Abstract
Bacteriophage genomic evolution has been largely characterized by rampant, promiscuous horizontal gene transfer involving both homologous and nonhomologous source DNA. This pattern has emerged through study of the tailed double-stranded DNA (dsDNA) phages and is based upon a sparse sampling of the enormous diversity of these phages. The single-stranded DNA phages of the family Microviridae, including φX174, appear to evolve through qualitatively different mechanisms, possibly as result of their strictly lytic lifestyle and small genome size. However, this apparent difference could reflect merely a dearth of relevant data. We sought to characterize the forces that contributed to the molecular evolution of the Microviridae and to examine the genetic structure of this single family of bacteriophage by sequencing the genomes of microvirid phage isolated on a single bacterial host. Microvirids comprised 3.5% of the detectable phage in our environmental samples, and sequencing yielded 42 new microvirid genomes. Phylogenetic analysis of the genes contained in these and five previously described microvirid phages identified three distinct clades and revealed at least two horizontal transfer events between clades. All members of one clade have a block of five putative genes that are not present in any member of the other two clades. Our data indicate that horizontal transfer does contribute to the evolution of the microvirids but is both quantitatively and qualitatively different from what has been observed for the dsDNA phages.
Bacteriophage genomes exhibit pervasive genetic mosaicism, appearing as patchworks of genetic modules that encode apparently exchangeable functional units such as the virion coat, the virion tail, or the genome integration proteins (1, 27, 46). As the proximate cause of this mosaicism, horizontal gene transfer is thought to play a large role in the evolution of bacteriophage genomes (1, 4, 21, 23, 39, 49). In phage, horizontal transfer can occur via both homologous and nonhomologous reassortment and appears to be limited primarily by selection against the disruption of functional interactions (23).
This picture of bacteriophage genome evolution emerged primarily from characterizations of double-stranded DNA (dsDNA) phage genomes. The dsDNA phages of the families Siphoviridae (e.g., λ), Podoviridae (e.g., T7), and Myoviridae (e.g., T4) have received the most attention because of their history as molecular biology model systems and their economic impact in dairy fermentation (3, 37). These families comprise the tailed phages, which are characterized by relatively large genomes (from just under 20 to hundreds of kilobases) and often by a lysogenic lifestyle. Both of these characteristics likely facilitate horizontal gene transfer, the former by minimizing constraints on gene acquisition or loss, the latter by increasing the opportunities for recombination between phages.
In contrast, the smaller genomes and lytic lifestyles of the icosahedral single-stranded DNA (ssDNA) and RNA phages are expected to limit the frequency and evolutionary success of horizontal transfer events. Packaging constraints on genome size are expected to limit the assimilation or loss of genetic material (43, 52). In addition, horizontal gene transfer between and within groups of strictly lytic phages would require fortuitous coinfection of a host, an event that would have to occur during the short time a lytic phage spends within an infected host. Thus, genome evolution in the icosahedral ssDNA and RNA phages is expected to proceed primarily vertically, with variation provided not through recombination but through the accumulation of mutations. At present, there are too few genome sequences available to address this expectation. Among the ssDNA phages, homologous recombination and mosaicism have been documented among the five sequenced filamentous phages typified by M13 (27), which are not lytic phages but not among the 12 sequenced icosahedral phages (2).
To further characterize the mechanisms that contribute to molecular evolution among the ssDNA phages, we investigated genome diversity among the Microviridae, the icosohedral ssDNA phages typified by φX174. The Microviridae have 4.5- to 6-kb circular genomes and a tailless icosahedral virion. They have been well studied in molecular biology (20) and in experimental evolution (5, 6, 9, 24, 53). The 12 previously sequenced microvirid phages were isolated on Escherichia coli, Salmonella, Bdellovibrio (2), Chlamydia, (15, 16, 28, 45), and Spiroplasma (41). The initial studies of microvirid genomes seemed to confirm differences in genome evolution from what has been observed in the dsDNA phages. Gene content was constant among closely related genomes, and no horizontal gene transfer was apparent (2).
In this study, we expanded the microvirid genome collection by isolating and sequencing the complete genomes of 42 new microvirid phages capable of infecting E. coli C, the standard laboratory host of φX174. Combining these sequences with the five previously sequenced E. coli phages (φX174, S13, G4, α3, and φK), we examined the broad patterns of evolution and diversity of microvirids isolated on a single host at a single temperature. In addition to providing an extensive characterization of microvirid diversity, our data provide the first characterization of the diversity existing within phage populations. Our extensive genome sampling enabled the identification of multiple horizontal gene transfer events that were not apparent in the previously available sparsely sampled genomes.
MATERIALS AND METHODS
Strains and culture conditions.
The phages φX174 and G4 used in this study were described previously (6, 24). These phages and their standard laboratory hosts Escherichia coli C and Salmonella enterica serovar Typhimurium LT2 strain IJ750 (xyl-404 metA22 metE551 galE719 trpD2 ilv-452 hsdLT6 hsdSA29 hsdSB121 fla-66 rpsL120 H1-b H2-e nix) were obtained from J. J. Bull. All phages and bacteria were grown in LB medium (10 g NaCl, 10 g Bacto tryptone, and 5 g yeast extract per liter) supplemented with 2 mM CaCl2 at 37°C and frozen in LB plus 25% glycerol at −80°C for long-term storage.
Environmental samples.
Environmental samples were taken from the following sources: University of Idaho Research Barns, Moscow, ID, in April and July 2001; Sewage Treatment Plant, Pullman, WA, in November, 2001; Waste Water Treatment Plant, Moscow, ID, in June 2002; and Mason Farm Waste Water Treatment Plant, Chapel Hill, NC, in March 2002.
Genomic hybridization identification of microvirid phages.
As a pilot study, barnyard samples were treated with chloroform, cleared of debris, and plated directly on E. coli C. All individual plaques were stored in 96-well microtiter plates. The genomes of these phage isolates were screened for homology to φX174 and G4 using a blotting method modified from Crill et al. (9). Briefly, phage was replica plated onto lawns of E. coli C and incubated overnight to allow plaque formation. Plaques on this plate were blotted onto a nylon membrane (Hybond-N1 membrane) in 0.4 M NaOH without pretreatment of the membrane or agar. After being blotted, phage DNA was UV cross-linked and baked onto the membrane. Hybridizations were carried out using PCR-amplified whole genomes of φX174 and G4 (separately) as probes.
Sucrose gradient enrichment.
Sewage samples were enriched for microvirid phages with a protocol derived from Godson (17). A total of 500 ml of raw sewage was treated with chloroform to kill any membrane-containing organisms and cleared of debris by centrifugation at 5,000 rpm. NaCl and polyethelene glycol were added to produce final concentrations of 1 M NaCl and 10% polyethelene glycol. Phages were then precipitated by centrifugation at 10,000 rpm for 20 min. The phage pellet was suspended in 1 ml suspension medium (31). A total of 400 μl of the resulting phage concentrate was layered on a 5 to 30% sucrose gradient, which was centrifuged at 24,000 rpm for 110 min at 4°C before fractionation. A sample of φX174 was run concurrently on a parallel gradient to determine which fractions contained microvirid phages. The identified fractions were plated on E. coli C, and 48 of the resulting plaques were saved for genome screening. Fractions from the North Carolina sample were also plated on S. enterica serovar Typhimurium, and 10 of the resulting plaques were saved.
PCR identification of microvirid phages.
Phages that were putatively identified as microvirids through a sucrose gradient or genomic hybridization were further screened for homology to the known microvirid E. coli phages using PCR. Two degenerate primer sets were developed based on conserved regions between the phages φX174, S13, G4, α3, and φK. Primer set 1 consisted of the following: UN1586, CAGAGTT(CT)TATCGCTTC(CA)ATGAC, and UN2180, AGG AGCAGGAAAGCGAGG. Primer set 2 consisted of UN3423, G(TAC)TT(CT)T(TA)(CT)GT(GT)CCTGAGCATGG, and UN4714, (AG)CC(TC)TGAAT(GA) GCAGA(CT)T(GT)AATACC. The numbering for each primer is based on the location of the targeted region in the G4 genome (GenBank accession no. AF454431). We attempted PCR amplification for each isolate with primer set 1 under stringent PCR conditions. Amplifications that failed were repeated under successively less-stringent conditions until nonspecific amplification occurred. This process was repeated using primer set 2 for the phages that did not amplify with primer set 1, and all of these phages again failed to amplify. Isolates that failed under both primer sets were not pursued further.
Molecular determination of genome size.
Phages were grown to high titer at 37°C on E. coli C in 250 ml of phage LB medium and concentrated with 10% polyethelene glycol. Genomic DNA was extracted with phenol-chloroform and run on a 0.7% agarose gel to determine size.
Sequencing and annotation.
Phage isolates were identified by sequencing the PCR product of primer set 1, a region consisting of approximately 600 bp, and given a name composed of two letters identifying their state of origin and a number. Duplicates sequences from the same environmental sample were excluded from genome sequencing to avoid sequencing multiple progeny released from a single host cell. The full genomes of isolates with distinct sequences in the region amplified by primer set 1 were amplified with primers designed either for closely related phages or from the sequences of the two products of degenerate PCR (primer sets 1 and 2). Sequencing primers were designed as needed. Sequences were assembled and proofread in SeqMan from the DNAStar Lasergene package (Madison, WI). Genes in the new isolates were identified by homology with the previously studied microvirids.
Phylogenetic methods.
Alignments for all phylogenetic analyses were generated using ClustalW (50) as implemented in the Megalign program of the DNAStar Lasergene package. Gene alignments were based upon amino acid sequences, and the genome alignment was based upon nucleotide sequences. Nucleotide sequences, with gaps excluded, were used for all phylogenetic analyses. Nucleotide substitution models were selected using DT-ModSel (34).
The genome phylogeny was estimated using Bayesian inference with MRBAYES, version 3.0 (22). Two independent searches were continued for 10 million generations, using the same nucleotide substitution model selected for the likelihood analysis, except that rates were not linked when six substitution types were used. The Metropolis-coupled Markov chain Monte Carlo temperature parameter was set to 0.2 with four chains per search. The first 1 million generations of each search were discarded as burn in, though parameter values typically converged much earlier. Convergence was checked by a visual examination of the parameter values plotted against generation number. The posterior probability of a clade was estimated as the percentage of trees after the burn in that included a particular clade. Two additional 1-million-generation runs for each analysis were performed to further confirm convergence.
Phylogenies of individual genes were estimated by maximum likelihood (ML). An initial tree generated with the neighbor-joining algorithm and logdet distances (29) was used for model parameter estimation in PAUP*, version 4.0b10 (47). Heuristic searches were performed on 10 random addition replicates with tree bisection-reconnection branch swapping. The best tree from this search (or one of the best trees if multiple trees with the same likelihood were recovered) was used to reestimate the parameters, and another identical search was performed. If a different tree topology was found, the process was repeated until consecutive searches yielded identical tree topologies.
Detecting horizontal gene transfer.
To look for evidence of horizontal gene transfer, we examined the evolutionary histories of individual genes. We were concerned only with deep-phylogeny recombination events, i.e., events typically considered as horizontal transfer that tend to produce phylogenetic incongruences across different gene phylogenies. We tested for incongruence between each gene tree and the full genome phylogeny by testing the monophyly of the three major clades present in the genome phylogeny (the φX174-like clade, the G4-like clade, and the α3-like clade). The existence of these three monophyletic groups was used as our null hypothesis, and if the maximum-likelihood phylogeny for a gene differed from this three-clade topology, we performed a parametric bootstrap analysis to determine whether this difference was significant (7, 19). Briefly, a test statistic for the actual data was calculated as the improvement in log likelihood in the maximum-likelihood phylogeny relative to the best tree conforming to the null hypothesis. This test statistic was compared to a null distribution that was approximated with 100 simulated sequence data sets generated with SEQ-GEN (38) on the constrained gene phylogeny. In this statistical analysis, P values correspond to the number of simulated data sets that produced larger improvements in log likelihoods than the actual data. Each simulated data set was analyzed in the same manner as the actual data set, except that only a single random addition replicate was performed. Ideally, we would have generated the null distribution using precisely the same analysis, but this approach was computationally prohibitive. Although the alternative tree search method is less thorough, simulated data are always cleaner than real data, ensuring that the less thorough search, nonetheless, finds the correct tree topology.
Detecting purifying selection.
To assess the functionality of putative open reading frames (ORFs), we performed likelihood ratio tests using PAML, version 3.14 (54), comparing a null model that assumes neutral evolution (i.e., dN/dS = 1) to a model in which the dN/dS ratio is free to vary. The likelihood ratio test compares negative twice the difference in log likelihood scores under the two models to a χ2 distribution with 1 df. Maximum-likelihood phylogenies for this analysis were estimated as described above for gene phylogenies.
Nucleotide sequence accession numbers.
Annotated genomic sequences can be found in GenBank with the following accession numbers: AY751298 and DQ079869 to DQ079909. The five previously sequenced microvirids are as follows: φX174, AF176034; φK, NC_001730; α3, NC_00133 and DQ085810; G4, AF454431; and S13, AF274751.
RESULTS
Sampling.
We used two methods to identify microvirid phages in environmental samples. The first method, in which phage genomes were screened for the ability to hybridize to φX174 and G4 genomic DNA, allowed us to measure the frequency of microvirids relative to other phages in the sample. By this method, we screened 579 individual phage isolates from the University of Idaho barnyards (soil, water, and cow, horse, and goat feces) and identified 20 phages that hybridized to microvirid genomic DNA (all isolates that hybridized to one microvirid genome also hybridized to the other). All 20 of these phages amplified with degenerate primer set 1 (see Materials and Methods); therefore, the frequency of microvirids in this environmental sample (assuming effective hybridization) was 3.5%. These 20 phages were composed of four distinct genotypes, i.e., four genotypes with different sequences in the amplified region (Table 1). One additional phage isolated from an Idaho wastewater sample was identified by this genome hybridization method.
TABLE 1.
Phage isolation from environmental samples
| Sample type and location (no. of sample) | Sampling methoda | Host used for isolation | No. confirmed/ no. screened by PCR | Identity of distinct genotypes |
|---|---|---|---|---|
| Barnyard | ||||
| University of Idaho | H | E. coli | 20/20 | ID2, -8, -11, -12 |
| Wastewater | ||||
| Moscow, ID | H | E. coli | 1/1 | ID1 |
| Moscow, ID | SG | E. coli | 1/1 | ID18 |
| Moscow, ID | SG | E. coli | 40/48 | ID22, -32, -34, -37, -41, -45, -52, -62 |
| Pullman, WA | SG | E. coli | 40/48 | WA2, -3, -4, -6, -10, -11, -13, -14, -45 |
| Chapel Hill, NC | SG | E. coli | 38/48 | NC1, -2, -3, -5, -6, -7, -10, -11, -13, -16, -19, -28, -29, -35, -37, -41 |
| S. enterica serovar Typhimurium | 10/10 | NC51, -56 | ||
| Total | 152/176 | 42 distinct genotypes |
H, identification of microvirids through genomic hybridization with φX174 and G4; SG, our sucrose gradient method.
In the second method, phages collected from Idaho, Washington, and North Carolina wastewater treatment plants were subjected to sucrose gradient centrifugation to separate phages by size. This method did not yield as much information about the makeup of our phage samples, but it identified microvirid phages more efficiently. Gradient fractions containing microvirid-sized virions were allowed to form plaques on either E. coli C or S. enterica serovar Typhimurium, and 155 of the plaque-forming phages were isolated. A total of 132 of these phages successfully amplified with the degenerate primer sets, and a total of 38 distinct genotypes were identified. The environmental origins and identities of these new phage isolates are described in detail in Table 1.
To determine whether the phages that failed to amplify with our degenerate primer sets were nonetheless microvirids, we estimated genome sizes of the 10 isolates from the North Carolina sample that did not amplify. All 10 phages were found to have genomes significantly larger than 10 kb, effectively excluding them from the Microviridae. Thus, our identification method does not appear to be biased against detecting phages that are distantly related to the five laboratory strains used to design the degenerate primers.
Genome properties.
We sequenced the genomes of the 42 new phage isolates and analyzed them together with the previously described phages φX174, S13, G4, α3, and φK. Genome sizes fall into three classes that are consistent with the genome phylogeny (described below). The phages most closely related to φX174 and S13 had the smallest and least-variable genome sizes, ranging from 5,386 to 5,387 bases. The genomes of phages related to G4 range in size from 5,486 to 5,577 bases, and the genomes of phages related to α3and φK range from 6,061 to 6,259 bases. Variability within each group was primarily due to insertions and deletions in intergenic regions.
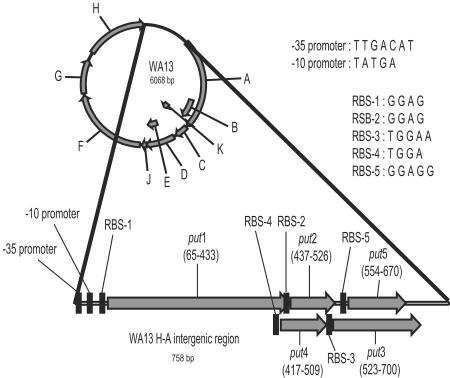
The genomes of the 42 new phage isolates all possessed readily identifiable homologues of the 11 genes common to φX174, S13, G4, α3, and φK (see Table 2 for a list of gene names and functions) (20). In addition, the phages most closely related to α3 and φK (see genome phylogeny below) possessed five additional ORFs in the H-A intergenic region (Fig. 1) and had multiple potential start sites for gene A*. Both of these genome characteristics were noted previously in α3 (26). We tentatively designated the five ORFs as put1 to -5, for putative genes 1 to 5. put1 was disrupted in the published α3 sequence (GenBank accession no. NC_001330) by a single base deletion which was not present in our laboratory strain of α3 (GenBank accession no. DQ085810). The functionality of these ORFs was suggested by the existence of a conserved transcription promoter at the 5′ end of the H-A intergenic region and by the existence of conserved ribosome binding sites preceding all five ORFs (Fig. 1). However, the upstream promoter and the majority of the H-A intergenic region are nonessential for growth in α3 (26).
TABLE 2.
Brief descriptions of the known microvirid proteins (20)
| Protein | Function |
|---|---|
| A | Phage DNA replication |
| A* | Unessential; may be involved in terminating host cell DNA replication |
| B | Internal scaffolding protein |
| C | Initiates production of single-stranded phage genomes and their concomitant packaging into procapsids |
| D | External scaffolding protein |
| E | Host cell lysis |
| F | Major capsid protein |
| G | Major spike protein |
| H | Minor spike protein; DNA pilot protein |
| J | DNA binding protein |
| K | Unessential; unknown function |
FIG. 1.
The H-A intergenic region for the phages clustering with α3 and φK encodes five conserved open reading frames. Gene A* was left off of the genome map because, as for α3, there are multiple potential start sites for this gene. Consensus sequences for ribosome binding sites and the promoter are provided and based upon only the new isolates. Gene positions are based on the sequence of WA13.
We further assessed the functionality of these five ORFs by testing for a significant deviation from a neutral model of evolution. If the region under consideration does not code for a functional protein, then evolution at nonsynonymous sites should not be constrained, and nonsynonymous substitutions should occur as often as synonymous substitutions. This scenario would produce a nonsynonymous-to-synonymous rate ratio (dN/dS) with a value of close to 1, and thus we would fail to reject neutrality. Maximum-likelihood phylogenies were estimated for the nine new isolates containing the extra open reading frames and used to evaluate the likelihood ratio test described above for all of the putative and previously described genes (Table 3). The sequences of α3 and φK were excluded, due to high levels of sequence divergence in some genes (see below).
TABLE 3.
Tests of neutrality for 5 putative genes and 10 previously characterized genes
| Gene | lnL(dN/dS = 1) | lnL(dN/dS = free) | dN/dS | −2lnΛ | Pa |
|---|---|---|---|---|---|
| put1 | −1,636.74 | −1,514.28 | 0.0857 | 244.90 | <0.001 |
| put2 | −213.02 | −208.12 | 0.2196 | 9.80 | <0.001 |
| put3 | −558.68 | −497.39 | 0.0453 | 122.58 | <0.001 |
| put4 | −205.52 | −205.06 | 1.6649 | 0.92 | 0.26 |
| put5 | −313.33 | −312.94 | 1.4360 | 0.79 | 0.30 |
| A | −3,751.12 | −3,504.24 | 0.0576 | 493.76 | <0.001 |
| B | −725.84 | −721.92 | 3.3423 | 7.85 | 0.003 |
| C | −544.12 | −487.63 | 0.0231 | 112.9882 | <0.001 |
| D | −1,125.70 | −1,016.24 | 0.0230 | 218.91 | <0.001 |
| E | −468.19 | −463.29 | 3.9191 | 9.80 | <0.001 |
| F | −3,836.09 | −3,402.21 | 0.0294 | 867.76 | <0.001 |
| G | −1,921.86 | −1,741.17 | 0.0634 | 361.39 | <0.001 |
| H | −3,159.144 | −2,914.78 | 0.0774 | 488.73 | <0.001 |
| J | −162.55 | −148.77 | 0.0380 | 27.57 | <0.001 |
| K | −292.26 | −292.17 | 0.8007 | 0.1734 | 0.88 |
Significance was ascertained by comparing negative twice the difference in log likelihood (−2lnΛ) to a χ2 distribution with 1 df.
put1, put2, put3, and most of the previously described genes show dN/dS ratios of significantly <1, indicating the action of purifying selection and confirming functionality. The genes with dN/dS ratios not significantly different from one (put4, put5, and K), or significantly >1 (B and E) were conspicuously the only genes that are entirely encoded within other genes (Fig. 1). These data suggest that a dN/dS value of 1 is not an appropriate neutral model of evolution within overlapping reading frames and thus provide little information regarding the functionality of put4 and put5, except that their observed dN/dS ratios were not unreasonable for overlapping reading frames.
Genome phylogeny.
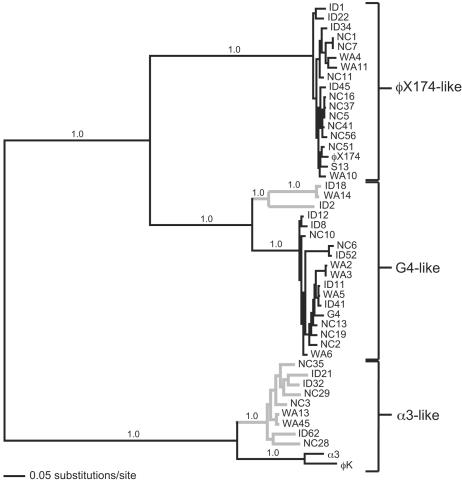
The phylogenetic relationship between our 42 new isolates and the five laboratory strains (φX174, S13, G4, α3, and φK) was estimated using Bayesian inference on the whole-genome alignment. We did not include the seven microvirids that were isolated on hosts other than E. coli (2, 16, 28, 40, 41, 45) in our analysis, because the degree of sequence divergence precludes any reliable alignment or phylogenetic analysis. The maximum a posteriori probability phylogeny (Fig. 2) delineates three well-defined clades, each supported by posterior clade probabilities of 1.0. The phylogenetic tree constructed using maximum likelihood (not shown) differs only in slight rearrangements near the terminal branches. Our new isolates formed three distinct monophyletic groups that cluster with the lab strains: 16 isolates grouped with φX174 and S13, forming the φX174-like clade; 17 isolates grouped with G4, forming the G4-like clade; and 9 isolates grouped with α3 and φK, forming the α3-like clade.
FIG. 2.
Maximum a posteriori probability phylogeny based upon a full genome alignment. Posterior probabilities are given above the relevant branches. At least three distinct clades are apparent and well supported, each including at least one of the previously sequenced laboratory strains. The tree is mid-point rooted for visual clarity.
A χ2 test, as implemented in PAUP*, indicated a significant deviation from equal base frequencies within our full data set (P ≪ 0.01), that was due to a difference in base frequencies among the G4-like clade. Exclusion of the G4-like group eliminates the discrepancy in base frequencies (P = 1.00). We examined the sensitivity of the estimated topology to this variation in base frequencies using the minimum evolution optimality criterion (25, 44) with logdet distances, a distance measure that is less sensitive to unequal base frequencies. The tree topology produced by this alternative method did not differ qualitatively from the Bayesian or maximum-likelihood topologies.
Gene phylogenies.
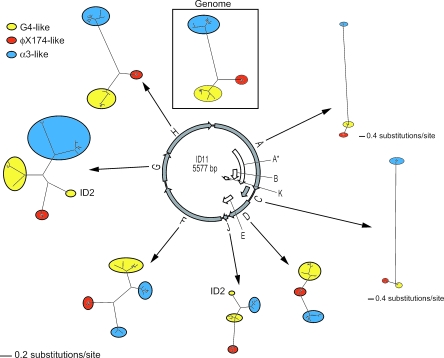
We estimated ML phylogenies for genes A, C, D, F, G, H, and J, excluding the remaining four genes (A*, B, E, and K) because they are encoded entirely within other genes (Fig. 3). Table 2 provides brief descriptions of the functions of the products of these genes. Although the genome phylogeny was well supported, the tree topologies among individual genes varied dramatically, suggesting that the underlying molecular evolution of individual genes was not well represented by the genome phylogeny. In particular, the φX174-like, G4-like, and α3-like clades present in the genome phylogeny (Fig. 2) did not form monophyletic groups in the ML phylogenies of genes A, D, F, G, and J, which together constitute the majority of the genome. Although genes A, F, G, and H showed significant deviation (P ≪ 0.01) from equal base frequencies, their tree topology estimates were not sensitive to this deviation (see above). Thus, the unequal base frequencies within particular genes do not explain their different topologies.
FIG. 3.
Gene phylogenies demonstrate complex evolutionary patterns in genome evolution. The models used for analyses are as follows: A, TIM + I + G; C, K80 + G; D, TrN + I + G; F, TrN + I + G; G, HKY + G; H, GTR + G; J, TrNef + I, as selected by DT-ModSel. Note that genes A and C are on different scales than the rest of the phylogenies.
To assess the significance of the apparent incongruence between gene and genome phylogenies, we performed a parametric bootstrap analysis on each of these gene phylogenies to determine whether the monophyly of these three clades could be rejected statistically. Genes F and G showed a significant deviation from the three clade topology of the genome phylogeny (gene F: −2lnΛ = 108.62; P < 0.01; gene G: −2lnΛ = 1.68; P = 0.01). Gene D showed a marginally nonsignificant deviation (−2LnΛ = 3.70; P = 0.06), and genes A and J did not deviate significantly from the three clade topology (gene A: −2LnΛ = 0.28; P = 0.75; gene J: −2LnΛ = 3.76; P = 0.11).
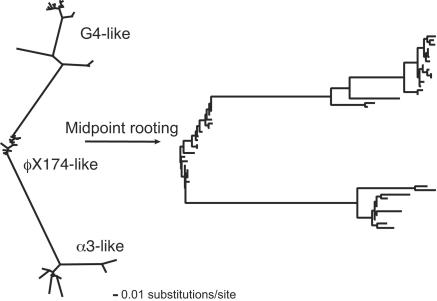
Deviation from the genome topology in gene F, which encodes the major capsid protein, resulted because the two sister taxa that comprise the α3-like clade in the genome phylogeny did not form sister taxa in the gene F phylogeny. The α3-like laboratory strains (α3 and φK) and the nine new α3-like isolates formed sister taxa in the genome phylogeny (shown as gray and black taxa in Fig. 2); however, the new α3-like isolates form a sister taxa to the G4-like clade in the gene F phylogeny (Fig. 3). Deviation from the genome topology in gene G, which encodes the major spike protein, was due to the movement of a single isolate, ID2, from within in the G4-like clade to a basal point along the φX174-like lineage. Although its deviation was marginally nonsignificant, the phylogeny for the gene encoding the internal scaffolding protein, gene D, is difficult to reconcile with the genome tree. The φX174-like group is not united into a single monophyletic group and both the α3-like and G4-like lineages emerged from within the φX174-like group (Fig. 4).
FIG. 4.
Gene D maximum-likelihood phylogeny places the ancestors of the α3-like group and the G4-like group within the φX174-like group. This and the slower rate of evolution of gene D suggest that the current incarnation of gene D for these other groups arose in the φX174-like group and subsequently spread to the other groups.
DISCUSSION
Our data refine the existing picture of diversity among the microvirid phages that infect E. coli. We established that microvirids represent a small minority of coliphages in at least one environment; only 3.5% of randomly sampled phages belonged to this family. Early investigations of the phages α3, φK, φX174, S13, and G4 and numerous unsequenced isolates used phenotypic characterizations to cluster phages into various numbers of groups. Host range suggested two groups; temperature range and strategy of DNA replication suggested three groups (18). However, group boundaries were indistinct; the variation was sufficiently large to lead Godson to suggest that “the [icosahedral] phages are better viewed as a continuous spectrum of differences of the same basic genome structure” (18). By intensively sampling microvirid phages capable of infecting a single strain of E. coli, we demonstrated that the microvirids do not vary continuously. In contrast, these phages form three distinct phylogenetic clades, each of which has at least one representative among the original five genomes. This phylogenetic clustering into three clades is further supported by genome size and gene content differences between the clades. Genome sizes delineate three distinct classes of phages in our samples, and these classes correspond exactly to the three clades apparent in the genome phylogeny (Fig. 2). These size differences are due, in part, to the five additional (putative) genes possessed by members of the α3-like clade.
Although our presentation of the data divided the sampled phages into only three distantly related monophyletic groups (Fig. 2), several other well-supported divisions exist. Both the G4-like and α3-like clades possess two monophyletic groups of phages that differ from each other by average pairwise uncorrected distances of >0.15 at the nucleotide level. Therefore, we now ask whether there are reasons to further subdivide the three major clades into smaller groups that better represent phage species. As proposed by Lawrence et al. (27), we attempt to identify biologically meaningful groups, or species, by examining the mechanisms that lead to cohesion within groups, including ecological isolation, periodic selection, and genetic exchange.
Ecological isolation and periodic selection.
Theoretical models demonstrate that low rates of recombination can generate sequence clusters corresponding to ecologically distinct groups (8). In these models, populations comprising different clusters persist and diverge only if they exist in niches that are sufficiently different to escape purging through periodic selection events. Following this logic, Palys et al. (35) suggested a phylogenetic species concept in which sequence clusters with ratios of average pairwise between-group to within-group distances of >2 are indicative of separate species. By this criterion, five ecologically distinct phage species can be identified in our collection. The φX174-like clade (Fig. 2) consists of one species that cannot be further split; the G4-like clade consists of at least two species (shown as gray and black clades in Fig. 2); and the α3-like clade consists of two species (also divided into gray and black clades in Fig. 2). Following the suggestion of Rohwer and Edwards (42) to name bacteriophage species after their first described member, we designate these microvirid species as φX174 like, G4 like, ID18 like, α3 like, and WA13 like.
Anecdotally, ecological differences between the G4-like and ID18-like sister taxa and between the α3-like and WA13-like sister taxa further suggest that the extent of divergence between these clades accurately describes species boundaries. The ID18-like phages ID18 and WA14 are capable of growth at lower temperatures than all but one of the G4-like phages; ID2 has a temperature range similar to the majority of the G4-like phages. The host strains used to isolate the α3-like phages, E. coli B and E. coli K-12 (18), do not support the growth of the WA13-like phages (data not shown).
Horizontal transfer.
Attempts to apply the biological species concept to microbes led to the suggestion that microbial species might, alternatively, be defined by the ability to undergo genetic exchange (13). By this definition, microbes whose genes produce incongruent phylogenies as a result of horizontal transfer would be considered members of the same species. Clearly, this definition is too restrictive to accurately describe microvirid species, as it would lump all of the microvirid coliphages into a single species despite the 40% uncorrected nucleotide sequence divergence between the most divergent clades. This is not the first case in which reticulate classification of bacteriophage species, i.e., a classification that allows recombination between species, has proven necessary (27).
Two or possibly three genes show a signature of horizontal transfer. The phylogenies of genes F and G, and possibly gene D, deviate from the three-clade topology of the genome phylogeny, suggesting that these genes have been horizontally transferred between clades. Despite the different topologies of the gene D, F, and G phylogenies, a similar conclusion can be drawn from all three events. In each case, horizontal transfer occurred between groups that we would classify as distinct species by a phylogenetic species concept. Thus, species boundaries do not present a complete barrier to genetic exchange among the microvirid coliphages.
By examining the outcomes of specific horizontal transfer events, we can gain further insight into the identity of species boundaries and the strength of the barrier to genetic exchange between microvirid species. For example, the evolutionary outcome of the gene F transfer confirms the identity of the WA13-like clade as a species that is distinct from the α3-like clade. The extent of divergence between these clades in the genome phylogeny suggests their existence as distinct species from a phylogenetic viewpoint. Conceptually, we can confirm their existence as species if they comprise clusters that are sufficiently different ecologically to escape purging through periodic selection events. The horizontal transfer of gene F confirms the species boundary between these two sister clades, because it was followed by a periodic selection event that caused the newly transferred gene to sweep through only one of these sister taxa. Regardless of whether the recipient clade was the α3-like clade or the WA13-like clade, it is evident that the transferred gene is present in every member of the recipient clade, because the division between these clades in the gene F phylogeny is identical to the division in the genome phylogeny.
The monophyly of the WA13-like and α3-like sister taxa in most genes in the genome suggests one of two alternative evolutionary histories for these taxa. First, the WA13-like and α3-like sister taxa may have existed as distinct species before the transfer of gene F. In this case, after the transfer, a selective sweep of the newly transferred gene was limited to only the recipient species. Alternatively, the transfer of gene F may have served as the initial trigger of divergence of these two sister taxa. As we have discussed above, populations comprising different clusters can persist and diverge only if they exist in niches that are sufficiently different to escape purging through periodic selection events. It is possible that the transfer of gene F allowed the two sister clades to occupy sufficiently different niches because gene F encodes the major capsid protein, a known determinant of host specificity in φX174 (9, 12) and because the host ranges of these phages are different. It is pertinent to this hypothesis that we failed to isolate close relatives of α3 and φK on E. coli C and that both φK and α3 were isolated on other E. coli host strains (18). Regardless of which evolutionary history is correct, the sweep of the transferred gene F through only one of the two sister taxa confirms that the WA13-like and α3-like clades are distinct species.
Unlike the transfer of gene F, which provides insight into the identity of species boundaries, the transfer of gene D (if confirmed) would suggest that these boundaries can be blurred. The phylogeny of gene D, which encodes the external scaffolding protein, has an unusual topology in which the ancestors of the G4-like, ID18-like, α3-like, and WA13-like clades appear to exist within the φX174-like clade (Fig. 4). The only firm conclusion that can be drawn from this topology is that during the time since the existence of the most recent common ancestor for all clades, gene D experienced selection for divergence in all of the phage groups except the φX174-like clade. Thus, unique fixation events appear to have occurred within all groups except for the φX174-like clade. Beyond this conclusion, we can only speculate on the evolutionary scenario generating this pattern.
Taken together with the absence of a gene D homologue among the microvirid phages isolated on hosts other than E. coli (2), data from the gene D phylogeny suggest that the current incarnation of gene D arose relatively recently in the φX174-like clade and spread through recombination into the other groups. Consistent with this scenario, gene D shows the least total divergence of any microvirid gene (Fig. 3), both in the region that overlaps with gene E and in the nonoverlapping region (unpublished observations). This low level of divergence could be construed as being indicative not of a reduced rate of evolution, but of a reduced period of time for evolution to occur.
This hypothesized spread of gene D throughout the microvirid coliphages does not match our expectation that the spread of transferred genes should be halted by species boundaries. Rather, the gene D topology may demonstrate that species boundaries do not always pose a barrier to horizontal transfer. The gene D event(s) may represent an example of a globally adaptive mutation, i.e., a mutation with beneficial effects that transcend specific ecologies. While these events are expected to reduce the rate of divergence between species, they should still allow the formation of sequence clusters corresponding to ecological distinct species (30).
The final transfer event observed in our collection, the transfer of gene G, affected only phage ID2. ID2 is a member of the ID18-like clade that appears to have acquired gene G from the φX174-like clade. Further sampling will be required to determine whether this recombinant genotype exists as a rare variant member of one of the sampled phage species or whether it is part of a distinct species that was under-sampled in our collection. As in the other two transfer events, the transfer affecting ID2 occurred between microvirid species. Our method was designed to detect horizontal transfer between species; thus, the events detected all acted to reduce the rate of divergence between phage species, maintaining similarities between microvirids.
Some patterns of gene evolution in our data differ from the genome phylogeny, but their significance and biological meaning cannot be ascertained with our current set of sequences. For example, the branch leading to the α3-like and WA13-like clades in the phylogeny of gene A is exceptionally long, and the divergence between the α3-like and the WA13-like clades in gene G is much higher than observed elsewhere in the genome (Fig. 3). These events may merely represent elevated rates of evolution, or they could be indicative of horizontal transfer involving unsampled clades. Gene J has a topology similar to that of gene D, but was not found to be significantly different than the genome phylogeny, possibly due to the short length of this gene.
Genome evolution in the Microviridae and in other DNA phages.
By intensively sampling from a single family of phages capable of infecting a single host strain, we characterized phage diversity among a number of species within a phage subfamily. We isolated a sufficiently large number of phages within individual clades to assert with confidence that more intensive sampling will not overturn the monophyly of those clades. Even so, the existence of microvirid coliphages that apparently inhabit ecological niches that are poorly represented in our sample (e.g., ID18, WA14, ID2, α3, and φK) suggests that we are at least missing close relatives of the sampled phages. It will not be surprising if additional species are identified by further sampling efforts.
Although we observed variance in gene content among the microvirid coliphages and there is further variance in gene content among the microvirids isolated on other hosts (2), this variance seems to result from a different phenomenon than in the dsDNA phages. Lysogeny allows phages to improve their fitness by improving their host's fitness. Thus, dsDNA phages often possess nonhomologous sequence that appears to provide an incentive for prophage retention (23). The lytic lifestyle of the microvirids eliminates this form of selection, and the strict constraints on genome size (43, 52) limit the acquisition or loss of DNA. Variation among the dsDNA phage genomes in the number and identity of encoded genes results both from the reassortment of functionally equivalent modules between genomes and from the gain or loss of genes. In contrast, there is no signature of modular reassortment or organization among the microvirids. Variation in gene content appears to result only from the gain or loss of certain genes; for our data set, this variation can be ascribed to a single event involving the H-A intergenic region in the ancestor of the WA13-like and α3-like phages.
It is suprising that modular organization was not apparent in the patterns of horizontal gene transfer in the microvirids. The modularity of the dsDNA phages likely facilitates the transfer of sets of interacting genes, minimizing the conflict of the new genetic material with its recipient genome and allowing successful exchange across highly divergent genomes. Yet with the microvirids we have observed horizontal transfer of genes across species despite strong interactions with other genes in the genome. For example, the F protein is known to physically interact with proteins B, D (10, 11), G (32, 33, 51), J (32, 33), and possibly A (14, 48), yet it appears to have been transferred independently. Such horizontal transfer events require coinfection by divergent phages. In an environment where coinfection of hosts by two divergent phages is common, selection for the ability to produce functional chimeric phages might make successful exchange of single genes more likely to produce functional genomes. If these recombinant phages are able to occupy a new ecological niche, subsequent evolution could produce the patterns of divergence in our data.
The difference in mechanisms of genome evolution between these phages is most apparent in the inability to delineate viral lineages among the dsDNA phages with a criterion that is based strictly on genetic distance (i.e., phylogenetics), whereas species were easily defined among the microvirid phages with a phylogenetic criterion. This difference results from a reduced rate of horizontal transfer among the ssDNA phages. Although horizontal transfer occurs among the Microviridae, it appears to result primarily from homologous recombination, and it has not occurred at a sufficiently high rate to obscure the species phylogeny when appropriate analytical methods are used.
The picture of diversity revealed in this study differs from that of dsDNA phages. Although we have sampled more intensively over smaller genetic distances, it is unlikely that similar sampling of dsDNA phages would reveal obvious phylogenetic clustering into phage species. It is apparent that the genetic diversity of the dsDNA phages infecting a single host is much greater than that of the Microviridae; thus, more sampling would be required to delineate any species clustering analogous to our observations. However, for the tailed dsDNA phages, no two isolates have yet been found to be genomically similar at levels comparable to our phage species (36), yet regions or genetic modules are often found with as little sequence divergence as found within microvirid species (23, 36), suggesting that at least some modules have been thoroughly sampled. Thus, it is unlikely that the different patterns of genome diversity to emerge from our study of ssDNA phages and previous studies of dsDNA phages result from a difference in sampling intensity.
While the microvirid genome phylogeny provides strong evidence of the existence of well-defined species, it also underscores potential complications in phylogenetic inference on large and complex data sets. It is particularly striking that the genome phylogeny is well supported, despite the significantly different topologies of multiple genes. The genome phylogeny gives no hint of the underlying complexities of genome evolution uncovered with gene-by-gene analysis. Thus, by averaging data from across the evolutionary histories of the individual genes, phylogenetic analyses of whole genomes appear likely to miss even strong signatures of horizontal transfer.
Acknowledgments
The work was supported by a grant from the National Institutes of Health (P20 RR16448). Analytical resources and support for D.R.R. were provided by the Idaho INBRE Program (NIH P20 RR16454). C.L.B. was supported by a grant from the National Science Foundation (DBI 0102079).
We thank Z. Abdo for assistance in the laboratory, B. Carstens and J. Sullivan for many helpful discussions, and J. J. Bull and J. Huelsenbeck for their participation in the first phage-collecting trip.
REFERENCES
- 1.Botstein, D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484-491. [DOI] [PubMed] [Google Scholar]
- 2.Brentlinger, K. L., S. Hafenstein, C. R. Novak, B. A. Fane, R. Borgon, R. McKenna, and M. Agbandje-McKenna. 2002. Microviridae, a family divided: isolation, characterization, and genome sequence of φMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovorus. J. Bacteriol. 184:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brüssow, H. 2001. Phages of dairy bacteria. Annu. Rev. Microbiol. 55:283-303. [DOI] [PubMed] [Google Scholar]
- 4.Brüssow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 5.Bull, J. J., M. R. Badgett, and H. A. Wichman. 2000. Big-benefit mutations in a bacteriophage inhibited with heat. Mol. Biol. Evol. 17:942-950. [DOI] [PubMed] [Google Scholar]
- 6.Bull, J. J., M. R. Badgett, H. A. Wichman, J. P. Huelsenbeck, D. M. Hillis, A. Gulati, C. Ho, and I. J. Molineux. 1997. Exceptional convergent evolution in a virus. Genetics 147:1497-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carstens, B. C., A. L. Stevenson, J. D. Degenhardt, and J. Sullivan. 2004. Testing nested phylogenetic and phylogeographic hypotheses in the Plethodon vandykei species group. Syst. Biol. 53:781-792. [DOI] [PubMed] [Google Scholar]
- 8.Cohan, F. M. 1994. The effects of rare but promiscuous genetic exchange on evolutionary divergence in prokaryotes. Am. Nat. 143:965-986. [DOI] [PubMed] [Google Scholar]
- 9.Crill, W. D., H. A. Wichman, and J. J. Bull. 2000. Evolutionary reversals during viral adaptation to alternating hosts. Genetics 154:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dokland, T., R. A. Bernal, A. Burch, S. Pletnev, B. A. Fane, and M. G. Rossman. 1999. The role of scaffolding proteins in the assembly of the small, single-stranded DNA virus φX174. J. Mol. Biol. 288:595-608. [DOI] [PubMed] [Google Scholar]
- 11.Dokland, T., R. McKenna, L. L. Ilag, B. R. Bowman, N. L. Incardona, B. A. Fane, and M. G. Rossmann. 1997. Structure of a viral procapsid with molecular scaffolding. Nature 389:308-313. [DOI] [PubMed] [Google Scholar]
- 12.Dowell, C. E., H. S. Jansz, and J. Zandberg. 1981. Infection of Eschericia coli K-12 by bacteriophage φX-174. Virology 114:252-255. [DOI] [PubMed] [Google Scholar]
- 13.Dykhuizen, D. E., and L. Green. 1991. Recombination in Escherichia coli and the definition of biological species. J. Bacteriol. 173:7257-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekechukwu, M. C., D. J. Oberste, and B. A. Fane. 1995. Host and φX174 mutations affecting the morphogenesis or stabilization of the 50S complex, a single-stranded DNA synthesizing intermediate. Genetics 140:1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everson, J. S., S. A. Garner, P. R. Lambden, B. A. Fane, and I. N. Clarke. 2003. Host range of chlamydiaphages φCPAR39 and Chp3. J. Bacteriol. 185:6490-6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garner, S. A., J. S. Everson, P. R. Lambden, B. A. Fane, and I. N. Clarke. 2004. Isolation, molecular characterisation and genome sequence of a bacteriophage (Chp3) from Chlamydophila pecorum. Virus Genes 28:207-214. [DOI] [PubMed] [Google Scholar]
- 17.Godson, G. N. 1974. Evolution of φX174. Isolation of four new φX-like phages and comparison with φX174. Virology 58:272-289. [DOI] [PubMed] [Google Scholar]
- 18.Godson, G. N. 1978. The other isometric phages, p. 103-112. In D. T. Denhardt, D. Dressler, and D. S. Ray (ed.), The single-stranded DNA phages. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Goldman, N., J. P. Anderson, and A. G. Rodrigo. 2000. Likelihood-based tests of topologies in phylogenetics. Syst. Biol. 49:652-670. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi, M., A. Aoyama, D. L. Richardson, and M. N. Hayashi. 1988. Biology of the bacteriophage φX174, p. 1-71. In R. Calendar (ed.), The bacteriophages, vol. 2. Plenum Press, New York, N.Y. [Google Scholar]
- 21.Hendrix, R. W. 2002. Bacteriophages: evolution of the majority. Theor. Popul. Biol. 61:471-480. [DOI] [PubMed] [Google Scholar]
- 22.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 23.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 24.Kichler Holder, K., and J. J. Bull. 2001. Profiles of adaptation in two similar viruses. Genetics 159:1393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kidd, K. K., and L. A. Sgaramella-Zonta. 1971. Phylogenetic analysis: concepts and methods. Am. J. Hum. Genet. 23:235-252. [PMC free article] [PubMed] [Google Scholar]
- 26.Kodaira, K.-I., K. Nakano, S. Okada, and A. Taketo. 1992. Nucleotide sequence of the genome of the bacteriophage α3: interrelationship of the genome structure and the gene products with those of the phages, φX174, G4 and φK. Biochim. Biophys. Acta 1130:277-288. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence, J. G., G. F. Hatfull, and R. W. Hendrix. 2002. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J. Bacteriol. 184:4891-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, B. L., J. S. Everson, B. Fane, P. Giannikopoulou, E. Vretou, P. R. Lambden, and I. N. Clarke. 2000. Molecular characterization of a bacteriophage (Chp2) from Chlamydia psittaci. J. Virol. 74:3464-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lockhart, P. J., M. A. Steel, M. D. Hendy, and D. Penny. 1994. Recovering evolutionary trees under a more realistic model of sequence evolution. Mol. Biol. Evol. 11:605-612. [DOI] [PubMed] [Google Scholar]
- 30.Majewski, J., and F. M. Cohan. 1999. Adapt globally, act locally: the effect of selective sweeps on bacterial sequence diversity. Genetics 152:1459-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.McKenna, R., L. L. Ilag, and M. G. Rossmann. 1994. Analysis of the single-stranded DNA bacteriophage φX174, refined at a resolution of 3.0 Å. J. Mol. Biol. 237:517-543. [DOI] [PubMed] [Google Scholar]
- 33.McKenna, R., D. Xia, P. Willingmann, L. L. Ilag, S. Krishnaswamy, M. G. Rossmann, N. H. Olson, T. S. Baker, and N. L. Incardona. 1992. Atomic structure of single-stranded DNA bacteriophage φX174 and its functional implications. Nature 355:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minin, V., Z. Abdo, P. Joyce, and J. Sullivan. 2003. Performance-based selection of likelihood models for phylogeny estimation. Syst. Biol. 52:1-10. [DOI] [PubMed] [Google Scholar]
- 35.Palys, T., L. K. Nakamura, and F. M. Cohan. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145-1156. [DOI] [PubMed] [Google Scholar]
- 36.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, J. William R. Jacobs, R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 37.Proux, C., D. van Sinderen, J. Suarez, P. Garcia, V. Ladero, G. F. Fitzgerald, F. Desiere, and H. Brüssow. 2002. The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J. Bacteriol. 184:6026-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rambaut, A., and N. C. Grassley. 1997. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 13:235-238. [DOI] [PubMed] [Google Scholar]
- 39.Ravin, V., N. Ravin, S. Casjens, M. E. Ford, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 299:53-73. [DOI] [PubMed] [Google Scholar]
- 40.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. Deboy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renaudin, J., M.-C. Pascarel, and J.-M. Bové. 1987. Spiroplasma virus 4: nucleotide sequence of the viral DNA, regulatory signals, and proposed genome organization. J. Bacteriol. 169:4950-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell, P. W., and U. R. Müller. 1984. Construction of bacteriophage φX174 mutants with maximum genome sizes. J. Virol. 52:822-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rzhetsky, A., and M. Nei. 1992. A simple method for estimating and testing minimum-evolution trees. Mol. Biol. Evol. 9:945-967. [Google Scholar]
- 45.Storey, C. C., M. Lusher, and S. J. Richmond. 1989. Analysis of the complete nucleotide sequence of Chp1, a phage which infects avian Chlamydia psittaci. J. Gen. Virol. 70:3381-3390. [DOI] [PubMed] [Google Scholar]
- 46.Susskind, M. M., and D. Botstein. 1978. Molecular genetics of bacteriophage P22. Microbiol. Rev. 42:385-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swofford, D. L. 1998. Phylogenetic analysis using parsimony* (PAUP*), version 4.0. Sinauer Associates, Sunderland, MA.
- 48.Tessman, E. S., and P. K. Peterson. 1976. Bacterial rep− mutations that block development of small DNA bacteriophages late in infection. J. Virol. 20:400-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tétart, F., C. Desplats, M. Kutateladze, C. Monod, H.-W. Ackermann, and H. M. Krisch. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J. Bacteriol. 183:358-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tonegawa, S., and M. Hayashi. 1970. Intermediates in the assembly of φX174. J. Mol. Biol. 48:19-42. [DOI] [PubMed] [Google Scholar]
- 52.van der Ende, A., R. Teertstra, and P. J. Weisbeek. 1982. Initiation and termination of the bacteriophage φX174 rolling circle DNA replication in vivo: packaging of plasmid single-stranded DNA into bacteriophage φX174 coats. Nucleic Acids Res. 10:6849-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wichman, H. A., M. R. Badgett, L. A. Scott, C. M. Boulianne, and J. J. Bull. 1999. Different trajectories of parallel evolution during viral adaptation. Science 285:422-424. [DOI] [PubMed] [Google Scholar]
- 54.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biol. Sci. 13:555-556. [DOI] [PubMed] [Google Scholar]