Abstract
During Bacillus subtilis sporulation, the transient engulfment defect of spoIIB strains is enhanced by spoVG null mutations and suppressed by spoVS null mutations. These mutations have opposite effects on expression of the motility regulon, as the spoVG mutation reduces and the spoVS mutation increases σD-directed gene expression, cell separation, and autolysis. Elevating σD activity by eliminating the anti-σ factor FlgM also suppresses spoIIB spoVG, and both flgM and spoVS mutations cause continued expression of the σD regulon during sporulation. We propose that peptidoglycan hydrolases induced during motility can substitute for sporulation-specific hydrolases during engulfment. We find that sporulating cells are heterogeneous in their expression of the motility regulon, which could result in phenotypic variation between individual sporulating cells.
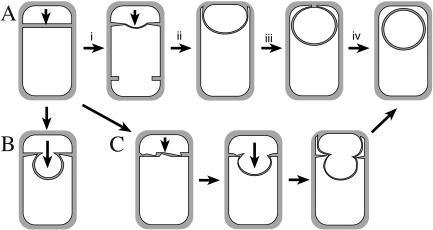
A key step in Bacillus subtilis sporulation is engulfment, which mediates a dramatic rearrangement of the two cells required for sporulation, from two adjacent daughter cells to a sporangium in which one daughter cell (the forespore) is completely enclosed within the cytoplasm of another (the mother cell). This transformation is mediated by the migration of the mother cell membrane around the forespore, which is ultimately released into the mother cell cytoplasm (Fig. 1A). Engulfment depends on three conserved proteins, SpoIID, SpoIIM, and SpoIIP (1, 5, 17, 30), and is facilitated by the less-conserved SpoIIB protein (19, 23). These proteins participate in septal thinning (1, 5, 17, 19, 23, 30), during which septal peptidoglycan is degraded by the SpoIID peptidoglycan hydrolase and perhaps other enzymes (1). While spoIID, spoIIM, and spoIIP mutants are completely defective in septal thinning and membrane migration (Fig. 1B), a spoIIB mutant shows slow and uneven septal thinning but ultimately completes engulfment and produces spores (Fig. 1C). Two additional proteins have been proposed to play a role in septal thinning: SpoVG, whose absence exacerbates the engulfment defect of spoIIB (19, 23), and SpoVS, whose absence suppresses a spoIIB spoVG double mutant, allowing engulfment to proceed (27). Here, we provide evidence that the spoVS mutation causes decreased expression of early-sporulation genes and increased expression of motility genes and that motility-specific genes, likely peptidoglycan hydrolases, can suppress septal thinning defects.
FIG. 1.
Engulfment in B. subtilis. (A) Asymmetric division produces the small forespore and large mother cell. Septal thinning (step i) commences at the septal midpoint and proceeds towards the edges, followed by membrane migration up (step ii) and around (step iii) the forespore, until the membrane meets and fuses (step iv) to release the forespore into the mother cell cytoplasm. (B) In the absence of SpoIID, SpoIIM, or SpoIIP, septal thinning is blocked, and the forespore ultimately bulges into the mother cell. (C) In the absence of SpoIIB, septal thinning is slowly and uneven, forming a transient bulge, although engulfment is ultimately completed. In the absence of both SpoIIB and SpoVG, membrane migration is blocked.
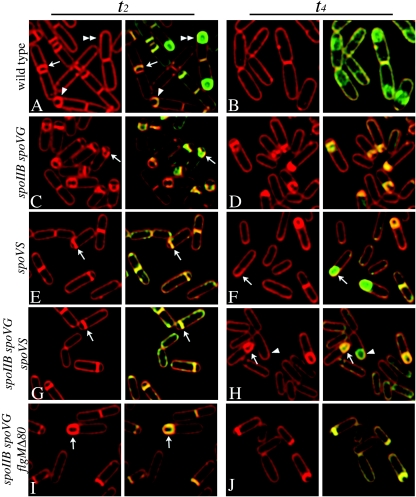
To investigate the mechanism of spoVS-mediated suppression of spoIIB spoVG, we used an in vivo membrane fusion assay that employs two membrane stains, the membrane-impermeable FM 4-64 and the membrane-permeable stain MitoTracker Green (strains of B. subtilis used in this study are shown in Table 1) (29). During engulfment in the wild type, the engulfing membrane smoothly curves around the forespore (Fig. 2A, arrow and arrowhead), with the septal membrane staining twice as brightly as other regions, due to the presence of two membranes (24). Ultimately, the migrating membrane meets and fuses to release the forespore into the mother cell cytoplasm. After membrane fusion, FM 4-64 is excluded from the forespore, resulting in a green forespore enclosed within the red mother cell (Fig. 2A, double arrowhead). In spoIIB spoVG, the growing forespore pushes through the unthinned septum, forming a bulge that is typical of septal thinning defective mutants (Fig. 2C to D). In the spoIIB spoVG spoVS triple mutant, sixfold-fewer bulges were observed than in the spoIIB spoVG mutant (6% versus 35% at t3.0) (Fig. 2G and H; Table 2), and engulfment was complete in 21% of spoVS spoIIB spoVG sporangia versus 0% of spoIIB spoVG sporangia at t4 (Fig. 2; Table 2). The spoVS mutation also weakly suppressed spoIID and spoIIM strains, reducing the frequency of bulges twofold (from 64% to 31% in spoIIM and from 45% to 21% in spoIID) but not supporting the completion of engulfment or increased spore production (see Fig. S1 and Table S1 in the supplemental material). These results suggest that the spoVS mutation is a general suppressor of septal thinning defects.
TABLE 1.
Bacillus subtilis strains used in this studya
| Strain | Genotype | Reference or source |
|---|---|---|
| PY79 | Wild type | 35 |
| KP52 | ΔspoIIB::erm spoVG::Tn917 | 19 |
| KP84 | spoIIAC-lacZ | 28 |
| KP174 | Δ(spoIIAA-AC)::spec | 2 |
| KP343 | ΔspoIIB::erm | 19 |
| KP548 | spoVG::Tn917::spec | 23 |
| KP535 | ΔspoVS::spec | 27 |
| KP787 | ΔspoIIB::erm spoVG::Tn917 ΔspoVS::spec | This study |
| KP798 | ΔsinR::neo | 6 |
| KP799 | ΔsinR::neo ΔspoVS::spec | This study |
| KP812 | flgMΔ80 | 21 |
| KP813 | sigD::pLM5 | 10 |
| KP814 | ΔspoIIB::erm spoVG::Tn917::spec | This study |
| KP815 | ΔspoIIB::erm spoVG::Tn917::spec flgMΔ80 | This study |
| KP818 | Phag(−UP)-lacZ | 6 |
| KP819 | flgMΔ80 Phag(−UP)-lacZ | This study |
| KP820 | sigD::pLM5 (cmr) Phag(−UP)-lacZ | This study |
| KP821 | ΔspoVS::spec, Phag(−UP)-lacZ | This study |
| KP822 | ΔsinR::neo Phag(−UP)-lacZ | This study |
| KP823 | spoVG::Tn917, Phag(−UP)-lacZ | This study |
| KP826 | ΔspoIIB::erm spoVG::Tn917 ΔspoVS::spec lytABC::neo | This study |
| KP827 | ΔspoIIB::erm spoVG::Tn917 ΔspoVS::spec lytD::tet | This study |
| KP828 | ΔspoIIB::erm spoVG::Tn917 ΔspoVS::spec lytABC::neo lytD::tet | This study |
| KP829 | flgMΔ80 sigD::pLM5 | This study |
| KP830 | ΔspoVS::spec sigD::pLM5 | This study |
| KP930 | spoIIAC-lacZ::tet rvtA11::spec | This study |
| KP931 | spoIIAC-lacZ::tet rvtA11::spec ΔcotE::cm ΔspoVS::spec | This study |
| KP932 | spoIIAC-lacZ::cm ΔabrB::tet | This study |
| KP933 | spoIIAC-lacZ::cat ΔabrB::tet, ΔspoVS::spec | This study |
| KP934 | spoIIAC-lacZ::cat flgMΔ80 | This study |
| KP936 | spoIIAC-lacZ::cat ΔspoVS::spec | This study |
| KP937 | spoIIAC-lacZ::cat ΔsinR::neo | This study |
| KP938 | spoIIAC-lacZ::cat spoVG::Tn917 | This study |
All strains are PY79 derivatives except those with flgMΔ80, which are JH642 derivatives.
FIG. 2.
Effect of the spoVS mutation on engulfment. Sporulation was induced by resuspension (33), and samples taken 2 h (t2) (A, C, E, G, and I) and 4 h (t4) (B, D, F, H, and J) later were stained with FM 4-64 (red) and MitoTracker Green (green) as previously described (24, 29). (A and B) Wild-type (PY79) sporangia after septation (arrow) and during engulfment (arrowhead). The septum stains approximately two times more brightly than the cytoplasmic membrane, because it contains two parallel membranes, while the engulfing membrane stains approximately three times more brightly, because it contains three membrane layers (Fig. 1A) (24). After membrane fusion (double arrowhead), FM 4-64 is excluded from the forespore (29), which is stained only with MitoTracker Green. (C and D) The spoIIB spoVG double mutant (KP52) shows sporangia with flat polar septa or bulges (arrow). (E and F) The spoVS mutant (KP535) is delayed in polar septation but engulfs normally (E, arrow) and completes membrane fusion (F, arrow). (G and H) The spoIIB spoVG spoVS triple mutant (KP787) exhibits few bulges (G, arrow) and completes membrane migration (H, arrow) and fusion (H, arrowhead). (I to J) The spoIIB spoVG flgMΔ80 triple mutant (KP815) shows few bulges and completes engulfment (I, arrow).
TABLE 2.
Effect of spoVS and flgM mutations on polar septation, engulfment, and spore formation
| Genotype | Spores/ml | % Sporangia (t2)a | % Sporangia with bulges (t3)b | % Sporangia fused (t4) |
|---|---|---|---|---|
| Wild type (PY79) | 4 × 108 | 55 (775)c | 0 (347) | 84 (248) |
| spoVS | 1 × 106 | 6 (1,342) | 0 (547) | 46 (623) |
| spoIIB | 2 × 108 | 45 (1,670)d | 37 (699) | 7 (345) |
| spoVG | 9 × 108 | 41 (391)d | 0 (301) | 40 (251) |
| spoIIB spoVG | 9 × 104 | 55 (1,211) | 35 (1,073) | 0 (401) |
| spoIIB spoVG spoVS | 2 × 106 | 1 (312) | 6 (1,270) | 21 (1,459) |
| spoIIB spoVG flgMΔ80 | 2 × 106 | 30 (573) | 5 (761) | 9 (474) |
| spoIIB spoVG spoVS lytABCe | 5 × 105 | NDf | 13 (615) | 35 (764) |
| spoIIB spoVG flgMΔ80 lytABCe | 3 × 105 | NDf | 14 (336) | 34 (126) |
Percent sporangia is determined by dividing the number of sporangia by the total number of total cells (in parentheses) in the fields scored.
Percent sporangia with bulges or fused is calculated by dividing the number of sporangia with the indicated phenotype by the total number of sporangia (in parentheses).
The number in parentheses indicates the total number of cells scored (vegetative cells and sporangia).
Data for percent sporangia at t2 are from Perez et al. (23) and are included for comparison only.
The additional deletion of lytD had no effect on suppression (data not shown).
ND, not determined.
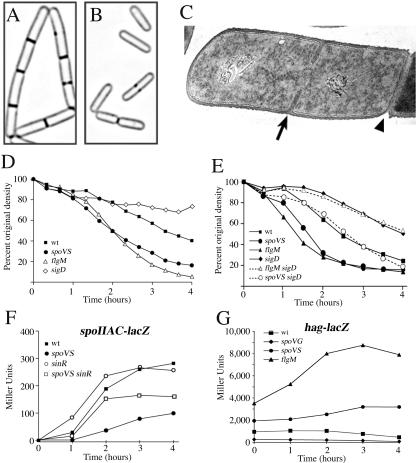
We noted that spoVS strains showed fewer polar septa than the wild type (Fig. 2E and F; Table 2). This was due to decreased expression of early sporulation genes such as spoIIAC (Fig. 3F) and spoIIE (see Fig. S3 in the supplemental material), which require the Spo0A∼P transcription factor that governs entry into sporulation (7). Expression of Spo0A∼P-dependent genes is regulated by kinases and phosphatases that modulate the level of phosphorylated Spo0A (22, 32) and by various transcription factors that inhibit Spo0A∼P-dependent gene expression (7-9). We found that the defect in polar septation and Spo0A∼P-dependent gene expression could be substantially rescued by a sinR mutation, which eliminates one of these transcription factors, but not by spo0Asad, which encodes a Spo0A protein active without phosphorylation (11), rvtA11, which encodes a Spo0A protein that can be phosphorylated by an alternative kinase (16), or abrB or spo0J-soj mutations, which eliminate repressors of Spo0A∼P-dependent gene expression (25, 34; see Fig. S3 in the supplemental material). These results suggest that the spoVS mutant has increased SinR activity.
FIG. 3.
Cell separation, autolysis, and Spo0A∼P- and σD-directed gene expression. (A and B) FM 4-64 membrane staining showing the wild type (PY79) growing in chains (A) and spoVS (KP535) growing as single cells (B). (C) Electron micrograph showing a vegetative PY79 cell dividing in the middle (arrow) and still connected to its sister cell by septal peptidoglycan in the progress of being split by autolytic enzymes (arrowhead); samples were prepared as previously described (23). (D and E) Sodium azide-induced autolysis during vegetative growth, measured by taking samples from a growing culture, adding sodium azide to a concentration of 0.05 M, and following the optical density at 600 nm over time at 37°C with continued aeration (3). The wild type (squares), sigD (open diamonds; KP813), spoVS (circles; KP535), and flgMΔ80 (open triangle; KP812) trains sare shown. (E) The following strains are shown: wild type (squares); sigD (filled diamonds; KP813); spoVS (circles; KP535); flgMΔ80 (triangle; KP812), flgM sigD (open triangle; KP829), and spoVS sigD (open circle; KP830). (F) Spo0A∼P-dependent expression of spoIIAC-lacZ in the wild type (squares; KP84), spoVS (circles; KP936), sinR (open circles; KP937), and spoVS sinR (open squares). Samples were harvested at the indicated time after the initiation of sporulation by resuspension at 37°C. (G) σD-directed expression of hag-lacZ during a resuspension sporulation in the wild-type (squares; KP818), spoVG (diamonds; KP823), spoVS (circle; KP821), and flgMΔ80 (triangle; KP819) strains.
SinR is a transcription factor that directly regulates biofilm formation (13), inhibits Spo0A∼P-dependent gene expression, and activates σD-dependent motility genes (4, 6, 13, 18, 26), perhaps indirectly (13). Thus, if the spoVS strain has elevated SinR activity, it should also show increased expression of motility genes. Indeed, the spoVS strain showed approximately twofold elevated expression of the flagellin gene (hag-lacZ) relative to the wild type, while the spoVG mutation (enhancer of spoIIB) showed approximately fivefold reduced σD activity (Fig. 3G). This suggests that SpoVS directly or indirectly governs SinR activity, similar to two other recently described proteins, YlbF and YmcA (13). Strains lacking these proteins share some phenotypes with spoVS strains, with continued SinR activity, a failure to form biofilms, and growth that is slightly slower than that of the wild type (spoVS data are not shown) (13).
It is unclear if SpoVS directly or indirectly modulates SinR or σD activity, and it remains possible that spoVS mutation has additional direct or indirect effects on gene expression. We therefore tested if elevated σD activity was sufficient to suppress spoIIB spoVG by inactivating the anti-sigma factor for σD, FlgM (21). As expected, flgM strains have increased σD-directed gene expression (Fig. 3G). Further, like the spoVS mutation, the flgM mutation increased spore production by spoIIB spoVG ∼20-fold (from 9 × 104 to 2 ×106) (Table 2), reduced the frequency of bulges ∼7-fold (from 35% to 5%), and increased the proportion of sporangia that completed the final step of engulfment, membrane fusion, from 0% to 9% (Fig. 2I and J; Table 2). Thus, increased σD activity is sufficient to suppress the engulfment defect of spoIIB spoVG to a similar extent as the spoVS mutation.
B. subtilis cells often grow in short chains of cells that remain connected by septal peptidoglycan (Fig. 3A and C), which ultimately is split by peptidoglycan hydrolases to allow daughter cell separation (31). We noted that spoVS strains rarely grew in chains (Fig. 3B), suggesting an increased activity of peptidoglycan hydrolases involved in cell separation. Indeed, the σD regulon includes genes that encode peptidoglycan hydrolases that mediate cell separation (14, 15, 20), which likely facilitates motility by the generation of single cells (3). The overexpression of peptidoglycan hydrolases in spoVS and flgM strains was confirmed by an autolysis assay, which measures the lysis of bacteria whose growth has been arrested (3, 12). Both strains lysed faster than the wild type; after 4 h, optical density at 600 nm was reduced by 60% in the wild type versus 84% in spoVS and 94% in flgM (Fig. 3D). The elevated autolysis of flgM and spoVS strains was eliminated by a sigD mutation, which inactivates σD (Fig. 3E), and by a lytABC mutation, which eliminates the major σD-directed autolysin (see Fig. S2 in the supplemental material). The lytABC mutation also reduced but did not eliminate spoVS- and flgM-mediated suppression of spoIIB spoVG, increasing bulges and reducing spore formation to ∼25% of suppressed levels (Table 2). We propose that spoVS and flgM suppress septal thinning defects by increasing expression of peptidoglycan hydrolases involved in cell separation and speculate that several such hydrolases are required for full suppression.
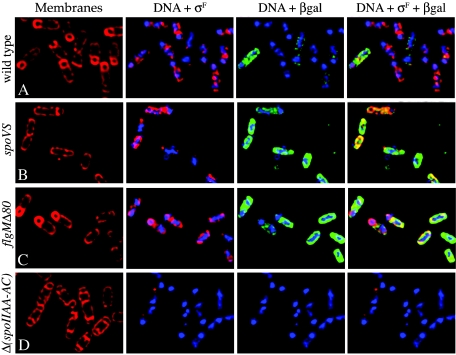
The increased σD activity in spoVS and flgM strains could be due to either an increased σD activity in all sporangia or an increased fraction of sporangia with σD activity. To test these possibilities, we used immunofluorescence microscopy to visualize Spo0A and σD activity in sporulating cultures. To detect Spo0A activity, we used antibodies to σF, the production of which requires Spo0A∼P; to detect σD activity, we used antibodies to β-galactosidase produced from the hag-lacZ fusion. After 2.5 h of sporulation, σD activity was observed in 16% of wild-type sporangia (cells containing a sporulation septum visualized by FM 4-64 membrane staining) (Fig. 4A; Table 3), compared to 36% of spoVS sporangia and 72% of flgMΔ80 sporangia (Fig. 4B to C; Table 3). Thus, spoVS and flgM strains have two to four times more sporangia expressing motility genes than the wild type, which would likely result in an increased number of sporangia with elevated levels of peptidoglycan hydrolases involved in cell separation. Clearly, even a wild-type population of sporulating cells displays heterogeneity in the activity of the motility regulon.
FIG. 4.
Immunofluorescence microscopy to examine Spo0A∼P- and σ D-directed gene expression. Sporulation was induced by resuspension, and samples were harvested at t2.5 and processed for immunofluorescence microcopy (23), staining with membrane stain FM 4-64 (red, left) and the DNA stain 4′,6′-diamidino-2-phenylindole
(DAPI; blue) and using antibodies specific for σF (magenta) and for β-galactosidase produced by hag-lacZ (green). The wild-type strain (KP818) (A), the spoVS strain (KP821) (B), the flgMΔ80 strain (KP819) (C), and the negative control strain KP174 Δ(spoIIAA-AC) which has a deletion in the gene encoding σF and no lacZ fusion (D) are shown.
TABLE 3.
Expression of σD and Spo0A∼P-directed genes at t2.5 of sporulation
| Genotype | Cell morphologya | Percentage in class | Percentage of cells in morphological class expressing:
|
|||
|---|---|---|---|---|---|---|
| σF | hag-lacZ | Both | Neither | |||
| Wild type (244)c | Vegetativeb | 41 | 46 | 10 | 4 | 39 |
| Sporulating | 59 | 81 | 0 | 16 | 3 | |
| spoVS (185)c | Vegetativeb | 85 | 9 | 46 | 27 | 18 |
| Sporulating | 15 | 57 | 0 | 36 | 7 | |
| flgMΔ80 (171)c | Vegetativeb | 71 | 2 | 69 | 26 | 2 |
| Sporulating | 29 | 26 | 0 | 72 | 2 | |
Vegetative cells do not contain a polar septum, while sporulating cells are defined as sporangia that contain a polar septum in any stage of engulfment.
Cells containing σF in the vegetative cell category likely have initiated sporulation, but have not yet synthesized the polar septum. Indeed, in many of these cells the chromosome was rearranged into the axial filament structure that precedes polar spetation.
Total cells scored.
These results demonstrate an unanticipated link between expression of the σD-directed motility regulon and the ability of sporulating cells to complete the phagocytosis-like process of engulfment during sporulation. Apparently, the continued expression of σD-directed genes during sporulation allows engulfment in the septal thinning defective spoIIB spoVG strain, perhaps by providing additional hydrolases that contribute to septal thinning. Because there is cell-to-cell variation in the level of motility gene expression in individual sporulating cells, these observations might explain why there is also cell-to-cell variation in the phenotypes of engulfment mutants, with variations in whether they divided at the second site in the mother cell and whether they have a flat or bulged septum (24). We anticipate the discovery of additional examples in which a cell's proteomic “memory” of past gene expression alters its behavior during subsequent transcriptional responses or developmental pathways.
Supplementary Material
Acknowledgments
We thank Stefan Aung, Dan Broder, Rich Losick, Jonathan Shum, and Joe Pogliano for providing comments on the manuscript; Marta Perego, John D. Helmann, and the Bacillus Genetics Stock Center (BGSC) for providing strains; and Rich Losick for providing σF-specific antibodies.
This work was supported by NIH grant GM-57405, and A.P. was supported by an NIH MARC predoctoral fellowship (GM-19570).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abanes-De Mello, A., Y. L. Sun, S. Aung, and K. Pogliano. 2002. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 16:3253-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arigoni, F., L. Duncan, S. Alper, R. Losick, and P. Stragier. 1996. SpoIIE governs the phosphorylation state of a protein regulating transcription factor sigma F during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 93:3238-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackman, S. A., T. J. Smith, and S. J. Foster. 1998. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology 144:73-82. [DOI] [PubMed] [Google Scholar]
- 4.Cervin, M. A., R. J. Lewis, J. A. Brannigan, and G. B. Spiegelman. 1998. The Bacillus subtilis regulator SinR inhibits spoIIG promoter transcription in vitro without displacing RNA polymerase. Nucleic Acids Res. 26:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frandsen, N., and P. Stragier. 1995. Identification and characterization of the Bacillus subtilis spoIIP locus. J. Bacteriol. 177:716-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fredrick, K., and J. D. Helmann. 1996. FlgM is a primary regulator of σD activity, and its absence restores motility to a sinR mutant. J. Bacteriol. 178:7010-7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grossman, A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477-508. [DOI] [PubMed] [Google Scholar]
- 8.Hamoen, L. W., G. Venema, and O. P. Kuipers. 2003. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149:9-17. [DOI] [PubMed] [Google Scholar]
- 9.Hamon, M. A., N. R. Stanley, R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 52:847-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helmann, J. D., L. M. Márquez, and M. J. Chamberlin. 1988. Cloning, sequencing, and disruption of the Bacillus subtilis σ28 gene. J. Bacteriol. 170:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ireton, K., D. Z. Rudner, K. J. Siranosian, and A. D. Grossman. 1993. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 7:283-294. [DOI] [PubMed] [Google Scholar]
- 12.Jolliffe, L. K., R. J. Doyle, and U. N. Streips. 1981. The energized membrane and cellular autolysis in Bacillus subtilis. Cell 25:753-763. [DOI] [PubMed] [Google Scholar]
- 13.Kearns, D. B., F. Chu, S. S. Branda, R. Kolter, and R. Losick. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55:739-749. [DOI] [PubMed] [Google Scholar]
- 14.Kuroda, A., M. H. Rashid, and J. Sekiguchi. 1992. Molecular cloning and sequencing of the upstream region of the major Bacillus subtilis autolysin gene: a modifier protein exhibiting sequence homology to the major autolysin and the spoIID product. J. Gen. Microbiol. 138:1067-1076. [DOI] [PubMed] [Google Scholar]
- 15.Lazarevic, V., P. Margot, B. Soldo, and D. Karamata. 1992. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J. Gen. Microbiol. 138:1949-1961. [DOI] [PubMed] [Google Scholar]
- 16.LeDeaux, J. R., and A. D. Grossman. 1995. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J. Bacteriol. 177:166-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Diaz, I., S. Clarke, and J. Mandelstam. 1986. spoIID operon of Bacillus subtilis: cloning and sequence. J. Gen. Microbiol. 132:341-354. [DOI] [PubMed] [Google Scholar]
- 18.Mandic-Mulec, I., L. Doukhan, and I. Smith. 1995. The Bacillus subtilis SinR protein is a repressor of the key sporulation gene spo0A. J. Bacteriol. 177:4619-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margolis, P. S., A. Driks, and R. Losick. 1993. Sporulation gene spoIIB from Bacillus subtilis. J. Bacteriol. 175:528-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Márquez, L. M., J. D. Helmann, E. Ferrari, H. M. Parker, G. W. Ordal, and M. J. Chamberlin. 1990. Studies of σD-dependent functions in Bacillus subtilis. J. Bacteriol. 172:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirel, D. B., P. Lauer, and M. J. Chamberlin. 1994. Identification of flagellar synthesis regulatory and structural genes in a σD-dependent operon of Bacillus subtilis. J. Bacteriol. 176:4492-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perego, M. 1998. Kinase-phosphatase competition regulates Bacillus subtilis development. Trends Microbiol. 6:366-370. [DOI] [PubMed] [Google Scholar]
- 23.Perez, A. R., A. Abanes-De Mello, and K. Pogliano. 2000. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in Bacillus subtilis. J. Bacteriol. 182:1096-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pogliano, J., N. Osborne, M. D. Sharp, A. Abanes-De Mello, A. Perez, Y. L. Sun, and K. Pogliano. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31:1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quisel, J. D., D. C. Lin, and A. D. Grossman. 1999. Control of development by altered localization of a transcription factor in B. subtilis. Mol. Cell 4:665-672. [DOI] [PubMed] [Google Scholar]
- 26.Rashid, M. H., and J. Sekiguchi. 1996. flaD (sinR) mutations affect SigD-dependent functions at multiple points in Bacillus subtilis. J. Bacteriol. 178:6640-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Resnekov, O., A. Driks, and R. Losick. 1995. Identification and characterization of sporulation gene spoVS from Bacillus subtilis. J. Bacteriol. 177:5628-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt, R., P. Margolis, L. Duncan, R. Coppolecchia, C. P. Moran, Jr., and R. Losick. 1990. Control of developmental transcription factor sigma F by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:9221-9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharp, M. D., and K. Pogliano. 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. USA 96:14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, K., M. E. Bayer, and P. Youngman. 1993. Physical and functional characterization of the Bacillus subtilis spoIIM gene. J. Bacteriol. 175:3607-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249-262. [DOI] [PubMed] [Google Scholar]
- 32.Sonenshein, A. L. 2000. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3:561-566. [DOI] [PubMed] [Google Scholar]
- 33.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strauch, M. A., G. B. Spiegelman, M. Perego, W. C. Johnson, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator AbrB of Bacillus subtilis is a DNA binding protein. EMBO J. 8:1615-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Youngman, P., J. B. Perkins, and R. Losick. 1984. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol. Gen. Genet. 195:424-433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.