Abstract
In Salmonella enterica, the isc operon contains genes necessary for the synthesis of Fe-S clusters and strains lacking this operon have severe defects in a variety of cellular processes. Other cellular loci that impact Fe-S cluster synthesis to a lesser extent have been described. The cyaY locus encodes a frataxin homolog, and it is shown here that lesions in this locus affect Fe-S cluster metabolism. When present in combination with other lesions, mutations in cyaY can result in a strain with more severe defects than those lacking the isc locus.
Iron sulfur (Fe-S) clusters are a component of various cellular proteins and have diverse roles in metabolism (reviewed in references 14, 15, and 17). Significant progress has been made in identifying the components and the biochemistry involved in the synthesis of Fe-S clusters (reviewed in reference 25). Many of these components are structurally and functionally conserved throughout all kingdoms of life, emphasizing the similarity of this process throughout biology (22, 40).
The small protein frataxin was first identified as the missing protein in patients with Friedreich's ataxia, a progressive cardio- and neurodegenerative disease resulting from abnormal iron homeostasis and oxidative damage (5-8, 27, 36). Subsequently, frataxin was suggested to have a role in Fe-S cluster assembly, due to the cooccurrence of frataxin with the Isc Fe-S cluster assembly proteins (22, 37) and the finding that loss of the frataxin homolog in Saccharomyces cerevisiae resulted in phenotypes indicative of defects in Fe-S cluster assembly (1, 12, 16).
Biochemical, structural, and biophysical studies with purified protein have determined that proteins of the frataxin family have an affinity for iron (4, 26, 28, 38). In particular, Bou-Abdallah et al. demonstrated that under anaerobic conditions, the bacterial ortholog CyaY forms a tetramer and binds two ferrous ions per monomer with weak affinity (2).
To our knowledge, there has been a single report addressing the in vivo consequences of a cyaY mutation in a prokaryotic system (24). The authors concluded that loss of CyaY did not detectably alter the metabolism of the cell (24) and suggested that CyaY in bacteria may have a different function than in eukaryotes. An alternative interpretation is that the function(s) of CyaY can be performed by other gene products in the cell. In our work on metabolic integration, a number of loci with weak and possibly indirect effects on the metabolism of Fe-S clusters in Salmonella enterica were identified (35). The defects associated with lesions in these genes are often detectable only in the presence of other lesions and/or under nonstandard growth conditions. This study addresses the possibility that lesions in cyaY could result in detectable phenotypes if present in combination with other specific lesions. We demonstrate that cyaY mutations in S. enterica can result in severe metabolic defects, which are exacerbated or only detected when other mutations are present.
Loss of CyaY affects Fe-S cluster metabolism in S. enterica.
An insertion mutation conferring kanamycin resistance was constructed in the cyaY gene of S. enterica by linear replacement followed by appropriate confirmation (11). Strains defective in up to three loci (apbC, yggX, and cyaY) were generated. YggX is a small protein (91 amino acids) implicated in the protection of the cell from oxidative stress (20) and has recently been shown to affect the consequences of other lesions impacting Fe-S cluster metabolism (35) and weakly bind iron (10). ApbC is a member of the MinD protein family (29, 34) and is implicated in Fe-S cluster metabolism in the three domains of life (23, 31, 33).
In several studies, the activity of succinate dehydrogenase (SDH), an Fe-S enzyme, has been used to assess the status of Fe-S cluster biosynthesis/repair (32-34). Table 1 shows that a cyaY lesion had no significant effect on the SDH activity in the strain (strain DM7644 versus strain DM8000). When both cyaY and yggX were defective (DM7643), SDH activity was significantly decreased, dropping to the level caused by a lesion in the isc locus (DM7220) (35). As previously reported (20), strains lacking only yggX were indistinguishable from the wild-type parent (data not shown). The addition of an apbC mutation to either the single (cyaY) or double (cyaY yggX) mutant had no further effect on the level of SDH (DM7642 and DM7641). Thus, by this assay, a lesion in cyaY was additive with a yggX lesion, and the resulting strain was unaffected by a lesion in apbC.
TABLE 1.
Loss of CyaY activity impacts the activity of the Fe-S cluster enzyme SDHa
| Strain | Relevant genotype | SDH activityb | Relative activityc |
|---|---|---|---|
| DM8000 | Wild type | 34,840 ± 159 | 1.00 |
| DM7220 | iscS | 10,804 ± 102 | 0.31 |
| DM7644 | cyaY | 32,645 ± 271 | 0.93 |
| DM7643 | cyaY yggX | 8,537 ± 232 | 0.24 |
| DM7642 | apbC cyaY | 31,809 ± 113 | 0.90 |
| DM7641 | yggX apbC cyaY | 9,679 ± 250 | 0.27 |
Strains were grown in nutrient broth to an optical density at 650 nm of 0.5. Crude cell-free extracts were generated by sonication as previously described (35). In all cases, loci indicated for relevant genotypes were inactivated by insertion.
SDH activity was measured as described previously (34) and is reported as a specific activity (ΔA600/min/mg protein [average ± standard deviation]).
Relative activity was obtained by dividing the activity of the indicated strain by the activity of the wild-type parent (DM8000).
Thiamine auxotrophy is exacerbated by lesions in cyaY.
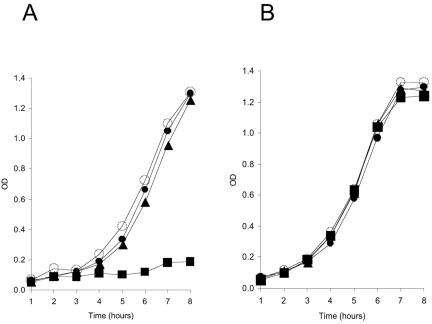
A class of mutant loci that indirectly inhibit thiamine synthesis via disruption of Fe-S cluster metabolism has been described previously (19, 34). In general, the thiamine auxotrophy of these mutant strains requires loss of the yggX locus and can be eliminated by addition of iron to the medium (35). Strains lacking cyaY, with or without a yggX mutation, had no detectable requirement for thiamine. However, under conditions where the apbC yggX double mutant was prototrophic (i.e., with excess iron in the medium), a lesion in cyaY eliminated growth in the absence of thiamine. A representative experiment in Fig. 1 shows that the combination of three lesions results in a strict requirement for thiamine. These data showed that only the triple mutant was defective in thiamine synthesis, implying that the function of just one of the three loci was needed for prototrophic growth in this medium. One interpretation of these results is that the CyaY protein is required to allow iron to suppress the lesion in yggX. This scenario is consistent with the general roles proposed for each of these proteins in the cell and the finding that each can interact with iron (2, 10).
FIG. 1.
The yggX apbC cyaY mutant has a stringent thiamine requirement. Overnight nutrient cultures were subcultured and grown at 37°C with shaking, as described in reference 35. Growth of strains was monitored over time by measurement of absorbance (optical density [OD] at 650 nm) with a Bausch and Lomb Spectronic 20. Strains DM7226 (wild type) (•), DM7306 (abpC yggX) (▴), DM7644 (cyaY) (○), and DM7641 (abpC yggX cyaY) (▪) were grown in (A) minimal medium supplemented with adenine (5 mM) and ferric chloride (20 μM) or (B) the same medium as described for panel A but with thiamine (50 μM).
Sensitivity to paraquat is increased by cyaY mutations.
Strains compromised in Fe-S cluster metabolism have increased sensitivity to the redox cycling compound paraquat (35). Data from growth experiments in Table 2 show that in the absence of paraquat all cultures reach a similar final density but that with ≥40 μM paraquat in the medium the strains reach different growth densities. The cyaY mutation alone did not significantly affect the sensitivity of a strain to paraquat, but in combination with a lesion in yggX, it resulted in dramatically increased sensitivity. An apbC mutation also increased the sensitivity of the cyaY strain to paraquat. The data in Table 2 indicated that when any two of the three loci tested (apbC, yggX, and cyaY) were defective, a similar increase in sensitivity to paraquat occurred. Lesions in all three loci further decreased growth of the strain when ≥40 μM paraquat was present. These results were consistent with a partially overlapping function of the YggX, CyaY, and ApbC proteins in the processes that impact sensitivity to superoxide.
TABLE 2.
Increased sensitivity to paraquat caused by a cyaY mutationa
| Strain | Relevant genotype | Final OD650 in NB containing paraquat atb:
|
||
|---|---|---|---|---|
| 0 μM | 40 μM | 80 μM | ||
| DM7226 | Wild type | 0.97 ± <0.01 | 0.92 ± 0.01 | 0.92 ± 0.03 |
| DM7225 | yggX | 0.98 ± 0.01 | 0.74 ± 0.12 | 0.25 ± 0.01 |
| DM7644 | cyaY | 1.03 ± 0.01 | 0.93 ± 0.02 | 0.74 ± 0.06 |
| DM7307 | apbC | 0.97 ± 0.01 | 0.73 ± 0.04 | 0.62 ± 0.01 |
| DM7642 | apbC cyaY | 1.00 ± 0.01 | 0.37 ± 0.01 | 0.33 ± 0.01 |
| DN7306 | yggX apbC | 0.98 ± <0.01 | 0.35 ± 0.01 | 0.32 ± <0.01 |
| DM7643 | yggX cyaY | 1.00 ± 0.01 | 0.34 ± 0.01 | 0.31 ± 0.02 |
| DM7641 | yggX apbC cyaY | 1.00 ± 0.01 | 0.22 ± 0.01 | 0.19 ± 0.01 |
Overnight cultures grown in rich media were subcultured with 150 μl added to 5 ml nutrient broth (NB) containing 0, 40, or 80 μM paraquat as indicated. Cultures were incubated with shaking at 37°C, and growth was monitored by measurement of optical density at 650 nm (OD650) with a Bausch and Lomb Spectronic 20. The final OD650 reported was after 24 h, since absorbance for all strains had reached a plateau by that time. In all cases, loci indicated for relevant genotypes were inactivated by insertion.
Values reflect the averages (±standard deviations) of two independent cultures.
Growth results detect a complex phenotype caused by a cyaY lesion.
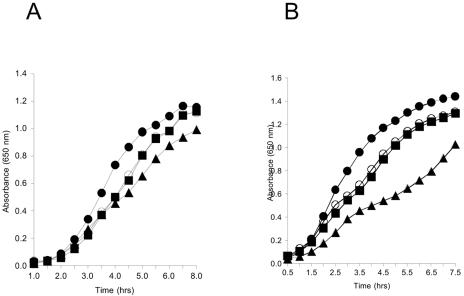
To quantify an apparent growth defect of strains lacking cyaY, yggX, and an additional locus (gshA, apbC, or apbE), growth in both nutrient broth and Luria broth (LB) was monitored. From these growth studies, the following conclusions could be made: when grown in Luria broth, and to a much lesser extent in nutrient broth, the triple mutants grew with a pattern distinct from that of the parental strains. A representative experiment is shown in Fig. 2. As illustrated, the severity of the defect was dependent on the number of mutations present in addition to the cyaY lesion. No growth defect was detected with a strain lacking both apbC and yggX, emphasizing the contribution of the cyaY lesion. Addition of various carbon sources to the medium (glucose, gluconate, or Casamino Acids) eliminated the unusual growth pattern, while addition of others (citrate, succinate, or fumarate) had no effect. A negative effect of acetate on the cell growth in LB was reminiscent of strains lacking NADH:ubiquinone oxidoreductase (EC 1.6.99.3; NADH dehydrogenase complex I) (30). A comparison of growth patterns of a strain lacking nuo (DM4489 [9]) and the triple mutant (cyaY yggX apbC) on a number of rich media proved that the growth patterns, while not identical, showed several similarities (data not shown).
FIG. 2.
Distinct growth pattern of a yggX apbC cyaY mutant in rich medium. Strains were grown and subcultured as described in the legend for Fig. 1. Growth in (A) nutrient broth and (B) Luria broth is shown for strains DM7641 (abpC yggX cyaY) (▴), DM7642 (apbC cyaY) (○), DM7643 (yggX cyaY) (▪), and DM7644 (cyaY) (•). hrs, hours.
The NADH dehydrogenase complex I (NDH-I) is a 14-subunit complex that contains nine Fe-S clusters of various arrangements (18, 21) and is the product of genes in the nuo locus (13). Enzyme assays tested whether the severity of the growth phenotype observed with LB correlated with reduced NDH-I activity. Data from a representative experiment are shown in Table 3, indicating that strains lacking cyaY had a reduced level of NDH-I activity. Under these conditions, a strain lacking nuo had undetectable activity (data not shown). However, the data eliminated a simple correlation between this activity and growth phenotype. An isc mutant (DM7220) did not have the distinct growth pattern of the triple (apbC cyaY yggX) mutant (data not shown), and yet the two strains had similarly low levels of NDH-I activity. Rather, the isc mutant had a reduced growth rate, as previously noted (32, 34). The strain with the second-most-severe defect in activity (apbC yggX) had no growth defect. Conversely, double mutants that lacked cyaY and one of the other loci of interest (e.g., apbC) showed an intermediate growth defect (Fig. 2) yet had enzyme activity similar to that of the cyaY mutant, which had no detectable growth defect.
TABLE 3.
Multiple lesions in combination with a cyaY mutation reduce Nuo activitya
| Strain | Relevant genotype | Nuo activityb | Relative activityc |
|---|---|---|---|
| DM8000 | Wild type | 23,508 ± 136 | 1.00 |
| DM7220 | iscS | 3,990 ± 397 | 0.17 |
| DM7644 | cyaY | 11,420 ± 518 | 0.48 |
| DM7642 | apbC cyaY | 13,631 ± 810 | 0.58 |
| DM7643 | cyaY yggX | 17,790 ± 513 | 0.76 |
| DM5986 | yggX apbC | 6,146 ± 1239 | 0.26 |
| DM7641 | yggX apbC cyaY | 3,375 ± 213 | 0.14 |
Strains were grown in nutrient broth to an optical density at 650 nm of 0.5. Crude cell-free extracts were generated by sonication as has been described previously (35). The activity of the iscS strain reflects the level of cluster synthesis maintained in a strain lacking the major Fe-S cluster biosynthetic operon. All loci listed for relevant genotypes were inactivated by insertion.
Nuo activity was measured as described previously, by using d-amino-NADH to distinguish it from the cytoplasmic activity encoded by the ndh gene (39), and is reported as a specific activity (ΔA600/min/mg protein [average ± standard deviation]).
Relative activity was obtained by dividing the activity of the relevant strain by the activity of the wild-type parent.
On tryptone swarm plates, strains lacking nuo fail to form the inner growth band that reflects chemotaxis to aspartate (30). Relevant strains were inoculated into tryptone swarm agar as described in reference 30. After incubation at 37°C for ∼5 h, of the seven strains tested, only DM4489 (nuo) was unable to form the interior ring of growth (data not shown). This result, consistent with the above findings, indicated that the defect in nuo was not the cause of the growth pattern detected for strains lacking cya in combination with other loci. Thus, a growth phenotype that is specific for a cyaY lesion and is amenable to genetic analysis was identified.
Endogenous Fe(II) levels are not altered in cyaY mutants.
Yeast mutants lacking the frataxin homolog Yfh1 displayed a 10-fold increase in mitochondrial iron levels (1), while analysis of Escherichia coli cyaY mutants showed no aberrant accumulation of endogenous iron (24). Transcription of the Fur reporter entB (as measured by β-galactosidase activity from a lac fusion) should reflect the amount of Fe(II) that the cell senses as available for cellular processes (reviewed in reference 3). A lac fusion in the entB gene was used to monitor Fur activity. When monitored for various stains, transcription of the entB fusion was unaffected by the presence of CyaY (data not shown). While not definitive, this result is consistent with the finding that in E. coli loss of cyaY did not alter the endogenous levels of Fe(II) (24).
Conclusions.
Similarly to the report of Li et al. with E. coli (24), these studies failed to define a significant phenotypic consequence of eliminating the cyaY locus in an otherwise wild-type strain. However, when strains multiply defective in loci involved in Fe-S cluster metabolism are used, phenotypes specific to cyaY lesions emerge. Here it was demonstrated that strains lacking cyaY in combination with one or two other loci are more defective than the parental strains when a number of parameters are monitored. The three loci tested (apbC, yggX, and cyaY) appear to interact differently, depending on the process being monitored, as judged by the different synergies displayed for different phenotypes.
By surveying the defects of various multiple mutant strains, we have defined conditions in which a single mutation (i.e., cyaY) can make a measurable difference in phenotype (i.e., when the parental background is apbC yggX). This result provides a phenotype that can be attributed to the lesion in the cyaY locus. As such, this work lays a foundation for future genetic and biochemical efforts to tease apart the physiological role of CyaY in the context of other gene products involved in Fe-S cluster metabolism in Salmonella enterica.
Results herein are consistent with a complex integration of the three loci investigated, and possibly others, in fine-tuning Fe-S metabolism of the cell. These data illustrate the synergy of complex cellular systems such as Fe-S cluster homeostasis and emphasize the need to consider multiple interacting loci when performing genetic and biochemical studies to dissect the system.
Acknowledgments
This work was supported by competitive grants GM47296 from the NIH and MCB0445654 from the NSF. Funds were also provided by a 21st Century Scientists Scholars Award from the J. M. McDonnell fund to D.M.D. Elizabeth Skovran was supported by a William H. Peterson predoctoral fellowship from the Department of Bacteriology.
We acknowledge the assistance of Inna Larsen in the preparation of the manuscript.
REFERENCES
- 1.Babcock, M., D. de Silva, R. Oaks, S. Davis-Kaplan, S. Jiralerspong, L. Montermini, M. Pandolfo, and J. Kaplan. 1997. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276:1709-1712. [DOI] [PubMed] [Google Scholar]
- 2.Bou-Abdallah, F., S. Adinolfi, A. Pastore, T. M. Laue, and N. D. Chasteen. 2004. Iron binding and oxidation kinetics in frataxin CyaY of Escherichia coli. J. Mol. Biol. 341:605-615. [DOI] [PubMed] [Google Scholar]
- 3.Braun, V. 2003. Iron uptake in Escherichia coli. Front. Biosci. 8:s1409-s1421. [DOI] [PubMed] [Google Scholar]
- 4.Bulteau, A. L., H. A. O'Neill, M. C. Kennedy, M. Ikeda-Saito, G. Isaya, and L. I. Szweda. 2004. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 305:242-245. [DOI] [PubMed] [Google Scholar]
- 5.Campuzano, V., L. Montermini, Y. Lutz, L. Cova, C. Hindelang, S. Jiralerspong, Y. Trottier, S. J. Kish, B. Faucheux, P. Trouillas, F. J. Authier, A. Durr, J. L. Mandel, A. Vescovi, M. Pandolfo, and M. Koenig. 1997. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 6:1771-1780. [DOI] [PubMed] [Google Scholar]
- 6.Campuzano, V., L. Montermini, M. D. Molto, L. Pianese, M. Cossee, F. Cavalcanti, E. Monros, F. Rodius, F. Duclos, A. Monticelli, F. Zara, J. Canizares, H. Koutnikova, S. I. Bidichandani, C. Gellera, A. Brice, P. Trouillas, G. De Michele, A. Filla, R. De Frutos, F. Palau, P. I. Patel, S. Di Donato, J. L. Mandel, S. Cocozza, M. Koenig, and M. Pandolfo. 1996. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271:1423-1427. [DOI] [PubMed] [Google Scholar]
- 7.Chamberlain, S., M. Koenig, A. Richter, F. Palau, and M. Pandolfo. 1993. Molecular analysis of the Friedreich's ataxia locus. Adv. Neurol. 61:193-204. [PubMed] [Google Scholar]
- 8.Chamberlain, S., J. Shaw, A. Rowland, J. Wallis, S. South, Y. Nakamura, A. von Gabain, M. Farrall, and R. Williamson. 1988. Mapping of mutation causing Friedreich's ataxia to human chromosome 9. Nature 334:248-250. [DOI] [PubMed] [Google Scholar]
- 9.Claas, K., S. Weber, and D. M. Downs. 2000. Lesions in the nuo operon, encoding NADH dehydrogenase complex I, prevent PurF-independent thiamine synthesis and reduce flux through the oxidative pentose phosphate pathway in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:228-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui, Q., M. P. Thorgersen, W. M. Westler, J. L. Markley, and D. M. Downs. Solution structure of YggX: a prokaryotic protein involved in Fe(II) trafficking. Proteins, in press. [DOI] [PubMed]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duby, G., F. Foury, A. Ramazzotti, J. Herrmann, and T. Lutz. 2002. A non-essential function for yeast frataxin in iron-sulfur cluster assembly. Hum. Mol. Genet. 11:2635-2643. [DOI] [PubMed] [Google Scholar]
- 13.Falk-Krzesinski, H. J., and A. J. Wolfe. 1998. Genetic analysis of the nuo locus, which encodes the proton-translocating NADH dehydrogenase in Escherichia coli. J. Bacteriol. 180:1174-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flint, D. H. 1996. Escherichia coli contains a protein that is homologous in function and N-terminal sequence to the protein encoded by the nifS gene of Azotobacter vinelandii and that can participate in the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase. J. Biol. Chem. 271:16068-16074. [PubMed] [Google Scholar]
- 15.Flint, D. H., and R. M. Allen. 1996. Iron-sulfur proteins with nonredox functions. Chem. Rev. 96:2315-2334. [DOI] [PubMed] [Google Scholar]
- 16.Foury, F. 1999. Low iron concentration and aconitase deficiency in a yeast frataxin homologue deficient strain. FEBS Lett. 456:281-284. [DOI] [PubMed] [Google Scholar]
- 17.Frazzon, J., and D. R. Dean. 2003. Formation of iron-sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Curr. Opin. Chem. Biol. 7:166-173. [DOI] [PubMed] [Google Scholar]
- 18.Friedrich, T., and B. Böttcher. 2004. The gross structure of the respiratory complex I: a Lego system. Biochim. Biophys. Acta 1608:1-9. [DOI] [PubMed] [Google Scholar]
- 19.Gralnick, J., E. Webb, B. Beck, and D. Downs. 2000. Lesions in gshA (encoding γ-l-glutamyl-l-cysteine synthetase) prevent aerobic synthesis of thiamine in Salmonella enterica serovar typhimurium LT2. J. Bacteriol. 182:5180-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gralnick, J. A., and D. M. Downs. 2003. The YggX protein of Salmonella enterica is involved in Fe(II) trafficking and minimizes the DNA damage caused by hydroxyl radicals: residue CYS-7 is essential for YggX function. J. Biol. Chem. 278:20708-20715. [DOI] [PubMed] [Google Scholar]
- 21.Hinchliffe, P., and L. A. Sazanov. 2005. Organization of iron-sulfur clusters in respiratory complex I. Science 309:771-774. [DOI] [PubMed] [Google Scholar]
- 22.Huynen, M. A., B. Snel, P. Bork, and T. J. Gibson. 2001. The phylogenetic distribution of frataxin indicates a role in iron-sulfur cluster protein assembly. Hum. Mol. Genet. 10:2463-2468. [DOI] [PubMed] [Google Scholar]
- 23.Lezhneva, L., K. Amann, and J. Meurer. 2004. The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 37:174-185. [DOI] [PubMed] [Google Scholar]
- 24.Li, D. S., K. Ohshima, S. Jiralerspong, M. W. Bojanowski, and M. Pandolfo. 1999. Knock-out of the cyaY gene in Escherichia coli does not affect cellular iron content and sensitivity to oxidants. FEBS Lett. 456:13-16. [DOI] [PubMed] [Google Scholar]
- 25.Mansy, S. S., and J. A. Cowan. 2004. Iron-sulfur cluster biosynthesis: toward an understanding of cellular machinery and molecular mechanism. Acc. Chem. Res. 37:719-725. [DOI] [PubMed] [Google Scholar]
- 26.Nair, M., S. Adinolfi, C. Pastore, G. Kelly, P. Temussi, and A. Pastore. 2004. Solution structure of the bacterial frataxin ortholog, CyaY: mapping the iron binding sites. Structure (Cambridge) 12:2037-2048. [DOI] [PubMed] [Google Scholar]
- 27.Pandolfo, M. 2002. The molecular basis of Friedreich ataxia. Adv. Exp. Med. Biol. 516:99-118. [DOI] [PubMed] [Google Scholar]
- 28.Park, S., O. Gakh, H. A. O'Neill, A. Mangravita, H. Nichol, G. C. Ferreira, and G. Isaya. 2003. Yeast frataxin sequentially chaperones and stores iron by coupling protein assembly with iron oxidation. J. Biol. Chem. 278:31340-31351. [DOI] [PubMed] [Google Scholar]
- 29.Petersen, L. A., and D. M. Downs. 1997. Identification and characterization of an operon in Salmonella typhimurium involved in thiamine biosynthesis. J. Bacteriol. 179:4894-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prub, B. M., J. M. Nelms, C. Park, and A. J. Wolfe. 1994. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J. Bacteriol. 176:2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy, A., N. Solodovnikova, T. Nicholson, W. Antholine, and W. E. Walden. 2003. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 22:4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz, C. J., O. Djaman, J. A. Imlay, and P. J. Kiley. 2000. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:9009-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skovran, E., and D. M. Downs. 2003. Lack of the ApbC or ApbE protein results in a defect in Fe-S cluster metabolism in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skovran, E., and D. M. Downs. 2000. Metabolic defects caused by mutations in the isc gene cluster in Salmonella enterica serovar Typhimurium: implications for thiamine synthesis. J. Bacteriol. 182:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skovran, E., C. T. Lauhon, and D. M. Downs. 2004. Lack of YggX results in chronic oxidative stress and uncovers subtle defects in Fe-S cluster metabolism in Salmonella enterica. J. Bacteriol. 186:7626-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong, A., J. Yang, P. Cavadini, C. Gellera, B. Lonnerdal, F. Taroni, and G. Cortopassi. 1999. The Friedreich's ataxia mutation confers cellular sensitivity to oxidant stress which is rescued by chelators of iron and calcium and inhibitors of apoptosis. Hum. Mol. Genet. 8:425-430. [DOI] [PubMed] [Google Scholar]
- 37.Yoon, T., and J. A. Cowan. 2004. Frataxin-mediated iron delivery to ferrochelatase in the final step of heme biosynthesis. J. Biol. Chem. 279:25943-25946. [DOI] [PubMed] [Google Scholar]
- 38.Yoon, T., and J. A. Cowan. 2003. Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 125:6078-6084. [DOI] [PubMed] [Google Scholar]
- 39.Zambrano, M. M., and R. Kolter. 1993. Escherichia coli mutants lacking NADH dehydrogenase I have a competitive disadvantage in stationary phase. J. Bacteriol. 175:5642-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng, L., V. L. Cash, D. H. Flint, and D. R. Dean. 1998. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273:13264-13272. [DOI] [PubMed] [Google Scholar]