Abstract
The thyX gene for thymidylate synthase of the Lyme borreliosis (LB) agent Borrelia burgdorferi is located in a 54-kb linear plasmid. In the present study, we identified an orthologous thymidylate synthase gene in the relapsing fever (RF) agent Borrelia hermsii, located it in a 180-kb linear plasmid, and demonstrated its expression. The functions of the B. hermsii and B. burgdorferi thyX gene products were evaluated both in vivo, by complementation of a thymidylate synthase-deficient Escherichia coli mutant, and in vitro, by testing their activities after purification. The B. hermsii thyX gene complemented the thyA mutation in E. coli, and purified B. hermsii ThyX protein catalyzed the conversion of dTMP from dUMP. In contrast, the B. burgdorferi ThyX protein had only weakly detectable activity in vitro, and the B. burgdorferi thyX gene did not provide complementation in vivo. The lack of activity of B. burgdorferi's ThyX protein was associated with the substitution of a cysteine for a highly conserved arginine at position 91. The B. hermsii thyX locus was further distinguished by the downstream presence in the plasmid of orthologues of nrdI, nrdE, and nrdF, which encode the subunits of ribonucleoside diphosphate reductase and which are not present in the LB agents B. burgdorferi and Borrelia garinii. Phylogenetic analysis suggested that the nrdIEF cluster of B. hermsii was acquired by horizontal gene transfer. These findings indicate that Borrelia spp. causing RF have a greater capability for de novo pyrimidine synthesis than those causing LB, thus providing a basis for some of the biological differences between the two groups of pathogens.
Spirochetes are a coherent group of bacteria with a unique morphology and cell structure (38). The point of divergence of spirochetes from other bacterial groups was deep in time, and the topology of spirochetes' branching from other phyla, such as proteobacteria and cyanobacteria, remains unresolved. Phylogenetic inference has been advanced with the availability of genome sequences of pathogenic spirochete species from the following three genera: Borrelia, including Borrelia burgdorferi (13, 19) and B. garinii (23), two agents of Lyme borreliosis (LB); Treponema, including Treponema pallidum (20), the agent of syphilis, and Treponema denticola (51), a mouth organism associated with periodontal disease; and two serovars of Leptospira interrogans (36, 44). Leptospires have free-living capacity and, not surprisingly, larger chromosomes, at ∼4.5 Mb, than the ∼1-Mb chromosomes of the obligately parasitic organisms T. pallidum (20) and B. burgdorferi (18). But the leptospires' branching from the common ancestor of Treponema and Borrelia appears to be nearly as ancient as the spirochetes' origin itself, and for the most part, leptospire sequences have been of limited use for further defining evolution within the spirochete clade.
The several Borrelia species can be divided into two major groups on the basis of DNA sequence and biological differences (2). One group includes all the agents of LB, such as B. burgdorferi and B. garinii, as well as related species of uncertain pathogenicity for humans. The second monophyletic group comprises several species that cause relapsing fever (RF), such as B. hermsii and B. turicatae, as well as some other species, such as B. miyamotoi. Lyme borreliosis is an illness with a more protracted course and a generally milder character than those experienced during relapsing fever (4). This clinical difference may be attributable in part to the much lower peak densities in the blood that are achieved by LB spirochetes than RF spirochetes (39, 47, 52). While RF spirochetes can be found in abundant numbers in blood smears from febrile patients or experimentally infected animals, LB agents in the blood are detectable only by culture or PCR assay (10). As suggested by Schwan et al. (49), these differences in the carrying capacities of the two types of Borrelia in the blood may be the consequence of differences in metabolic capacities of LB and RF agents.
The characteristics of the spirochetes in their vectors also may differentially correlate with the metabolic capabilities of RF and LB species (49). The tick vectors of RF are soft ticks whose feeding times on a host are measured in minutes rather than the days typical of the hard tick vectors of LB. As expected for this limited period for achieving transmission, RF spirochetes are already positioned in the tick's salivary glands at attachment, and they soon enter the host through the tick's saliva as it feeds. In unfed hard ticks, B. burgdorferi is mainly inside the midgut and does not migrate to the salivary glands until a day or two after being exposed to blood during feeding (7, 46). There is thus more time for Borrelia spp. causing LB to adjust to the vertebrate environment before they actually enter the host (50).
The genomes of Borrelia spp. are distinguished from those of other spirochetes and most other bacteria by having linear chromosomes and linear as well as circular plasmids (18, 41). Approximately 40% of the B. burgdorferi genome is carried on plasmids (13, 19). The linear plasmids vary in size from 5 to 180 kb and have covalently closed hairpins for telomeres (5, 12). Borrelia organisms maintain on their plasmids the genes for most of the major lipoprotein surface antigens, but the plasmids also carry some metabolism genes, such as guaA and guaB, which are part of the purine biosynthetic pathway (32). The purine biosynthetic genes purA and purB are on the chromosomes of B. hermsii and other RF species, but their absence from LB species and their nucleotide sequences suggest that they were acquired through horizontal gene transfer (6).
What genes there are for de novo pyrimidine synthesis in B. burgdorferi and B. garinii are located on the chromosomes of these LB agents (19, 23). An exception is an orthologue of thyX, which is located near the telomere of a 54-kb linear plasmid of B. burgdorferi (19). For B. garinii, a thyX orthologue was not included in the annotation of the genome (23), but an examination of position 55200 to the end of the sequence at position 55560 of a 56-kb linear plasmid (accession number NC_006129) reveals an open reading frame encoding a 119-amino-acid polypeptide with 86% sequence identity with the N-terminal half of the B. burgdorferi thyX gene product. In other species of organisms, the thyX product, ThyX, is thymidylate synthase X (EC 2.1.1.148), which catalyzes the methylation of dUMP to yield the DNA precursor dTMP. Sequences homologous to thyX have been found in several groups of bacteria, including cyanobacteria, actinobacteria, and epsilon-proteobacteria such as Helicobacter pylori, but they are also present in some double-stranded DNA viruses of bacteria and at least one eukaryote, the slime mold Dictyostelium discoides (30, 35). Other groups of bacteria, including gamma-proteobacteria such as Escherichia coli and firmicutes such as Bacillus subtilis, usually have instead a thyA gene, which encodes thymidylate synthase A (ThyA; EC 2.1.1.45) and is not homologous to thyX (11, 30). ThyX proteins are homotetrameric and flavin adenine dinucleotide (FAD) dependent, while ThyA proteins are homodimeric and FAD independent in their catalysis (33, 35).
Using B. hermsii genomic DNA as a hybridization probe of a B. burgdorferi DNA array, we had previously found evidence that B. hermsii has a thyX gene (54). This finding prompted our further study of the RF species B. hermsii, first to confirm the presence of a thyX orthologue in this species and then, if present, to characterize it for further insight into thyX origins in spirochetes and to investigate the function of the thyX gene product. In the course of the study, we identified in B. hermsii other pyrimidine biosynthesis genes that were not present in LB species genomes.
MATERIALS AND METHODS
Bacterial strains.
B. burgdorferi B31 (ATCC 35210) was grown at 34°C in tightly capped tubes of Barbour-Stoenner-Kelly broth medium (BSK) (3). B. hermsii HS1 (45) was grown in either BSK or Dulbecco's modified Eagle's medium (Invitrogen, San Diego, Calif.) supplemented with 4% fraction V of bovine serum albumin (US Biochemical Corp., Cleveland, Ohio), 1.1% gelatin, and 1.5 mM N-acetylglucosamine (SDMEM). Spirochetes were counted with a Petroff-Hauser chamber by phase-contrast microscopy, and cultivated cells were harvested by centrifugation at 2,000 × g for 20 min at 20°C. For some experiments, B. hermsii cells were recovered from citrated blood of infected CB17 scid mice by centrifugation of the citrated blood at 100 × g for 5 min as described previously (40). E. coli K-12 strain χ2913 (ΔthyA572 recA56 srlA::Tn10) (14), which was provided by D. V. Santi of the University of California at San Francisco, was grown at 37°C in M9 minimal medium supplemented with 0.1 mM CaCl2, 2 mM MgSO4, 0.2% Difco Casamino Acids, and 50 μg/ml thymidine. E. coli strains TOP10 (Invitrogen) and SURE (Stratagene, La Jolla, Calif.) were grown at 37°C in Luria-Bertani medium, with or without 100 μg/ml carbenicillin.
Extraction and labeling of DNA.
Plasmid DNA was isolated from E. coli with a High Pure plasmid isolation kit (Roche Diagnostics, Mannheim, Germany), and total DNAs were isolated from B. hermsii and B. burgdorferi with a DNeasy tissue kit (QIAGEN Science, Valencia, Calif.). DNAs were labeled with [α-32P]dATP (Perkin-Elmer, Boston, Mass.) by random priming using the Megaprime DNA labeling system (Amersham Biosciences, Piscataway, N.J.), as described previously (54).
Recombinant B. hermsii genomic library and screening.
A genomic library in the plasmid pUC18 with inserts of 4 to 8 kb of B. hermsii strain HS1 DNA was described previously and was stored frozen in 50% glycerol in Luria-Bertani broth at −20°C in 384-well microtiter plates (42). Colony hybridization with labeled DNA probes was carried out as described previously (54).
PCRs.
PCRs were carried out in Roche PCR buffer with 1.5 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate, 3 units Taq polymerase (Roche Diagnostics), and 0.1 μM (each) of forward and reverse primers. The forward and reverse primers for amplification were as follows: 5′-GTACGGATCCTATGCACGATCATGTAAAGGC-3′ and 5′-GCTGCTGCAGTTATAAATTAAGTTTTTCTTTTAACCT-3′ for B. hermsii thyX; 5′-GTACGGATCCTTTGAATAAAGAATATAAAATTTTG-3′ and 5′-GCACCTGCAGTACTAATACACACTTACAAT-3′ for B. burgdorferi thyX (accession number AE000790); 5′-GACGCATATGATGAAACAGTATTTAGAACT-3′ and 5′-GACGCATATGTTAGATAGCCACCGGCGCTT-3′ for E. coli K-12 thyA (U00096); 5′-GTTGATTTCATCTGTAAGTTGCTCAATT-3′ and 5′-ACTTGCTGTTCAATCTGGTAATGG-3′ for B. hermsii flaB; 5′-AGCTAAGAGTAATGATGGCAAT-3′ and 5′-ATTTATCACCTTTAGCCATTCT-3′ for the 28-kb linear plasmid lp7E of B. hermsii (Z11876); and 5′-CAGATGGTCTTACTGCTGAAGC-3′ and 5′-CAGCAACAACCTTTTCCTTTAG-3′ for the 53-kb linear plasmid lp53 of B. hermsii (L24911). The PCR conditions were as follows: 1 cycle of 95°C for 4 min; 40 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min; and 1 cycle of 72°C for 7 min. For use as probes, the PCR products were labeled with [α-32P]dATP by the random priming method (Amersham Biosciences). For cloning of the fragments, the PCR products were ligated into the TOPO TA cloning vector, and the products were transformed into E. coli TOP10 cells (Invitrogen).
Pulsed-field gel electrophoresis and Southern blot hybridization.
Unsheared genomic DNAs were prepared in agarose plugs as described previously (18). In brief, 109 B. burgdorferi cells from a culture or 109 B. hermsii cells freshly harvested from mouse plasma were embedded in 80 μl 1% (wt/vol) SeaPlaque low-melting-temperature agarose (BioWhittaker Molecular Applications, Rockland, Maine). The agarose plugs were incubated for 24 h in 150 mM NaCl-50 mM Tris-HCl (pH 8.0) buffer with 1% sodium dodecyl sulfate and 1 mg/ml proteinase K at 50°C and then were washed with 10 mM Tris-HCl-1 mM EDTA (pH 8.0) buffer three times for 30 min each at 20°C. Electrophoresis in 1% GenePure LE agarose (ISC BioExpress, Kaysville, Utah) in a CHEF Mapper apparatus (Bio-Rad, Hercules, Calif.) was carried out in 45 mM Tris-45 mM borate-1 mM EDTA (pH 8.3) buffer at 4°C at 6 V/cm, with a pulsed-field angle of 120° and switch times that were linearly ramped between 0.35 and 89 s over 18 h; the gels were stained with ethidium bromide. Southern blot analysis was carried out as described previously (54).
DNA sequencing and analysis.
Sequence analysis over both strands was carried out by using custom oligonucleotides and a CEQ 8000 automated DNA sequencer (Beckman Coulter, Fullerton, CA) as described previously (54). Nucleotide and protein databases were searched with the BLASTX, BLASTP, and TBLASTX algorithms (31). A sequence of 13,218 bp containing the thyX, nrdI, nrdE, and nrdF genes, as well as their flanking regions, of B. hermsii strain HS1 was obtained. For phylogenetic reconstruction, the homologous thyX, nrdE, and nrdF sequences of representative bacteria, viruses, or eukaryotes for which genome sequences were available, as well as selected organisms without full genome sequences, were obtained. The accession numbers of the sequences are given in the figure legends. Amino acid sequences were aligned using Clustal X, version 1.83, and this alignment (http://spiro.mmg.uci.edu/data) was the basis for a codon-based, gapped nucleotide alignment produced with CodonAlign, version 2.0 (http://www.sinauer.com/hall/index.php). Alignments of concatenated sequences were then edited manually with MacClade, version 4.06 (Sinauer Associates). To minimize effects of base composition bias across taxa, the third positions were excluded, and the evolutionary model of Galtier and Gouy was applied (21) using PHYLO_WIN phylogenetic analysis software (http://pbil.univ-lyon1.fr/software/phylowin.html). Positions with gaps were ignored. Phylograms showing nodes with bootstrap support values under distance (neighbor-joining) and maximum likelihood criteria were constructed.
Quantitative reverse transcriptase PCR.
B. hermsii cells in logarithmic growth phase were harvested and resuspended at a density of 3 × 106 cells per ml of BSK or 1.0 × 107 cells per ml of SDMEM. In the case of SDMEM, the cells were washed twice in SDMEM to minimize carryover of BSK components. The cell suspensions were incubated at 37°C for 20 h and then harvested as described above. Total RNA was immediately extracted from the pellets, using TriReagent (Molecular Research Center, Cincinnati, OH). The RNAs were treated twice with 2 units of RNase-free DNase I (Ambion, Austin, TX) at 37°C for 30 min and then reverse transcribed with random hexamers and a Transcriptor First Strand cDNA synthesis kit (Roche Diagnostics). Reactions with 200 ng substrate RNA were carried out in a 20-μl volume containing 50 mM Tris (pH 8.5)-30 mM KCl-8 mM MgCl2 with a 1 mM concentration of each deoxynucleotide, 20 units RNase inhibitor, 60 μM random hexamers, and 10 units reverse transcriptase at 25°C for 10 min, followed by 50°C for 1 h. As a measure of DNA contamination in each sample, the reverse transcriptase was omitted in matched reactions. Real-time quantitative PCRs with the cDNA or plasmid DNA samples were carried out using a Rotor-Gene RG-3000 apparatus (Corbett Research, San Francisco, CA). TaqMan minor-groove binding probes were obtained from Applied Biosystems, Foster City, CA. The VIC-labeled probe, forward primer, and reverse primer for the B. hermsii thyX gene were 5′-TCATGTTAAAGCTCC-3′, 5′-CTGGTCTTATTGACTATTTGATTCGAA-3′, and 5′-CATCCATTGCCTTGCAACAA-3′, respectively. The forward and reverse primers for the flaB gene of B. hermsii were those described above; the 6-carboxyfluorescein (FAM)-labeled probe for flaB was 5′-AACCTCTGTCTGCATC-3′. PCRs were carried out in triplicate in 25-μl volumes with quantitative PCR Master Mix Plus containing HotGoldStar DNA polymerase (Eurogentec, San Diego, CA) and a 0.2 μM concentration of each primer and probe. The PCR conditions were as follows: 94°C for 10 min and then 50 cycles of 15 s at 94°C followed by 1 min at 58°C. There were four replicates of each growth condition, and from each tube three cDNA reactions were carried out. The PCR results for the three cDNA samples were then averaged to yield a single value for each of the replicates. In a preliminary study, both PCR assays were negative with B. burgdorferi RNA.
Complementation assay.
The full-length coding sequences of the thyX genes of B. hermsii and B. burgdorferi were amplified by PCR as described above and ligated between the PstI and BamHI sites of pUC18. The thyA gene of E. coli K-12 was similarly amplified and ligated into pUC18 at the NdeI site. A plasmid with the Lactobacillus casei thyA gene cloned into the PstI and BamHI sites of pUC18 was provided by D. V. Santi of the University of California, San Francisco (14). The different plasmid constructs were then transformed into the thyA mutant strain χ2913 and plated on Luria-Bertani agar medium with 50 μg/ml carbenicillin and 50 μg/ml thymidine (Sigma, St. Louis, MO) (35). The different transformants were grown overnight in M9 minimal medium with 50 μg/ml thymidine and 50 μg/ml carbenicillin and then washed three times with unsupplemented M9 minimal medium before inoculation at a final concentration of 5 × 106 cells per ml into M9 minimal medium with carbenicillin alone. Cultures were incubated in duplicate at 37°C on a rotating shaker set at 250 rpm, and cell growth was measured spectrophotometrically by changes in transmittance, as measured by the optical density at 600 nm.
In vitro thymidylate synthase assay.
Full-length thyX genes were amplified by PCR, using the conditions described above and the following forward and reverse primers: 5′-CGGGATCCATGCACGATCATGTAAAG-3′ and 5′-AACTGCAGTTATAAATTAAGTTTTTC-3′ for B. hermsii and 5′-CGGGATCCATGAATAAAGAATATAA-3′ and 5′-GGGGTACCTCAATCAATTCCTAGCTT-3′ for B. burgdorferi. The products were ligated between the BamHI and KpnI sites for B. burgdorferi thyX and the BamHI and PstI sites for B. hermsii thyX in the His tag-encoding plasmid vector pQE80L (QIAGEN Science) and transformed into E. coli SURE competent cells. The transformants were grown in Luria-Bertani medium at 37°C, and recombinant protein expression was induced when the culture reached an optical density at 600 nm of 0.4 by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM. After another 2 h of cultivation, the cells were harvested by centrifugation at 4,300 × g for 20 min at 4°C. His-tagged proteins were purified by Ni-nitrilotriacetic acid affinity chromatography (QIAGEN Science) as described previously (35). The thymidylate synthase activity of the recombinant proteins was measured by using tritiated dUMP as the substrate, as described by Leduc et al. (29). The reaction was performed at 37°C for 30 min in a total volume of 50 μl of 50 mM HEPES (pH 7.5)-10 mM MgCl2-10% glycerol-0.5 mM methylenetetrahydrofolate (CH2H4 folate; Merck Eprova, Switzerland)-1.5 mM FAD-1.2 mM β-NADPH-0.6 mM β-NADH-20 μM dUMP (Sigma). The amounts of His-tagged ThyX proteins in the reaction were 1.3 μg for B. hermsii and 3 μg for B. burgdorferi, and the specific activities of [5-3H]dUMP (Moravek Biochemicals, Brea, CA) were 0.15 Ci/mmol for B. hermsii ThyX and 0.3 Ci/mmol for B. burgdorferi ThyX. Reactions were terminated by the addition of activated charcoal at 10% (wt/vol) and 2% trichloroacetic acid. Released 3H ions in the supernatant were measured by scintillation counting as previously described (29).
Nucleotide sequence accession number.
The sequence of 13,218 bp containing the thyX, nrdI, nrdE, and nrdF genes, as well as their flanking regions, of B. hermsii strain HS1 was assigned GenBank accession number AY623744.
RESULTS
B. hermsii thyX locus and adjacent genes.
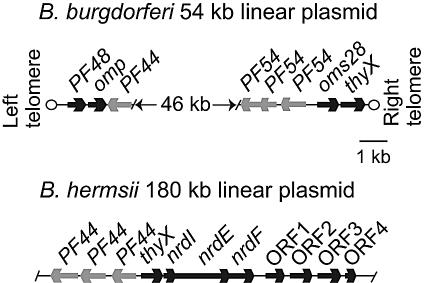
We amplified by PCR the B. burgdorferi thyX gene from total DNA and used it in turn to probe a genomic library of B. hermsii. Three hybridizing clones were isolated, and their inserts were sequenced. The overlapping fragments comprised a single sequence of 13,218 bp with 11 open reading frames (ORFs) of at least 300 nucleotides on the two strands, as well as incomplete ORFs at each end of the 13.2-kb fragment. A physical map of the B. hermsii sequence along with selected regions of the 54-kb linear plasmid of B. burgdorferi (19) is shown in Fig. 1. The overall GC content of the combined sequence was 32%; there were no direct or inverted repeats of 10 or more nucleotides.
FIG. 1.
Physical maps of the thyX locus and flanking regions of the 180-kb linear plasmid of Borrelia hermsii and the 54-kb linear plasmid of Borrelia burgdorferi. The solid arrows correspond to the lengths of the open reading frames and the presumed direction of transcription. PF numbers indicate the paralogous family designations for B. burgdorferi genes (13); the product of omp is a hypothetical outer membrane protein.
Possible functions were provisionally assigned to 4 of the 11 ORFs of B. hermsii, namely, thyX, nrdI, nrdE, and nrdF, on the basis of sequence identity and similarity. They are located on the same strand and in the middle of the fragment (Fig. 1). At the left end of the fragment (not shown) is a partial ORF that is similar to the portal protein genes of bacteriophages of firmicutes and Fusobacterium nucleatum (protein family 05133 [http://www.sanger.ac.uk/Software/Pfam/]). Upstream of thyX, on the opposite strand, are three ORFs with 36 to 41% pairwise amino acid identities with each other and with 31 to 33% identities with the “S2” proteins of LB Borrelia spp. (17). Genes for S2 in B. burgdorferi B31 are located on the lp54 plasmid, the same plasmid bearing thyX in the opposite orientation, but in B. burgdorferi, thyX and the S2 gene are ∼50 kb apart. The S2-like genes of LB Borrelia spp. were assigned to paralogous family number 44 (13).
The thyX orthologue of B. hermsii is 75% identical in nucleotide sequence to that of B. burgdorferi. The GC contents of the two genes are 32 to 34%, which is similar to the overall GC content of B. burgdorferi (13, 19). In contrast to the thyX gene of B. burgdorferi, which is a terminal ORF on the 54-kb linear plasmid (19; J. Aron and S. Casjens, personal communication), the thyX gene of B. hermsii is 30 bp upstream of three other closely spaced or overlapping ORFs on the same strand, with a GC content of ∼33%. The putative gene products are components of both the pyrimidine biosynthesis and salvage pathways: they are the alpha (NrdE) and beta (NrdF) subunits of ribonucleoside diphosphate reductase and an associated ribonucleoprotein (NrdI) in the complex. The nrdI, nrdE, and nrdF genes were not reported for the genomes of B. burgdorferi and B. garinii (19, 23). This was confirmed for this study by TBLASTX searches of the deposited chromosome and plasmid sequences of B. burgdorferi and B. garinii, using the nucleotide sequences of the nrd genes. The gene order of thyX-nrdI-nrdE-nrdF was only identified for B. hermsii. The codon usages and GC skews of the thyX and nrdIEF genes were similar to the corresponding values calculated for the remaining sequence of the 13-kb fragment. There were consensus ribosomal binding sequences preceding the thyX, nrdI, nrdE, and nrdF genes, but a consensus σ70-type, σ54-type, or σ38-type promoter was not discernible within 150 nucleotides upstream of the thyX start codon.
At the right end of the fragment and following the nrdF gene on the same strand are three complete ORFs and one partial ORF. The three complete ORFs have ∼30% pairwise identity with each other over their aligned lengths, with BLASTP E values of <10−7, but they are unlike any other protein in the database (E value, >0.1). The partial ORF at the end also appears to be homologous to this family of hypothetical proteins (E value, <10−4).
Location of B. hermsii thyX gene.
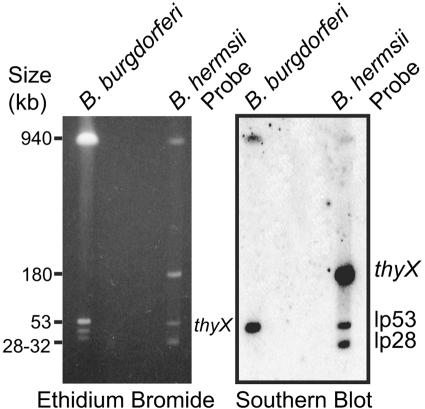
To identify the location of the B. hermsii thyX gene, we separated the linear chromosome and several linear and circular plasmids by pulsed-field gel electrophoresis and carried out Southern blot analysis. The chromosome and plasmids of B. burgdorferi were also included in the gel. The left panel of Fig. 2 shows the ethidium bromide-stained gel. There were major bands of approximately 920, 180, 53, and 30 kb, as expected, for B. hermsii and 920, 54, 32, and 28 kb for B. burgdorferi (18). The blot of this gel was sequentially probed first with the labeled, PCR-amplified thyX gene of B. hermsii and then with similarly prepared probes for a 28-kb linear plasmid and the 53-kb linear plasmid of B. hermsii. The thyX probe bound to the 180-kb plasmid of B. hermsii and, by cross-hybridization, to the 54-kb plasmid of B. burgdorferi. The two other probes that were subsequently hybridized with the blot bound as expected to the 28-kb and 53-kb plasmids in the gel.
FIG. 2.
Southern blot analysis of replicons of B. hermsii and B. burgdorferi separated by pulsed-field gel electrophoresis and sequentially hybridized with radiolabeled probes for thyX, the lp53 linear plasmid, and the lp28 linear plasmid of B. hermsii. An ethidium bromide-stained gel is shown on the left, and an autoradiograph is shown on the right. Sizes (in kb) of linear plasmids of B. hermsii and of size standards are indicated.
Phylogenetic analyses.
We compared the thyX, nrdE, and nrdF sequences of B. hermsii with a selection of corresponding orthologues from other organisms that represented the diversity of these genes. The nucleotide sequences were aligned on a codon basis with the corresponding B. hermsii sequences. The nrdE and nrdF sequences are tandemly arrayed in all the selected organisms used for the comparison, and accordingly, the aligned sequences of these genes were concatenated for the analysis. To adjust for differences in base composition across the taxa, we excluded the third position of each aligned codon and used the algorithm of Galtier and Gouy (21).
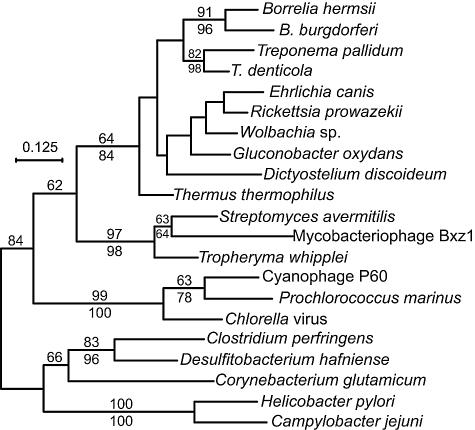
Figure 3 shows a phylogram of the thyX genes. Although the thyA gene, much studied in the gamma-proteobacterium E. coli and the firmicute B. subtilis, is considered the usual thymidylate synthase gene among bacteria, the cumulative bacterial genomes sequenced to date indicate that the thyX gene is at least as widely distributed as thyA among bacterial phyla. The figure shows that the B. hermsii and B. burgdorferi genes are monophyletic. There is moderate support for a node with two other spirochetes, T. pallidum and T. denticola, besides the two Borrelia species, and for a deeper node encompassing the spirochetes, some alpha-proteobacteria, Dictyostelium discoideum, and the thermophilic bacterium Thermus thermophilus. The borrelial genes are distant from the thyX genes of the epsilon-proteobacteria H. pylori and Campylobacter jejuni as well as those of some clostridia and the actinobacterium Corynebacterium glutamicum.
FIG. 3.
Phylogram of codon-aligned nucleotide sequences of thyX genes from the following organisms (with accession numbers): Borrelia burgdorferi (NC_001857), B. hermsii (AY623744), Campylobacter jejuni (NC_003912), Chlorella virus (NC_000852), Clostridium perfringens (BA000016), Corynebacterium glutamicum (BX927153), cyanophage P60 (AF338467), Desulfitobacterium hafniense (NZ_AAAW03000212), Dictyostelium discoideum (M27713), Ehrlichia canis (NZ_AAEJ01000001), Gluconobacter oxydans (NC_006677), Helicobacter pylori (NC_000921), mycobacteriophage Bxz1 (AY129337), Prochlorococcus marinus (AE017161), Rickettsia prowazekii (NC_000963), Streptomyces avermitilis (BA000030), Thermus thermophilus (NC_005835), Treponema denticola (NC_002967), T. pallidum (AE001268), Tropheryma whipplei (AE016852), and a Wolbachia endosymbiont of Drosophila melanogaster (NC_002978). The third position of each codon and positions with gaps were excluded from the analysis. Nodes with bootstrap values with >61% support by maximum likelihood (100 replicates; shown above the line) or neighbor-joining (1,000 replicates; shown below the line) distance criteria are indicated. Bar, nucleotide distance.
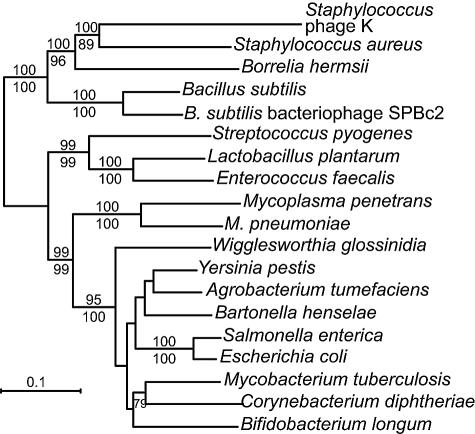
In contrast to the thyX genes of B. hermsii and B. burgdorferi, the nrdEF genes of B. hermsii clustered with nrdEF genes of the firmicutes Staphylococcus and Bacillus and some of their viruses (Fig. 4). The same clustering was noted in phylograms of nrdE and nrdF genes examined separately (data not shown). The arthropod-borne Ehrlichia, Rickettsia, and Wolbachia species whose genomes have been sequenced to date do not have nrdEF orthologues. Agrobacterium tumefaciens and Bartonella henselae, the alpha-proteobacteria known to have nrdEF genes, are represented in a more distant lineage which is shared with gamma-proteobacteria and actinobacteria.
FIG. 4.
Phylogram of codon-aligned concatenated nucleotide sequences of nrdE and nrdF genes from the following bacteria and viruses (with accession numbers): Agrobacterium tumefaciens (AE008981), Bacillus subtilis (NC_000964), B. subtilis bacteriophage SPBc2 (AF020713), Bartonella henselae (BX897699), Bifidobacterium longum (AE014295), Borrelia hermsii (AY623744), Corynebacterium diphtheriae (NC_002935), Enterococcus faecalis (NC_004668), Escherichia coli (U00096), Lactobacillus plantarum (NC_004567), Mycobacterium tuberculosis (NC_000962), Mycoplasma penetrans (BA000026), Mycoplasma pneumoniae (AE000050), Salmonella enterica (NC_006511), Staphylococcus aureus (NC_002758), Staphylococcus phage K (NC_005880), Streptococcus pyogenes (AE010059), Wigglesworthia glossinidia (BA000021), and Yersinia pestis (AE017136). Nucleotide positions with gaps and the third base of each codon in the aligned sequences were excluded from the analysis. Bootstrap values of nodes with >71% support by maximum likelihood (100 replicates; shown above the line) or neighbor-joining (1,000 replicates; shown below the line) distance criteria are indicated. Bar, nucleotide distance.
Expression of B. hermsii thyX gene.
Expression of the B. hermsii thyX gene was examined under the following two in vitro conditions: complete BSK medium, which includes supplemental pyrimidines and purines as well as rabbit serum (3), and SDMEM, which contained bovine serum albumin, gelatin, and N-acetylglucosamine but not serum and was based on Dulbecco's modified Eagle's medium, which lacks purines or pyrimidines. The lack of purines in SDMEM was confirmed by the inability of an E. coli purA mutant strain, H1238 (purA54 thr fhuA argF relA spoT argI), to grow in the medium (data not shown). Transcription of thyX and the constitutively expressed flaB gene was measured with probe-based real-time PCR of cDNAs produced with random hexamers; copy numbers were estimated from a standard curve produced with cloned thyX and flaB genes of B. hermsii.
BSK medium supported a threefold increase in cell density over 20 h. There was no detectable growth of the spirochetes in SDMEM, and both suspensions were harvested at cell densities of 107 per ml. In the absence of reverse transcriptase, there was no detectable PCR product when thyX and flaB primers and probes were used, but there were products with both reactions when reverse transcriptase was added. By quantitative PCR, the mean (95% confidence intervals) ratio of thyX to flaB cDNA was 0.020 (0.017 to 0.024) in SDMEM and 0.022 (0.019 to 0.025) in BSK medium (P = 0.4 by a two-tailed t test). The experiment showed that thyX was transcribed under two different in vitro conditions, but in smaller amounts than the constitutively expressed flaB gene (9, 37).
Complementation of thyA mutation.
A thyA mutant of E. coli was transformed with expression plasmids encoding the thyX gene of B. hermsii or B. burgdorferi and, as positive controls, the thyA genes of E. coli and L. casei. E. coli transformed with the plasmid vector without an insert served as the negative control. The thyA mutant host had previously been used to demonstrate complementation by the thyX gene of H. pylori (35). In a preliminary study, expression of the recombinant ThyX protein was detectable and measurable by Coomassie blue staining of sodium dodecyl sulfate-polyacrylamide gels and was similar in quantity for E. coli transformants with either the B. hermsii or B. burgdorferi thyX gene (data not shown).
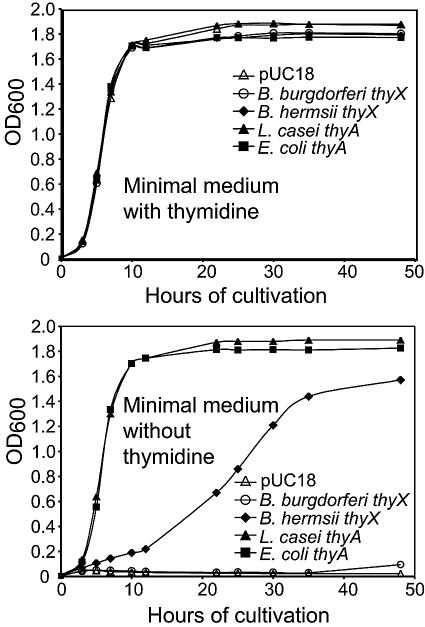
All transformants were grown in minimal medium with or without thymidine, and then their growth was monitored by a spectrophotometer. Figure 5 shows the growth of the different isolates under nonselective and selective conditions. In the presence of supplemental thymidine, all isolates grew at approximately the same rate. In the absence of thymidine, E. coli cells with the B. burgdorferi thyX gene, like E. coli with the plasmid vector alone, did not detectably grow under either aerobic (Fig. 5) or anaerobic conditions (data not shown). In contrast, E. coli cells with the B. hermsii thyX gene grew in the absence of thymidine, albeit with a longer generation time (∼10 h) than cells bearing the thyA gene of either E. coli or L. casei, which had generation times of ∼2 h. By 50 h of incubation of the broth cultures, the average cell density of B. hermsii thyX-expressing cells was 86% as high as the densities of L. casei thyA- and E. coli thyA-expressing cells (Fig. 5).
FIG. 5.
Growth in the presence and absence of thymidine of a thyA mutant of Escherichia coli transformed with the plasmid vector pUC18 without an insert (open triangles) or with an insert of thyA of E. coli (solid squares), thyA of Lactobacillus casei (solid triangles), thyX of Borrelia burgdorferi (open circles), or thyX of B. hermsii (solid diamonds). Growth was determined by measurement of the optical density at 600 nm by a spectrophotometer.
Thymidylate synthase activity.
The failure of the B. burgdorferi thyX gene to complement the thyA mutation in E. coli was further investigated by purifying the recombinant gene products and subjecting the proteins to an assay for thymidylate synthase activity (35). The amount of protein expression in E. coli SURE cells and the purification yields of the ThyX proteins of B. hermsii and B. burgdorferi were comparable, as assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). The assay's end point was the amount of free 3H ions in the supernatant after precipitation of dUMP and dTMP with trichloroacetic acid. The results are shown in Fig. 6. The B. hermsii thyX gene product had thymidylate synthase activity as high as 2.51 pmol 3H+/μg/min, and its activity remained linear for at least 20 min. This specific activity was approximately twofold higher than that achieved with purified ThyX of H. pylori in the same assay (35). In contrast, the in vitro activity of the B. burgdorferi protein was barely detectable above the background (Fig. 6).
FIG. 6.
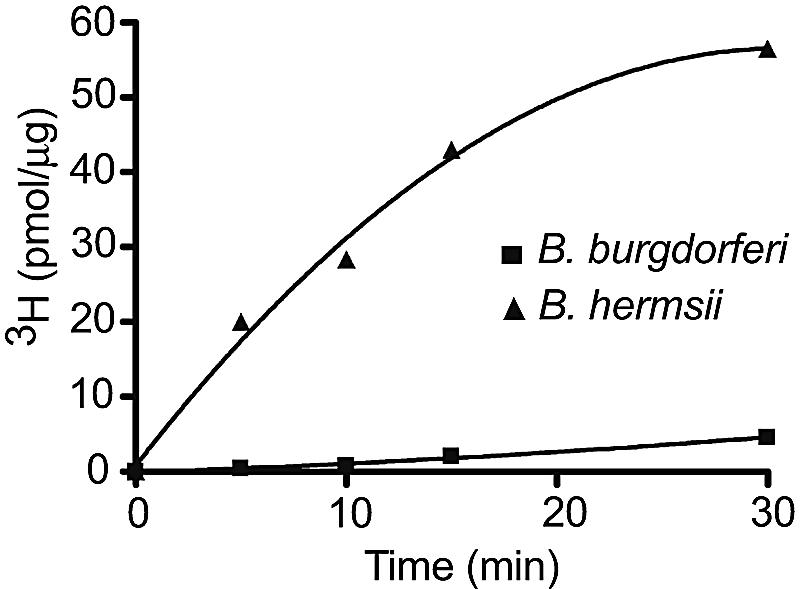
In vitro thymidylate synthase activities of purified recombinant ThyX proteins of B. hermsii and B. burgdorferi. Activity was measured as released 3H ions, in pmol/μg (y axis), from the deprotonation of C-5 of the dUMP ring over time (x axis).
Alignment of deduced proteins.
Partial amino acid sequences of the gene products of the thyX orthologues of B. burgdorferi, B. garinii, and B. hermsii as well as of the ThyX proteins of other selected organisms are shown in Fig. 7. All the Borrelia sp. proteins have the ThyX motif of RHRX7S (positions 90 to 100 of B. hermsii ThyX) that is common to these proteins at their conserved amino-terminal domain (30, 35). The gene products of B. hermsii and B. burgdorferi share the following residues in common with other ThyX orthologues (B. hermsii ThyX numbering): H65, E70, F76, R90, H91, R92, S100, Y103, E168, R171, L174, P175, L197, R198, E206, R208, A211, and P223 (29). However, in the B. burgdorferi and B. garinii ThyX proteins, a cysteine residue substitutes for the conserved arginine at position 102 in B. hermsii ThyX, at position 86 in H. pylori ThyX (29), and at comparable positions in the ThyX proteins of other organisms, including Treponema spp.
FIG. 7.
Amino acid alignment of partial ThyX proteins of the following organisms (with accession numbers): Borrelia burgdorferi (NP_045749), B. garinii (CP000015), B. hermsii (AAT68707), Chlorella virus (NP_049030), cyanophage P60 (NP_570363), Dictyostelium discoideum (XP_641335), Ehrlichia canis (ZP_00211002), Helicobacter pylori (NP_224139), Magnetococcus sp. (ZP_00605242), mycobacteriophage Bxz1 (NP_818164), Prochlorococcus marinus (NP_874667), Rickettsia prowazekii (NP_220685), Streptomyces avermitilis (NP_823693), Thermotoga maritima (NP_228259), Thermus thermophilus (YP_004706), Treponema denticola (NP_970925), T. pallidum (AAC65955), Tropheryma whipplei (NP_787744), and a Wolbachia endosymbiont of Drosophila melanogaster (NP_966910). Numbers indicate the positions of the amino acids aligned according to the B. burgdorferi sequence. The consensus amino acids are shown at the bottom. The cysteine-for-arginine substitution in B. burgdorferi and B. garinii at position 91 is highlighted in gray.
DISCUSSION
The thyX gene was first discovered in the eukaryote D. discoideum (15), but the great majority of subsequent examples of thyX have been described for bacterial species or their viruses (30, 34). For some of these species, the thyX gene product's function as a thymidylate synthase was demonstrated by genetic complementation and/or a biochemical assay (16, 22, 24, 25, 33, 35), but in most cases the thyX orthologues in bacterial genomes were identified on the basis of sequence identity alone. The latter situation was the case for a thyX-like sequence in B. burgdorferi's genome (19) as well as for similar sequences identified in the genomes of T. pallidum (20) and T. denticola (51). Using a B. burgdorferi genome array hybridized with DNA from B. hermsii, we obtained evidence that the RF agent has a thyX gene as well (54). Further microarray experiments indicated that the thyX gene of B. burgdorferi and the putative thyX gene of B. hermsii were transcribed both in vitro and in vivo (37, 54).
For the present study, we confirmed that B. hermsii has a thyX orthologue by identifying it in a genomic library and then sequencing the gene and ∼13 kb of its flanking regions. As was the case for B. burgdorferi and B. garinii, the thyX gene of B. hermsii was carried on a plasmid, but in the RF species it was carried on the 180-kb linear plasmid instead of the 54- to 56-kb plasmids. The thyX gene is the first gene located so far on these large linear plasmids of B. hermsii and other RF species (18). Upstream of the B. hermsii thyX gene are three ORFs that are homologous to the PF44 genes, which are also located on the lp54 plasmid of B. burgdorferi. This is an indication that at least part of the 180-kb linear plasmid of RF species has a common lineage with the lp54-type plasmids of LB species.
Unlike the B. burgdorferi thyX gene, which is the rightmost ORF on the linear plasmid, the B. hermsii thyX gene was flanked on its right by several other ORFs, none of which were similar to sequences in the B. burgdorferi or B. garinii genome (Fig. 1). These additional genes included what may be an operon comprising the nrdI, nrdE, and nrdF genes. The nrdE and nrdF genes encode the large and small subunits, respectively, of ribonucleoside diphosphate reductase, an enzyme that catalyzes the de novo synthesis of deoxyribonucleotides required for DNA replication and repair (43). The nrdI gene encodes a small protein that is also part of the enzyme complex (53). On the basis of their deduced amino acid sequences, the NrdI, NrdE, and NrdF proteins of B. hermsii most closely resemble components of the class Ib ribonucleoside diphosphate reductase complex of bacteria (26-28). To investigate the function of the nrdEF genes of B. hermsii, they were cloned in tandem and expressed in E. coli, but the transformant was not able to detectably complement a nrdAB mutation in the host strain (unpublished findings). The gene products of nrdAB and nrdEF are in two different classes of ribonucleoside diphosphate reductase enzymes with limited sequence similarity (28), and it is possible that the B. hermsii proteins were not functional in the E. coli enzyme complex.
Using quantitative reverse transcriptase PCR assays with specific primers and probes for thyX and flaB, we confirmed our previous microarray result that the thyX gene of B. hermsii was expressed (54). In the present study, this was shown during in vitro cultivation in both complete medium, which provides a source of purines and pyrimidines, and a more defined medium lacking purines and pyrimidines. The expression of thyX of B. hermsii may be constitutive: under both conditions, the relative level of expression of the B. hermsii thyX gene was about 1/50 that of flaB, which is a highly expressed gene that encodes a major structural protein of spirochetal flagella (54).
Unlike the ribonucleoside diphosphate reductase complex, which may be dependent on proper interactions of the different subunits for full or even partial enzyme activity, the single polypeptide of ThyX appears to be sufficient for thymidylate synthase activity in complementation and in vitro assays in studies of other organisms (28). Accordingly, we examined the activities of the recombinant ThyX proteins of B. burgdorferi and B. hermsii in providing complementation of a thyA mutant of E. coli and then in an in vitro assay of thymidylate synthase activity. Although the ThyA thymidylate synthase is not discernibly homologous to ThyX, recombinant ThyX proteins of H. pylori and C. jejuni complemented thyA mutations in E. coli in other studies (22, 35). Here we showed that recombinant ThyX of B. hermsii had thymidylate synthase activity in both types of experiments. In contrast, recombinant ThyX of B. burgdorferi had very weak activity in the in vitro assay and was not capable of substituting for ThyA in an E. coli mutant.
Although B. burgdorferi ThyX appeared to be well expressed in E. coli, it is possible that the recombinant protein's lack of activity was attributable to misfolding or other posttranslational events that inexplicably did not affect the recombinant protein of B. hermsii under similar conditions. A more plausible explanation for the outcomes of the function experiments was found in the deduced amino acid sequence of the B. burgdorferi polypeptide. While the ThyX protein of B. burgdorferi, as well as that of B. garinii, has the motif RHRX7S that serves to define ThyX proteins (35), there is a substitution of a cysteine for the highly conserved arginine at position 91 in the B. burgdorferi and B. garinii proteins (Fig. 7). This arginine corresponds to an arginine at position 90 of Thermotoga maritima ThyX (33) and position 86 of H. pylori ThyX (29). Crystallographic studies of the T. maritima ThyX protein suggest that hydrogen bonding occurs between this arginine and the dUMP substrate (33). We propose that replacement of a charged side chain-containing amino acid with an uncharged one might abolish the interaction between the ThyX protein and the dUMP substrate. It is possible that the LB ThyX proteins have functional thymidylate synthase activity in spirochetes, but they also may have other roles in cells or be examples of plasmid genes that are undergoing degradation in Borrelia spp. (13). Whatever its role, if any, the thyX gene of B. burgdorferi is not essential for in vitro growth, at least under usual laboratory conditions: an isolate of B. burgdorferi strain B31 without the lp54 plasmid or its genes grew at the same rate as wild-type B31 in broth medium (48).
The nrdIEF genes were adjacent to the thyX gene on the 180-kb linear plasmid of B. hermsii, which suggests a common origin for these genes involved in pyrimidine metabolism. But we propose that the nrdIEF cluster, and perhaps other genes on the 180-kb plasmid, were acquired by a Borrelia ancestor at a time other than that of the acquisition of the thyX gene. The thyX genes of Borrelia spp. are in what may be a monophyletic cluster containing treponemes as well as some alpha-proteobacteria, notably some arthropod-borne species of Ehrlichia, Rickettsia, and Wolbachia (Fig. 3). None of these other species, including T. pallidum and T. denticola, have discernible orthologues of the nrdE or nrdF gene in their genomes. On the other hand, the nrdEF genes of B. hermsii are in a monophyletic cluster with species and their bacteriophages of the Bacillales group of Firmicutes, most or all of which have thyA genes and not thyX genes providing thymidylate synthase function to the cells (Fig. 4). Although the phylogenetic evidence of separate events for the acquisition of thyX and nrdIEF genes is compelling, we cannot at this time distinguish between selective acquisition by RF Borrelia spp. of the nrdIEF genes and selective loss of this operon by an ancestor of LB Borrelia spp. after the divergence of the RF group.
The serial propagation of Borrelia species outside their arthropod and vertebrate hosts requires a complex growth medium such as BSK, which is based on a tissue culture medium containing deoxyribonucleotides as well as amino acids and vitamins (3, 52). Besides possessing a thyX gene, B. burgdorferi is known to have some genes that are capable of interconversion of pyrimidine and purine intermediates, such as uridine kinase (udk) (8), cytidylate kinase (cmk), nucleoside diphosphate kinase (ndk), thymidylate kinase (tmk), and thymidine kinase (tdk) (1, 19, 20). On the basis of the cross-species hybridization study of B. hermsii with a B. burgdorferi array, we concluded that B. hermsii has cmk, ndk, tmk, and tdk genes. From the results of the hybridization study, we could not detect the presence of a udk gene in B. hermsii using genomic DNA or cDNAs as probes, but evidence of a udk gene in the B. hermsii chromosome was found in its sequence (T. Schwan et al., unpublished data).
Figure 8 summarizes partial pathways for dTTP and dCTP biosynthesis, as well as our inferences about the presence of the genes for these pathways in B. burgdorferi and B. hermsii. B. hermsii apparently has all the genes for these pathways, while B. burgdorferi, lacking nrdEF genes and a functional thyX gene, does not. The LB agents B. burgdorferi and B. garinii would be unable to convert UDP to dUDP and then dUMP to dTMP. While both groups of Borrelia pathogens presumably could use their thymidine kinases in a salvage pathway to convert thymidine provided by their arthropod and vertebrate hosts to dTMP, only B. hermsii, and possibly other RF species, appears to be fully capable of synthesizing pyrimidines de novo. These supplemental capabilities in B. hermsii may permit greater deoxynucleoside triphosphate synthesis when only low concentrations of precursors are present in a host and, consequently, higher peak densities in the blood. Although there may be additional or alternative explanations for the differences between RF and LB spirochetes in the burdens in the blood of their hosts, the greater capability of B. hermsii for pyrimidine and purine biosynthesis may be sufficient.
FIG. 8.
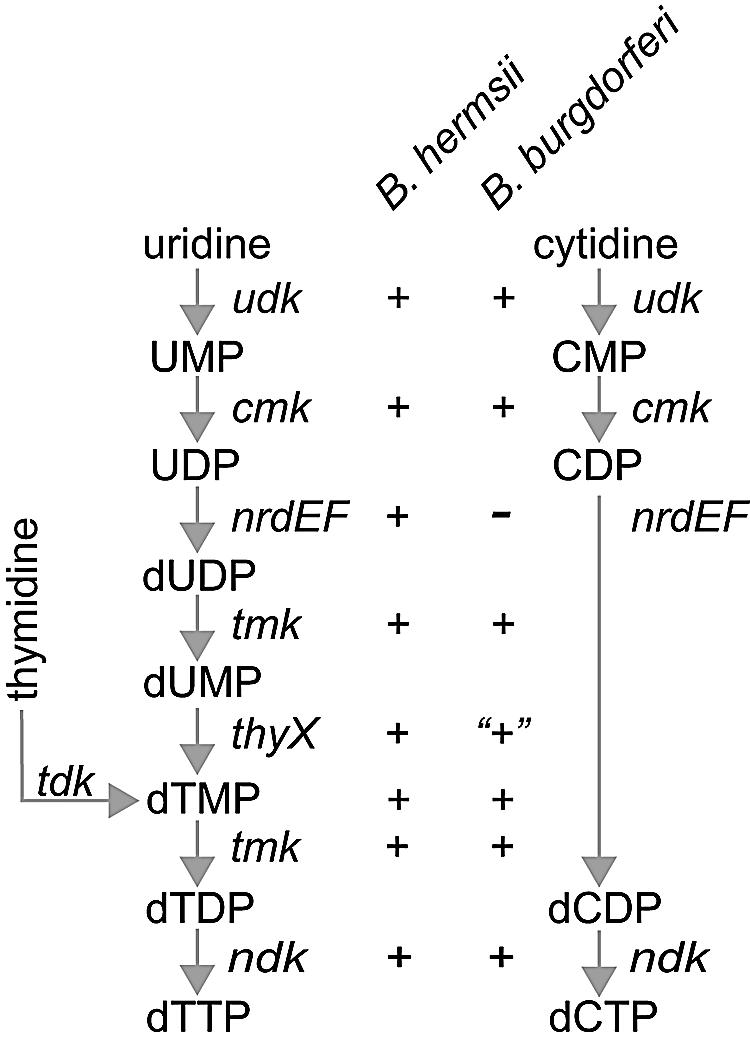
Genes for pyrimidine salvage pathways for synthesis of dTTP and dCTP and their inferred presence in (+) or absence from (−) Borrelia burgdorferi and B. hermsii. The genes are uridine kinase (udk; EC 2.7.1.48), cytidylate kinase (cmk; EC 2.7.4.14), ribonucleoside diphosphate reductase (nrdEF; EC 1.17.4.1), thymidylate kinase (tmk; EC 2.7.4.9), thymidine kinase (tdk; EC 2.7.1.21), nucleoside diphosphate kinase (ndk; EC 2.7.4.6), and thymidylate synthase (thyX; EC 2.1.1.148). The lack of function of the B. burgdorferi thyX gene product in complementation and in vitro assays is indicated by quotation marks.
Acknowledgments
We thank Daniel Santi for providing the E. coli strain; Sherwood Casjens, John Aron, and Tom Schwan for sharing their results before publication; and Hung Le, April Phillips, and Hany Mattaous for assistance with experiments.
This work was supported by NIH grant AI24424 to A.G.B. H.M. is supported by research funds from INSERM (BioAvenir program), CNRS (Fundamental Microbiology Program), and the Bettencourt-Schueller Foundation. S.S. is supported by an INSERM Young Researcher grant.
REFERENCES
- 1.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 2.Barbour, A. G. 2001. Borrelia: a diverse and ubiquitous genus of tick-borne pathogens, p. 153-173. In W. M. Scheld, W. A. Craig, and J. M. Hughes (ed.), Emerging infection, vol. 5. ASM Press, Washington, D.C. [Google Scholar]
- 3.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour, A. G. 1999. Relapsing fever and other Borrelia infections, p. 535-546. In R. L. Guerrant, D. H. Walker, and P. F. Weller (ed.), Tropical infectious diseases—principles, pathogens, and practice, vol. 1. Churchill Livingstone, Philadelphia, Pa. [Google Scholar]
- 5.Barbour, A. G., and C. F. Garon. 1987. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science 237:409-411. [DOI] [PubMed] [Google Scholar]
- 6.Barbour, A. G., A. D. Putteet-Driver, and J. Bunikis. 2005. Horizontally acquired genes for purine salvage in Borrelia spp. causing relapsing fever. Infect. Immun. 73:6165-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benach, J. L., J. L. Coleman, R. A. Skinner, and E. M. Bosler. 1987. Adult Ixodes dammini on rabbits: a hypothesis for the development and transmission of Borrelia burgdorferi. J. Infect. Dis. 155:1300-1306. [DOI] [PubMed] [Google Scholar]
- 8.Boursaux-Eude, C., D. Margarita, A. M. Gilles, O. Barzu, and I. Saint Girons. 1997. Borrelia burgdorferi uridine kinase: an enzyme of the pyrimidine salvage pathway for endogenous use of nucleotides. FEMS Microbiol. Lett. 151:257-261. [DOI] [PubMed] [Google Scholar]
- 9.Brooks, C. S., P. S. Hefty, S. E. Jolliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 71:3371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunikis, J., and A. G. Barbour. 2002. Laboratory testing for suspected Lyme disease. Med. Clin. N. Am. 86:311-340. [DOI] [PubMed] [Google Scholar]
- 11.Carreras, C. W., and D. V. Santi. 1995. The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem. 64:721-762. [DOI] [PubMed] [Google Scholar]
- 12.Casjens, S. 1999. Evolution of the linear DNA replicons of the Borrelia spirochetes. Curr. Opin. Microbiol. 2:529-534. [DOI] [PubMed] [Google Scholar]
- 13.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 14.Climie, S. C., C. W. Carreras, and D. V. Santi. 1992. Complete replacement set of amino acids at the C-terminus of thymidylate synthase: quantitative structure-activity relationship of mutants of an enzyme. Biochemistry 31:6032-6038. [DOI] [PubMed] [Google Scholar]
- 15.Dynes, J. L., and R. A. Firtel. 1989. Molecular complementation of a genetic marker in Dictyostelium using a genomic DNA library. Proc. Natl. Acad. Sci. USA 86:7966-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan, H. Z., G. McClarty, and R. C. Brunham. 1991. Biochemical evidence for the existence of thymidylate synthase in the obligate intracellular parasite Chlamydia trachomatis. J. Bacteriol. 173:6670-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, S., S. Das, T. Lam, R. A. Flavell, and E. Fikrig. 1995. A 55-kilodalton antigen encoded by a gene on a Borrelia burgdorferi 49-kilobase plasmid is recognized by antibodies in sera from patients with Lyme disease. Infect. Immun. 63:3459-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferdows, M. S., P. Serwer, G. A. Griess, S. J. Norris, and A. G. Barbour. 1996. Conversion of a linear to a circular plasmid in the relapsing fever agent Borrelia hermsii. J. Bacteriol. 178:793-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. C. Venter, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 20.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, J. C. Venter, et al. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 21.Galtier, N., and M. Gouy. 1995. Inferring phylogenies from DNA sequences of unequal base compositions. Proc. Natl. Acad. Sci. USA 92:11317-11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giladi, M., G. Bitan-Banin, M. Mevarech, and R. Ortenberg. 2002. Genetic evidence for a novel thymidylate synthase in the halophilic archaeon Halobacterium salinarum and in Campylobacter jejuni. FEMS Microbiol. Lett. 216:105-109. [DOI] [PubMed] [Google Scholar]
- 23.Glockner, G., R. Lehmann, A. Romualdi, S. Pradella, U. Schulte-Spechtel, M. Schilhabel, B. Wilske, J. Suhnel, and M. Platzer. 2004. Comparative analysis of the Borrelia garinii genome. Nucleic Acids Res. 32:6038-6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graziani, S., Y. Xia, J. R. Gurnon, J. L. Van Etten, D. Leduc, S. Skouloubris, H. Myllykallio, and U. Liebl. 2004. Functional analysis of FAD-dependent thymidylate synthase ThyX from Paramecium bursaria Chlorella virus-1. J. Biol. Chem. 279:54340-54347. [DOI] [PubMed] [Google Scholar]
- 25.Griffin, J., C. Roshick, E. Iliffe-Lee, and G. McClarty. 2005. Catalytic mechanism of Chlamydia trachomatis flavin-dependent thymidylate synthase. J. Biol. Chem. 280:5456-5467. [DOI] [PubMed] [Google Scholar]
- 26.Jordan, A., E. Aragall, I. Gibert, and J. Barbe. 1996. Promoter identification and expression analysis of Salmonella typhimurium and Escherichia coli nrdEF operons encoding one of two class I ribonucleotide reductases present in both bacteria. Mol. Microbiol. 19:777-790. [DOI] [PubMed] [Google Scholar]
- 27.Jordan, A., I. Gibert, and J. Barbe. 1994. Cloning and sequencing of the genes from Salmonella typhimurium encoding a new bacterial ribonucleotide reductase. J. Bacteriol. 176:3420-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jordan, A., E. Pontis, M. Atta, M. Krook, I. Gibert, J. Barbe, and P. Reichard. 1994. A second class I ribonucleotide reductase in Enterobacteriaceae: characterization of the Salmonella typhimurium enzyme. Proc. Natl. Acad. Sci. USA 91:12892-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leduc, D., S. Graziani, G. Lipowski, C. Marchand, P. Le Marechal, U. Liebl, and H. Myllykallio. 2004. Functional evidence for active site location of tetrameric thymidylate synthase X at the interphase of three monomers. Proc. Natl. Acad. Sci. USA 101:7252-7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leduc, D., S. Graziani, L. Meslet-Cladiere, A. Sodolescu, U. Liebl, and H. Myllykallio. 2004. Two distinct pathways for thymidylate (dTMP) synthesis in (hyper)thermophilic Bacteria and Archaea. Biochem. Soc. Trans. 32:231-235. [DOI] [PubMed] [Google Scholar]
- 31.Madden, T. L., R. L. Tatusov, and J. Zhang. 1996. Applications of network BLAST server. Methods Enzymol. 266:131-141. [DOI] [PubMed] [Google Scholar]
- 32.Margolis, N., D. Hogan, K. Tilly, and P. A. Rosa. 1994. Plasmid location of Borrelia purine biosynthesis gene homologs. J. Bacteriol. 176:6427-6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathews, I. I., A. M. Deacon, J. M. Canaves, D. McMullan, S. A. Lesley, S. Agarwalla, and P. Kuhn. 2003. Functional analysis of substrate and cofactor complex structures of a thymidylate synthase-complementing protein. Structure (Cambridge) 11:677-690. [DOI] [PubMed] [Google Scholar]
- 34.Myllykallio, H., D. Leduc, J. Filee, and U. Liebl. 2003. Life without dihydrofolate reductase FolA. Trends Microbiol. 11:220-223. [DOI] [PubMed] [Google Scholar]
- 35.Myllykallio, H., G. Lipowski, D. Leduc, J. Filee, P. Forterre, and U. Liebl. 2002. An alternative flavin-dependent mechanism for thymidylate synthesis. Science 297:105-107. [DOI] [PubMed] [Google Scholar]
- 36.Nascimento, A. L., A. I. Ko, E. A. Martins, C. B. Monteiro-Vitorello, P. L. Ho, D. A. Haake, S. Verjovski-Almeida, R. A. Hartskeerl, M. V. Marques, M. C. Oliveira, C. F. Menck, L. C. Leite, H. Carrer, L. L. Coutinho, W. M. Degrave, O. A. Dellagostin, H. El-Dorry, E. S. Ferro, M. I. Ferro, L. R. Furlan, M. Gamberini, E. A. Giglioti, A. Goes-Neto, G. H. Goldman, M. H. Goldman, R. Harakava, S. M. Jeronimo, I. L. Junqueira-de-Azevedo, E. T. Kimura, E. E. Kuramae, E. G. Lemos, M. V. Lemos, C. L. Marino, L. R. Nunes, R. C. de Oliveira, G. G. Pereira, M. S. Reis, A. Schriefer, W. J. Siqueira, P. Sommer, S. M. Tsai, A. J. Simpson, J. A. Ferro, L. E. Camargo, J. P. Kitajima, J. C. Setubal, and M. A. Van Sluys. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186:2164-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paster, B. J., and F. E. Dewhirst. 2000. Phylogenetic foundation of spirochetes. J. Mol. Microbiol. Biotechnol. 2:341-344. [PubMed] [Google Scholar]
- 39.Pennington, P. M., C. D. Allred, C. S. West, R. Alvarez, and A. G. Barbour. 1997. Arthritis severity and spirochete burden are determined by serotype in the Borrelia turicatae-mouse model of Lyme disease. Infect. Immun. 65:285-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pennington, P. M., D. Cadavid, and A. G. Barbour. 1999. Characterization of VspB of Borrelia turicatae, a major outer membrane protein expressed in blood and tissues of mice. Infect. Immun. 67:4637-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plasterk, R. H., M. I. Simon, and A. G. Barbour. 1985. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature 318:257-263. [DOI] [PubMed] [Google Scholar]
- 42.Putteet-Driver, A. D., J. Zhong, and A. G. Barbour. 2004. Transgenic expression of RecA of the spirochetes Borrelia burgdorferi and Borrelia hermsii in Escherichia coli revealed differences in DNA repair and recombination phenotypes. J. Bacteriol. 186:2266-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reichard, P. 1993. From RNA to DNA, why so many ribonucleotide reductases? Science 260:1773-1777. [DOI] [PubMed] [Google Scholar]
- 44.Ren, S. X., G. Fu, X. G. Jiang, R. Zeng, Y. G. Miao, H. Xu, Y. X. Zhang, H. Xiong, G. Lu, L. F. Lu, H. Q. Jiang, J. Jia, Y. F. Tu, J. X. Jiang, W. Y. Gu, Y. Q. Zhang, Z. Cai, H. H. Sheng, H. F. Yin, Y. Zhang, G. F. Zhu, M. Wan, H. L. Huang, Z. Qian, S. Y. Wang, W. Ma, Z. J. Yao, Y. Shen, B. Q. Qiang, Q. C. Xia, X. K. Guo, A. Danchin, I. Saint Girons, R. L. Somerville, Y. M. Wen, M. H. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888-893. [DOI] [PubMed] [Google Scholar]
- 45.Restrepo, B. I., T. Kitten, C. J. Carter, D. Infante, and A. G. Barbour. 1992. Subtelomeric expression regions of Borrelia hermsii linear plasmids are highly polymorphic. Mol. Microbiol. 6:3299-3311. [DOI] [PubMed] [Google Scholar]
- 46.Ribeiro, J. M., T. N. Mather, J. Piesman, and A. Spielman. 1987. Dissemination and salivary delivery of Lyme disease spirochetes in vector ticks (Acari: Ixodidae). J. Med. Entomol. 24:201-205. [DOI] [PubMed] [Google Scholar]
- 47.Sadziene, A., A. G. Barbour, P. A. Rosa, and D. D. Thomas. 1993. An OspB mutant of Borrelia burgdorferi has reduced invasiveness in vitro and reduced infectivity in vivo. Infect. Immun. 61:3590-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadziene, A., D. D. Thomas, and A. G. Barbour. 1995. Borrelia burgdorferi mutant lacking Osp: biological and immunological characterization. Infect. Immun. 63:1573-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwan, T. G., J. M. Battisti, S. F. Porcella, S. J. Raffel, M. E. Schrumpf, E. R. Fischer, J. A. Carroll, P. E. Stewart, P. Rosa, and G. A. Somerville. 2003. Glycerol-3-phosphate acquisition in spirochetes: distribution and biological activity of glycerophosphodiester phosphodiesterase (GlpQ) among Borrelia species. J. Bacteriol. 185:1346-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seshadri, R., G. S. Myers, H. Tettelin, J. A. Eisen, J. F. Heidelberg, R. J. Dodson, T. M. Davidsen, R. T. DeBoy, D. E. Fouts, D. H. Haft, J. Selengut, Q. Ren, L. M. Brinkac, R. Madupu, J. Kolonay, S. A. Durkin, S. C. Daugherty, J. Shetty, A. Shvartsbeyn, E. Gebregeorgis, K. Geer, G. Tsegaye, J. Malek, B. Ayodeji, S. Shatsman, M. P. McLeod, D. Smajs, J. K. Howell, S. Pal, A. Amin, P. Vashisth, T. Z. McNeill, Q. Xiang, E. Sodergren, E. Baca, G. M. Weinstock, S. J. Norris, C. M. Fraser, and I. T. Paulsen. 2004. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc. Natl. Acad. Sci. USA 101:5646-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stoenner, H. G., T. Dodd, and C. Larsen. 1982. Antigenic variation of Borrelia hermsii. J. Exp. Med. 156:1297-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torrents, E., I. Roca, and I. Gibert. 2003. Corynebacterium ammoniagenes class Ib ribonucleotide reductase: transcriptional regulation of an atypical genomic organization in the nrd cluster. Microbiology 149:1011-1020. [DOI] [PubMed] [Google Scholar]
- 54.Zhong, J., and A. G. Barbour. 2004. Cross-species hybridization of a Borrelia burgdorferi DNA array reveals infection- and culture-associated genes of the unsequenced genome of the relapsing fever agent Borrelia hermsii. Mol. Microbiol. 51:729-748. [DOI] [PubMed] [Google Scholar]