D-X-X-X-D motifs of the “DDDD” phosphohydrolase superfamily (39) that are also found in known MpgPs were present in the putative MpgP from M. burtonii (Table 1). The highest homology was found between this gene product and the MpgP domain of the bifunctional MpgS/MpgP complex from “D. ethenogenes” (44%). The putative MpgP protein was encoded by an 819-bp gene, yielding a 272-amino-acid polypeptide with a calculated molecular mass of 23.6 kDa (Table 1), and was functionally characterized as glucosyl-3-phosphoglycerate phosphatase (GpgP).
TABLE 1.
Amino acid identities between MpgP proteins, GpgP proteins, and homologues from different organismsa
| Organismb | Length (aa) of protein | % Identity |
|---|---|---|
| Archaea | ||
| M. burtonii* | 272 | 100 |
| P. horikoshii* | 243 | 38 |
| A. pernix | 281 | 19 |
| Bacteria | ||
| “D. ethenogenes”* | 274 | 44 |
| R. marinus | 277 | 40 |
| M. degradans | 277 | 37 |
| E. carotovora | 277 | 34 |
| T. thermophilus* | 259 | 34 |
| P. marinus | 267 | 34 |
| Silicibacter sp. | 265 | 30 |
| Synechococcus sp. | 264 | 29 |
The proteins examined were MpgPs from P. horikoshii OT3, A. pernix K1, “D. ethenogenes”195, R. marinus, and T. thermophilus HB27, GpgP from M. burtonii, and homologues from Microbulbifer degradans 2-40 (ZP_00317722), Erwinia carotovora subsp. atroseptica SCRI1043 (CAG75151), Prochlorococcus marinus subsp. pastoris MD4 (NC_893077), Silicibacter sp. strain TM 1040 (ZP_00620955), and Synechococcus sp. strain WH 8102 (NP_898523).
*, catalyzes the dephosphorylation of GPG and MPG.
A gene encoding a putative glucosyltransferase (ZP_00561711) was also encountered immediately upstream of gpgP. One small motif (DXD) involved in the binding of an NDP-sugar molecule (43) and another motif only found in MpgS proteins (L/N-A/S-G-E-X-A-M/L/F) were present in this putative glucosyltransferase. However, the protein had no overall homology with known MpgS proteins. The glucosyltransferase had homologues in some prokaryotes, namely, Prochlorococcus marinus subsp. pastoris strain MD4 (40% amino acid identity), Synechococcus sp. strain WH8102 (39%), Erwinia carotovora subsp. atroseptica (38%), Microbulbifer degradans (38%), Rhodopirellula baltica SH1 (38%), Silicibacter sp. strain TM1040 (37%), and Halobacterium sp. strain NRC-1 (32%) (Fig. 1). Homologues of the GpgP protein from M. burtonii were also found in these genomes, except those of R. baltica and Halobacterium sp., where putative GpgPs were absent (Fig. 1). The glucosyltransferase gene from M. burtonii contained 1,215 bp coding for a polypeptide with 404 amino acids and a calculated molecular mass of 43 kDa which was functionally characterized as glucosyl-3-phosphoglycerate synthase (GpgS).
FIG. 1.
Genomic organization and flanking regions of gpgS and gpgP genes from M. burtonii (NCBI accession number for gpgS, ZP_00561711; accession number for gpgP, ZP_00561712) and their homologues in Prochlorococcus marinus subsp. pastoris MD4 (NP_893079 and NC_893077), Synechococcus sp. strain WH 8102 (NP_898525 and NP_898523), Microbulbifer degradans 2-40 (ZP_00317723 and ZP_00317722), Erwinia carotovora subsp. atroseptica SCRI1043 (CAG75152 and CAG75151), Rhodopirellula baltica SH1 (CAD71863), Silicibacter sp. strain TM1040 (ZP_00620956 and ZP_00620955), and Halobacterium sp. (NP_279383). Arrows represent identified or putative GpgS proteins (black), identified or putative GpgP proteins (gray), and unknown open reading frames (open) and their orientations. The percentages of amino acid identity between homologous sequences are indicated inside the arrows. GPP, putative atypical UDP-glucose pyrophosphorylase (ZP_00561714); GH, putative glucosyl hydrolase (ZP_00562149); RBS, ribosomal binding site.
Different genetic organizations of putative gpgS and gpgP genes were found in bacteria and archaea, including an isolated gpgS gene without gpgP, consecutive gpgS and gpgP genes with the same orientation in an operon-like organization, and opposing gpgS and gpgP genes, either contiguous or separated by a putative sucrose phosphorylase gene (Fig. 1). In M. burtonii, gpgS and gpgP are found in opposing directions, with two unidirectional divergent promoters between them. We detected possible ribosome binding sites and two sequences that almost perfectly match consensus archaeal TATA boxes in the 241-bp region between gpgS and gpgP (Fig. 1).
The alignment of all GpgS homologues revealed high sequence conservation and two highly conserved motifs, F-R-Y-P/A-L-A/S-G-E-F/C and D/E-W-G-L-E-X-G-X-L (data not shown). The GpgP sequences also showed two highly conserved motifs, T/S-D-L-D-G-T/S-L-X-D-H and P-F-I-X-E-N-G (data not shown).
Identification of a gene encoding GpgS in Methanohalophilus portucalensis.
A 700-bp fragment coding for 196 amino acids was amplified from M. portucalensis DNA, which had high homology with the GpgS gene from M. burtonii (58%). A highly conserved motif, FRYP/ALA/SGEF/C, of GpgS homologues was also present (accession number DQ148457).
Cloning, functional overexpression of gpgS and gpgP from M. burtonii in E. coli, and purification of recombinant enzymes.
PCR amplification of gpgS and gpgP yielded products with the expected gene sizes and sequences. Activity assays carried out with GpgS- and GpgP-containing E. coli cell extracts showed synthesis and dephosphorylation of GPG, respectively, and the negative control extracts from E. coli with the empty vector did not. Extra bands of 43 and 24 kDa were visualized in SDS-PAGE gels of cell extracts of GpgS-containing and GpgP-containing E. coli clones induced with IPTG. GpgS was purified about 30-fold, to a specific activity of 11.6 nmol/min · mg (Table 2). This preparation had a major protein band with a molecular mass of 43 kDa corresponding to GpgS and two minor bands with higher molecular masses. Attempts to further improve the purity of the protein led to a loss of activity. The GST-GpgS fusion protein eluted from the GSTprep affinity column had no activity after enterokinase digestion. GpgP was purified to homogeneity.
TABLE 2.
Purification of recombinant GpgS from Methanococcoides burtonii
| Purification step | Amt of protein (mg) | Sp act (nmol/min · mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|
| Cell extract | 1,236 | 0.43 | 100 | 1 |
| Q Sepharose I chromatography | 193 | 2.12 | 76 | 5 |
| Q Sepharose II chromatography | 88 | 3.18 | 46.3 | 8 |
| Mono Q chromatography | 29 | 6.89 | 37 | 17 |
| Resource Q chromatography | 11.6 | 11.60 | 25 | 29 |
Catalytic properties of GpgS.
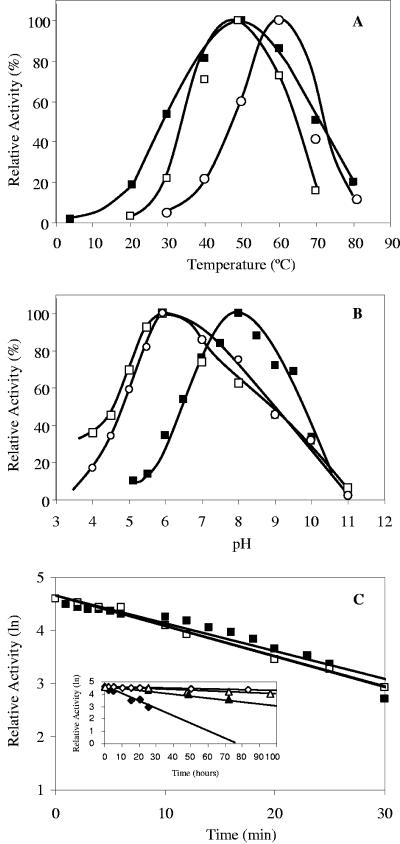
Of the substrates examined, only GDP-glucose and 3-PGA served as substrates for GpgS, which exhibited Michaelis-Menten kinetics at 30 and 50°C (Table 3); MPG synthesis was not detected. The enzyme was active between 4 and 80°C, with maximum activity around 50°C (Fig. 2A). The activity of the enzyme at 50°C was maximal between pH 7.5 and pH 8.5 (Fig. 2B). GpgS was strictly dependent on divalent cations in the following order of efficiency: Mn2+ > Co2+ > Mg2+. The maximum activation of the enzyme was obtained with 1.0 mM of Mn2+ or Co2+. Other cations tested, such as K+, Ca2+, Sr2+, Cu2+, Ba2+, and Zn2+, did not stimulate GpgS activity at any concentration. The enzyme had a half-life of 13.8 min at 50°C, 11.5 h at 23°C, and 51.3 h at 4°C (Fig. 2C).
TABLE 3.
Biochemical properties and kinetic parameters for the substrates of GpgS and GpgP from M. burtonii and MpgP from T. thermophilus
| Organism | Enzyme | Temp (°C) | Substrate | Km (mM)a | Vmax (μmol/min · mg protein)a |
|---|---|---|---|---|---|
| M. burtonii | GpgS | 30 | GDP-Glu | 1.75 ± 0.43 | ND |
| 30 | 3-PGA | 0.54 ± 0.20 | ND | ||
| 50 | GDP-Glu | 3.70 ± 0.05 | ND | ||
| 50 | 3-PGA | 1.60 ± 0.10 | ND | ||
| GpgP | 30 | GPG | 0.08 ± 0.01 | 0.10 ± 0.01 | |
| 30 | MPG | — | — | ||
| 50 | GPG | 0.27 ± 0.02 | 0.58 ± 0.04 | ||
| 50 | MPG | 0.28 ± 0.03 | 0.25 ± 0.03 | ||
| T. thermophilus | MpgP | 30 | GPG | 0.10 ± 0.01 | 0.29 ± 0.06 |
| 30 | MPG | 0.11 ± 0.02 | 0.15 ± 0.03 | ||
| 50 | GPG | 0.36 ± 0.04 | 1.01 ± 0.05 | ||
| 50 | MPG | 0.37 ± 0.03 | 0.50 ± 0.02 | ||
| 70 | GPG | 0.79 ± 0.03 | 46.30 ± 0.58 | ||
| 70 | MPG | 0.83 ± 0.01 | 46.70 ± 0.55 | ||
| 90 | GPG | 1.10 ± 0.06 | 178.57 ± 2.53 | ||
| 90 | MPG | 0.90 ± 0.02 | 156.25 ± 1.72 |
ND, not determined; —, not detected. The values are means ± standard deviations of two independent experiments.
FIG. 2.
Temperature (A) and pH (B) dependence of M. burtonii recombinant GpgS (▪) and GpgP (□) with GPG and with MPG (○) as the substrate. (C) Thermostabilities of recombinant GpgS (filled symbols) and GpgP (open symbols) at 50°C. The inset shows the thermostabilities at 4 (▴ and ▵) and 23°C (⧫ and ⋄). The data are the mean values of two independent experiments.
Catalytic properties of GpgP from M. burtonii and MpgP from T. thermophilus.
Of the substrates examined, GpgP dephosphorylated both GPG and MPG, but with slightly different temperature profiles. GpgP had maximum activity with GPG at 50°C, with the activity becoming undetectable below 20°C and above 70°C (Fig. 2A). The maximum activity of GpgP with MPG occurred at 60°C, but activity was almost undetectable at 30°C (Fig. 2A). The optimum pH range for the activity of the enzyme was between 5.5 and 6.5 for GPG and MPG alike, with lower degrees of activity at pH 4.0 and pH 10.0 (Fig. 2B). GpgP had a half-life of 12 min at 50°C, 165 h at 23°C, and 198 h at 4°C (Fig. 2C). GpgP exhibited Michaelis-Menten kinetics, with identical Km values for both substrates and a slightly higher apparent Vmax with GPG than with MPG at 50°C. At 30°C, GpgP also exhibited Michaelis-Menten kinetics with GPG (Table 3).
At 50°C, divalent cations were absolutely required for GpgP activity with GPG as the substrate. However, when MPG was the substrate, 3% activity could be detected without cations, and the preferred metal ion with both substrates was Co2+ (10 mM). At 30°C, GpgP retained 37% of its enzyme activity towards GPG and 12% activity towards MPG in a reaction mixture without cations compared to the activity with 10 mM K+ and Co2+. Potassium chloride, in the concentration range of 10 to 100 mM, stimulated enzyme activity about 40% at 50°C, but only when MPG was the substrate. However, at 30°C, K+ had a stimulatory effect towards GPG, but not with MPG as the substrate. Mg2+ and, to a lesser extent, Ni2+ could replace Co2+. Other cations, i.e., Ca2+, Sr2+, Ba2+, Cu2+, and Zn2+, had no effect on GpgP activity at any concentration. Enzyme activity was not detected with Mn2+ when GPG was used, while Mn2+ had a strong stimulatory effect (85% of the maximal activity with Co2+) with MPG. The recombinant MpgP enzyme from T. thermophilus HB27 (13) catalyzed the dephosphorylation of both MPG and GPG, showed Michaelis-Menten kinetics at 30, 50, 70, and 90°C (Table 3).
Identification of GPG by NMR.
The TOCSY spectrum revealed two spin systems, presenting the characteristics of a hexose moiety and a glycerate moiety. The COSY and HMQC spectra allowed the assignment of all proton and carbon signals (Table 4). An analysis of the proton-proton scalar coupling patterns determined the configuration of the hexose as α-glucopyranose. The NOESY spectrum revealed a correlation between H1 of glucose and H2 of the glycerate moiety, thus establishing the glycosidic bond as an α (1,2) linkage. The 31P-NMR spectra showed a resonance in the region of the phosphomonoesters. A proton spectrum with irradiation of this phosphorous signal caused a collapse of some multiplicities in the signal assigned to position 3 of the glycerate moiety. As a result, the structure of the product was established as (1,2)-α-glucopyranosyl-3-phosphoglycerate.
TABLE 4.
NMR parameters of α-glucosyl-3-phosphoglycerate (300 K, pH 9.0)
| Moiety | 13C δ (ppm)a |
1H parameter
|
31P δ (ppm)a | |
|---|---|---|---|---|
| δ (ppm)a | JH,H (Hz)a | |||
| Glucosyl | ||||
| C1 | 99.79 | 4.97 | 3J1,2 = 3.5 | |
| C2 | 74.35 | 3.53 | 3J2,3 = 9.7 | |
| C3 | 75.74 | 3.85 | 3J3,4 = 9.8 | |
| C4 | 72.87 | 3.32 | 3J4,5 = 9.8 | |
| C5 | 74.67 | 3.92 | 3J5,6a = 6.2 | |
| C6(a,b) | 63.67 | 3.90/3.67 | 2J6a,6b = 12.5 | |
| Glyceryl | ||||
| C1 | 179.69 | |||
| C2 | 79.75 | 4.10 | ND | |
| C3(a,b) | 67.99 | 4.00 | ND | 4.42 |
1H and 13C signals were referenced to internal 3-(trimethylsilyl)propanesulfonic acid (sodium salt), while 31P signals were referenced to 85% phosphoric acid (external). ND, not determined.
Analysis of compatible solutes from M. burtonii.
The growth rate of M. burtonii on trimethylamine was not affected by the absence of Trypticase and NH4Cl from the medium; the organism had generation times of 20 and 47 h at 1.8 and 3% NaCl, respectively. No growth was observed when methanol was used as the sole source of energy instead of trimethylamine. At a low salinity (1.8% NaCl), M. burtonii accumulated α-glutamate and aspartate. When the salinity was raised to 3% NaCl, α-glutamate and Nɛ-acetyl-β-lysine were the major solutes, along with small amounts of glycine betaine.
DISCUSSION
MG behaves as a compatible solute during the osmotic adaptation of several (hyper)thermophiles (35). On the other hand, the role of GG as a compatible solute has only been unequivocally demonstrated for Erwinia chrysanthemi strain 3937, where GG increased concomitantly with the NaCl concentration, but only in nitrogen-limited medium (18). In at least one Ectothiorhodospira sp., trehalose becomes a preferred organic osmolyte instead of ectoine under nitrogen-limiting conditions (16, 42). We were not able to detect GG in M. burtonii because the organism did not grow on methanol instead of trimethylamine, and therefore we could not decrease the levels of nitrogen to those that might be required for the accumulation of GG instead of aspartate, α-glutamate, Nɛ-acetyl-β-lysine, and glycine betaine. However, GG accumulates as a minor solute in the closely related but extremely halophilic methanogen Methanohalophilus portucalensis strain FDF-1 (33), which normally accumulates Nɛ-acetyl-β-lysine, glycine betaine, and β-glutamine in response to osmotic stress but also possesses a highly homologous gpgS gene.
We propose that the pathway for the synthesis of GG involves two steps (Fig. 3), where GDP-glucose and d-3-phosphoglycerate are converted by glucosyl-3-phosphoglycerate synthase (GpgS) into glucosyl-3-phosphoglycerate (GPG), which is then dephosphorylated to GG by glucosyl-3-phosphoglycerate phosphatase (GpgP). These two enzymes are encoded by two contiguous but divergently oriented genes in M. burtonii, designated gpgS and gpgP, respectively. The organization of gpgS and gpgP differs in other organisms that harbor them, unlike the standard operon-like organization of mpgS and mpgP (4, 12, 13). Consensus sequences for unidirectional divergent promoters, like those found between gpgS and gpgP from M. burtonii, have been reported often, indicating that these regions are not accidental rarities but represent a specific type of gene organization allowing a high degree of freedom of gene regulation (11). A putative regulatory protein may interact with the operator between the divergent genes to either activate or repress transcription in both or only one direction (3). The divergent organization of GG genes also offers the advantage of a self-contained unit which can be translocated to new locations without the loss of autonomy because the promoters are internal (44). This would be of major importance for the aptness of these genes if lateral transfer should occur.
FIG. 3.
Proposed two-step pathway for the synthesis of glucosylglycerate in M. burtonii.
Of the substrates examined, GDP-glucose and d-3-phosphoglycerate were the only ones used by GpgS from M. burtonii, which, like MpgS, retains the configuration of the glycosidic bond, indicating that GpgS could be included in glycosyltransferase family 55 (4, 8, 12, 13, 14). The GpgS enzyme from the psychrotolerant M. burtonii had maximum activity at 50°C, but like the MpgS domain of the bifunctional MpgS/MpgP complex from “D. ethenogenes,” it was progressively unstable as the temperature was raised above 4°C (14).
GpgP from M. burtonii dephosphorylated GPG and MPG, but the temperature for maximal activity shifted from 50°C with GPG to 60°C with MPG. We also verified that the MpgP proteins from T. thermophilus, P. horikoshii, and “D. ethenogenes” (12, 13, 14) dephosphorylate not only MPG but also GPG, indicating that these phosphatases recognize a common determinant in these substrates, probably the glycerylphosphate moiety. The apparent Vmax values of GpgP and MpgP from T. thermophilus were slightly higher at low temperatures towards GPG than towards MPG, while the Km values for both substrates were similar. These results could indicate a slightly higher catalytic efficiency for GPG than MPG at low growth temperatures, but no conclusions can be drawn from the results obtained.
The observation that KCl stimulates GpgP was striking because it is the first report of an MPG/GPG phosphatase stimulated by KCl in vitro (4, 12, 13). However, this effect was observed primarily with GPG. It is well documented that K+ is central in the signaling cascade between salt stress sensing and osmoadaptation in prokaryotes in general and archaea in particular (32). It could be argued that osmotic stress could cause a posttranslational activation of GpgP in vivo via K+ influx. The physiological significance of the observation that potassium stimulates the dephosphorylation of GPG could perhaps indicate that the formation of GG is osmotically regulated.
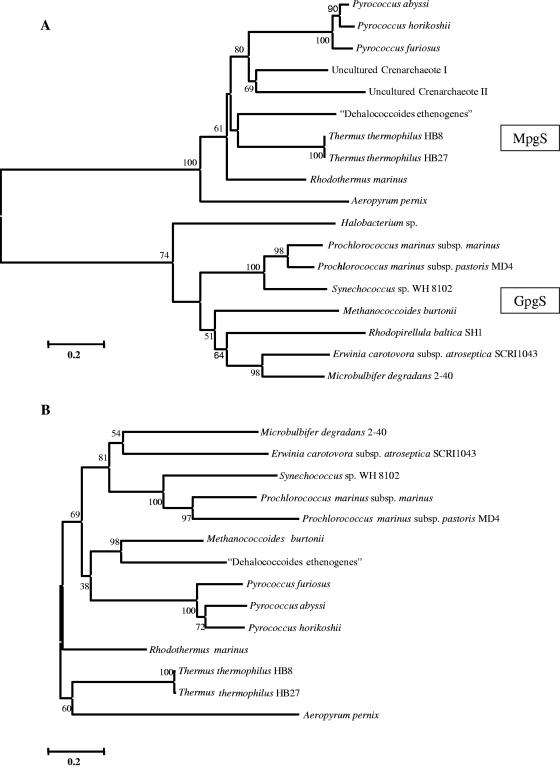
An unrooted phylogenetic tree constructed based on the amino acid sequences of known MpgPs, GpgPs, and their homologues forms one cluster, reflecting the high identity among them (Fig. 4). The higher sequence similarities of the “D. ethenogenes” and Pyrococcus MpgPs with the GpgP from M. burtonii could be explained by lateral gene transfer, as proposed recently for MpgS and MpgP of “D. ethenogenes” (14). The nucleotide sequences of the gpgS genes indicate that the codon usage of M. burtonii gpgS is distinctly archaeal, since some codons rarely used in bacteria are common in those genes (29).
FIG. 4.
Unrooted phylogenetic trees based on available amino acid sequences of GpgS, MpgS, GpgS, and MpgS homologues (A) and GpgP, MpgP, and GpgP homologues (B). The Clustal X program (40) was used for sequence alignment, and the MEGA2 program (24) was used to generate the phylogenetic tree. The significance of the branching order was evaluated by bootstrap analysis of 1,000 computer-generated trees. The bootstrap values are indicated. Bar, 0.2 changes/site.
All of the recombinant proteins of the MpgS cluster, with the exception of the crenarchaeotal proteins, which have not been examined, synthesize MPG (4, 12, 13, 14). The majority of the organisms represented in the GpgS cluster are, however, not known to accumulate GG, nor are the majority of these proteins known to catalyze the synthesis of GPG. Nevertheless, the high homology between these putative GpgS proteins indicates a common function.
This study demonstrates that the phosphatases responsible for the hydrolysis of MPG and GPG are functionally interchangeable and are encoded by homologous genes. However, the unrelated synthases that synthesize MPG and GPG have different sequences with only small motifs in common, possibly reflecting a very ancient divergence. The data gathered here lead to the hypothesis that the genes coding for MpgS and GpgS have diverged from a common ancestor with the adaptation of organisms to environments with different temperatures. This is corroborated, to some extent, by the observation that the accumulation of MG is more common in organisms that grow at high temperatures (35), while organisms with the genes for the synthesis of GG, which could presumably accumulate this solute, grow at low temperatures. MpgS and GpgS might have evolved from the common ancestor protein, possibly capable of MPG and GPG synthesis at different rates, by gene duplication (28, 30). For example, mannosylglycerate synthase from R. marinus is able to synthesize, with comparable catalytic efficiencies, both MG and GG at 25°C from GDP-mannose or GDP-glucose and glycerate, but the synthesis of MG is significantly favored over that of GG at 65°C (15, 27). These data suggest a preference of the R. marinus mannosylglycerate synthase for mannose over glucose precursors for the synthesis of compatible solutes at higher temperatures.
The results obtained in this work lead us to consider that the different synthases producing GPG and MPG arose via divergent evolution from a common ancestor, while the hydrolysis of those compounds to their respective compatible solutes is catalyzed by homologous phosphatases. Future research will clarify the origins and evolution of the corresponding genes.
Acknowledgments
This work was supported by the European Commission 6th Framework Programme contract COOP-CT-2003-508644, FEDER, and FCT, Portugal (POCTI//BIO/42331/2001 and POCI2010/010.6/A005/2005). J. Costa, N. Empadinhas, and P. Lamosa acknowledge scholarships from FCT (SFRH/BD/9681/2002, SFRH/BPD/14828/2003, and SFRH/BPD/11511/2002).
The technical assistance of Ana I. Mingote is gratefully acknowledged. Sequence data from M. burtonii were produced by the U.S. Department of Energy Joint Genomic Institute (http://www.jgi.doe.gov/).
REFERENCES
- 1.Alarico, S., N. Empadinhas, C. Simões, Z. Silva, A. Henne, A. Mingote, H. Santos, and M. S. da Costa. 2005. Distribution of genes for the synthesis of trehalose and mannosylglycerate in Thermus spp. and direct correlation with halotolerance. Appl. Environ. Microbiol. 71:2460-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames, B. N. 1966. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8:115-118. [Google Scholar]
- 3.Beck, C. F., and R. A. J. Warren. 1988. Divergent promoters, a common form of gene organization. Microbiol. Rev. 52:318-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borges, N., J. D. Marugg, N. Empadinhas, M. S. da Costa, and H. Santos. 2004. Specialized roles of the two pathways for the synthesis of mannosylglycerate in osmoadaptation and thermoadaptation of Rhodothermus marinus. J. Biol. Chem. 279:9892-9898. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brown, A. D. 1976. Microbial water stress. Bacteriol. Rev. 40:803-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cánovas, D., N. Borges, C. Vargas, A. Ventosa, J. Nieto, and H. Santos. 1999. Role of N-γ-acetyldiaminobutyrate as an enzyme stabilizer and an intermediate in the biosynthesis of hydroxyectoine. Appl. Environ. Microbiol. 65:3774-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328:307-317. [DOI] [PubMed] [Google Scholar]
- 9.Curatti, L., E. Folco, P. Desplats, G. Abratti, V. Limones, L. Herrera-Estrella, and G. Salerno. 1998. Sucrose-phosphate synthase from Synechocystis sp. strain PCC6803: identification of the spsA gene and characterization of the enzyme expressed in Escherichia coli. J. Bacteriol. 180:6776-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Costa, M. S., H. Santos, and E. A. Galinski. 1998. An overview of the role and diversity of compatible solutes in Bacteria and Archaea. Adv. Biochem. Eng. Biotechnol. 61:117-153. [DOI] [PubMed] [Google Scholar]
- 11.Davies, G., and B. Henrissat. 1995. Structures and mechanisms of glycosyl hydrolases. Structure 3:835-859. [DOI] [PubMed] [Google Scholar]
- 12.Empadinhas, N., J. D. Marugg, N. Borges, H. Santos, and M. S. da Costa. 2001. Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. Biochemical and genetic characterization of key enzymes. J. Biol. Chem. 276:43580-43588. [DOI] [PubMed] [Google Scholar]
- 13.Empadinhas, N., L. Albuquerque, A. Henne, H. Santos, and M. S. da Costa. 2003. The bacterium Thermus thermophilus, like hyperthermophilic archaea, uses a two-step pathway for the synthesis of mannosylglycerate. Appl. Environ. Microbiol. 69:3272-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Empadinhas, N., L. Albuquerque, J. Costa, S. H. Zinder, M. A. S. Santos, H. Santos, and M. S. da Costa. 2004. A gene from the mesophilic bacterium Dehalococcoides ethenogenes encodes a novel mannosylglycerate synthase. J. Bacteriol. 186:4075-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flint, J., E. Taylor, M. Yang, D. N. Bolam, L. E. Tailford, C. Martinez-Fleites, E. J. Dodson, B. G. Davis, H. J. Gilbert, and G. J. Davies. 2005. Structural dissection and high-throughput screening of mannosylglycerate synthase. Nat. Struct. Mol. Biol. 12:608-614. [DOI] [PubMed] [Google Scholar]
- 16.Galinski, E. A., and R. Herzog. 1990. The role of trehalose as a substitute for nitrogen-containing compatible solutes (Ectothiorhodospira halochloris). Arch. Microbiol. 153:607-613. [Google Scholar]
- 17.Giæver, H. M., O. B. Styrvold, I. Kaasen, and A. R. Strøm. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J. Bacteriol. 170:2841-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goude, R., S. Renaud, S. Bonnassie, T. Bernard, and C. Blanco. 2004. Glutamine, glutamate, and α-glucosylglycerate are the major osmotic solutes accumulated by Erwinia chrysanthemi strain 3937. Appl. Environ. Microbiol. 70:6535-6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemann, M., U. Effmert, T. Kerstan, A. Schoor, and N. Erdmann. 2001. Biochemical characterization of glucosylglycerol-phosphate synthase of Synechocystis sp. strain PCC 6803: comparison of crude, purified, and recombinant enzymes. Curr. Microbiol. 43:278-283. [DOI] [PubMed] [Google Scholar]
- 20.Hensel, R., and H. König. 1988. Thermoadaptation of methanogenic bacteria by intracellular ion concentration. FEMS Microbiol. Lett. 49:75-79. [Google Scholar]
- 21.Karsten, U., J. A. West, G. C. Zuccarello, R. Engbrodt, A. Yokoyama, Y. Hara, and J. Brodie. 2003. Low molecular weight carbohydrates of the Bangiophycidae (Rhodophyta). J. Phycol. 39:584-589. [Google Scholar]
- 22.Kollman, V. H., J. L. Hanners, R. E. London, E. G. Adame, and T. E. Walker. 1979. Photosynthetic preparation and characterization of 13C-labeled carbohydrates in Agmenellum quadruplicatum. Carbohydr. Res. 73:193-202. [Google Scholar]
- 23.Koonin, E. V., and R. L. Tatusov. 1994. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J. Mol. Biol. 244:125-132. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 25.Lehmacher, A., A. B. Vogt, and R. Hensel. 1990. Biosynthesis of cyclic 2,3-diphosphoglycerate. Isolation and characterization of 2-phosphoglycerate kinase and cyclic 2,3-diphosphoglycerate synthetase from Methanothermus fervidus. FEBS Lett. 272:94-98. [DOI] [PubMed] [Google Scholar]
- 26.Martins, L. O., and H. Santos. 1995. Accumulation of mannosylglycerate and di-myo-inositol-phosphate by Pyrococcus furiosus in response to salinity and temperature. Appl. Environ. Microbiol. 61:3299-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martins, L. O., N. Empadinhas, J. D. Marugg, C. Miguel, C. Ferreira, M. S. da Costa, and H. Santos. 1999. Biosynthesis of mannosylglycerate in the thermophilic bacterium Rhodothermus marinus. Biochemical and genetic characterization of a mannosylglycerate synthase. J. Biol. Chem. 274:35407-35414. [DOI] [PubMed] [Google Scholar]
- 28.Miller, B. G., and R. T. Raines. 2004. Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases. Biochemistry 43:6387-6392. [DOI] [PubMed] [Google Scholar]
- 29.Nalezkova, M., A. de Groot, M. Graf, P. Gans, and L. Blanchard. 2005. Overexpression and purification of Pyrococcus abyssi phosphopantetheine adenylyltransferase from an optimized synthetic gene for NMR studies. Protein Expr. Purif. 39:296-306. [DOI] [PubMed] [Google Scholar]
- 30.O'Brien, P. J., and D. Herschlag. 1999. Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 6:91-105. [DOI] [PubMed] [Google Scholar]
- 31.Rainey, F. A., N. Ward-Rainey, R. M. Kroppenstedt, and E. Stackebrandt. 1996. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int. J. Syst. Bacteriol. 46:1088-1092. [DOI] [PubMed] [Google Scholar]
- 32.Roberts, M. F. 2004. Osmoadaptation and osmoregulation in archaea: update 2004. Front. Biosci. 9:1999-2019. [DOI] [PubMed] [Google Scholar]
- 33.Robertson, D. E., M. Lai, R. P. Gunsalus, and M. F. Roberts. 1992. Composition, variation, and dynamics of major osmotic solutes in Methanohalophilus strain FDF1. Appl. Environ. Microbiol. 58:2438-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Santos, H., and M. S. da Costa. 2002. Compatible solutes of organisms that live in hot saline environments. Environ. Microbiol. 4:501-509. [DOI] [PubMed] [Google Scholar]
- 36.Seely, R. J., R. D. Krueger, and D. E. Fahrney. 1983. Cyclic-2,3-diphosphoglycerate levels in Methanobacterium thermoautotrophicum reflect inorganic phosphate availability. Biochem. Biophys. Res. Commun. 116:1125-1128. [DOI] [PubMed] [Google Scholar]
- 37.Silva, Z., S. Alarico, A. Nobre, R. Horlacher, J. Marugg, W. Boos, A. I. Mingote, and M. S. da Costa. 2003. Osmotic adaptation of Thermus thermophilus RQ-1: lesson from a mutant deficient in synthesis of trehalose. J. Bacteriol. 185:5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva, Z., M. M. Sampaio, A. Henne, A. Bohm, R. Gutzat, W. Boos, M. S. da Costa, and H. Santos. 2005. The high-affinity maltose/trehalose ABC transporter in the extremely thermophilic bacterium Thermus thermophilus HB27 also recognizes sucrose and palatinose. J. Bacteriol. 187:1210-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thaller, M. C., S. Schippa, and G. M. Rossolini. 1998. Conserved sequence motifs among bacterial, eukaryotic, and archaeal phosphatases that define a new phosphohydrolase superfamily. Protein Sci. 7:1647-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomson, K.-S. 1983. Purification of UDP galactose:sn-glycerol-3-phosphate-d-galactosyltransferase from Poterioochromonas malhamensis. Biochim. Biophys. Acta 759:154-159. [Google Scholar]
- 42.Trüper, H. G., and E. A. Galinski. 1990. Biosynthesis and fate of compatible solutes in extremely halophilic phototrophic eubacteria. FEMS Microbiol. Rev. 75:247-254. [Google Scholar]
- 43.Wiggins, C. A. R., and S. Munro. 1998. Activity of the yeast MNN1 alpha-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc. Natl. Acad. Sci. USA 95:7945-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, M., J. Hay, and W. T. Ruyechan. 2004. The DNA element controlling expression of the varicella-zoster virus open reading frame 28 and 29 genes consists of two divergent unidirectional promoters which have a common USF site. J. Virol. 78:10939-10952. [DOI] [PMC free article] [PubMed] [Google Scholar]