Abstract
The Shigella flexneri transcription activator MxiE is produced by transcriptional slippage from two overlapping open reading frames. By using plasmids encoding a mxiD-lacZ translational fusion, we showed that transcriptional slippage in mxiE influences both transcription and translation of the downstream mxiD gene encoding an essential component of the type III secretion apparatus.
Bacteria belonging to Shigella spp. are responsible for shigellosis in humans. They use a virulence plasmid-encoded type III secretion (TTS) system to induce their entry into epithelial cells (7). Twenty adjacent mxi and spa genes encode the TTS apparatus (TTSA) and the transcription activator MxiE, an AraC family member (1). MxiE controls transcription of a set of genes encoding proteins that transit through the TTSA, and its activity is regulated by TTSA activity (3, 8). MxiE production is dependent upon transcriptional slippage in the region of overlap between the 59-codon open reading frame (ORF) mxiEa containing the translation start site and the 214-codon ORF mxiEb encoding the DNA binding domain (9). Transcriptional slippage involves incorporation, by the RNA polymerase, of nontemplated nucleotides into the mRNA (2, 5, 10). The mxiE slippage site consists of a run of 9 A's and its efficiency is ≈30%, i.e., one out of three mRNA molecules contains 10 U's. This additional nucleotide places mxiEb in the mxiEa reading frame, permitting translation of mxiEab and production of MxiE (9). The reason why MxiE is encoded by transcriptional slippage is not known. The mxiEb 3′ region overlaps the 5′ region of the downstream mxiD gene encoding a TTSA component. Since transcriptional slippage directly controls translation of the mRNA downstream from the slippage site and (i) adjacent genes are subject to translational coupling (6) and (ii) premature translation termination within a gene reduces transcription of downstream genes (4), we investigated the consequences of transcriptional slippage in mxiE on translation and transcription of mxiD.
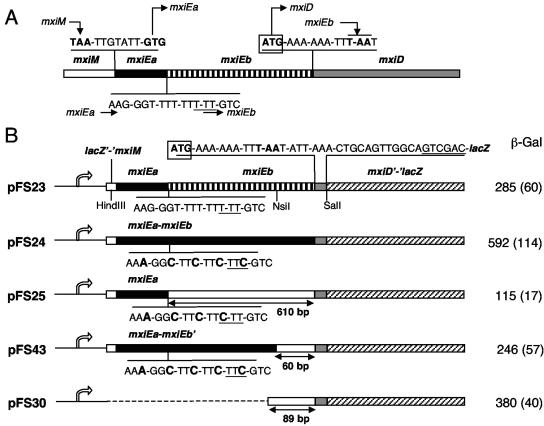
Due to the virulence plasmid instability, the slippage site could not be mutagenized at the mxiE locus to analyze expression of mxiD encoded by the virulence plasmid. Instead, we used low-copy-number plasmids carrying an mxiD-lacZ translational fusion. pFS23, pFS24, and pFS25 (Fig. 1) contain a DNA fragment encompassing the last 17 codons of mxiM, the entire mxiEa and mxiEb ORFs, and the first 7 codons of mxiD cloned between the HindIII and SalI sites of pFS10, i.e., between a lac promoter and codon 15 of lacZ (9). These plasmids differ by the slippage site sequence, which affects translation efficiency of mxiEb (compared to mxiEa). pFS23 contains the wild-type slippage site (mxiEb translation at 30% efficiency), pFS24 contains a mutated site such that mxiEb is in the mxiEa reading frame (mxiEb translation at 100% efficiency), and pFS25 contains a mutated site such that mxiEb is not translated (mxiEb translation at 0% efficiency). Each plasmid was introduced into the Shigella flexneri wild-type strain M90T-Sm (which does not contain a lacZ gene), and β-galactosidase activity was assayed in bacteria harvested in the exponential phase of growth at 37°C in tryptocasein soy broth.
FIG. 1.
Inserts carried by plasmids bearing mxiD-lacZ fusions and β-galactosidase activities assayed in S. flexneri strains harboring these plasmids. (A) The mxiM-mxiEa and mxiEb-mxiD intergenic regions are indicated above and the mxiE slippage site is below a schematic map of the mxiMED region (not shown to scale). Start and stop codons are in boldface type, the mxiD start codon is boxed, and arrows indicate reading frames. (B) On the left, the arrow corresponds to the lac promoter, and the small open box corresponds to the first codons of the α-peptide coding sequence, up to the HindIII site of pSU19, fused in frame to the last 17 codons of mxiM (lacZ′-′mxiM); on the right, the striped box corresponds to the first 7 codons of mxiD fused in frame to codon 15 of lacZ (mxiD′-′lacZ). The slippage site sequences are shown below the map of each plasmid, with mutated nucleotides indicated in boldface type. Underlined nucleotides indicate the mxiEb reading frame. The box corresponding to mxiEb is striped to indicate that mxiEb is expressed with 30% efficiency compared to mxiEa, filled to indicate that mxiEb is in the mxiEa reading frame, and empty to indicate that mxiEb or its 3′ region is not translated. Numbers below double-headed arrows indicate the length of the mxiEb region that is not translated. β-Galactosidase activities assayed in derivatives of M90T-Sm harboring each plasmid are indicated on the right. Activities are expressed in Miller units and correspond to mean values of triplicate assays made from at least four independent cultures; standard deviations are indicated in parentheses.
Strains harboring pFS24 (mxiEb always translated) and pFS25 (mxiEb not translated) contained 592 and 115 U of β-galactosidase activity, respectively, indicating that translation of mxiEb leads to a fivefold increase in the expression of MxiD-LacZ. When mxiEb is not translated, decreased expression of MxiD-LacZ might be due to the absence of a translational coupling between mxiEb and mxiD, decreasing translation of mxiD-lacZ, and/or to a premature transcription termination over the 610-bp mxiEb ORF, decreasing transcription of mxiD-lacZ. To test these hypotheses, we constructed pFS43 by treating pFS24 (mxiEb always translated) with NsiI and mung bean nuclease, resulting in the deletion of an 8-bp fragment around the NsiI site that introduced a stop codon in mxiEb 60 bp upstream from mxiD. In pFS43, translational coupling, if any, between mxiEb and mxiD should be abolished, and the mxiEb 3′ untranslated region should be too short to induce a polar effect on mxiD-lacZ transcription. The strain harboring pFS43 produced 246 U of β-galactosidase activity, i.e., 2.4 times less than the strain harboring pFS24. This indicates that reduced translation of the 3′ region of mxiEb resulted in a concomitant reduction in expression of MxiD-LacZ, suggesting translational coupling of mxiEb and mxiD expression. Moreover, the strain harboring pFS43 (carrying mxiEb with a 3′ region of 60 nucleotides that cannot be translated) contained 2.1 times more β-galactosidase activity than the strain harboring pFS25 (carrying the 610-bp mxiEb ORF that cannot be translated). Since there is no translational coupling between mxiEb and mxiD carried by pFS25 and pFS43, this result suggests that lack of translation of mxiEb induces transcription termination over the 610-bp mxiEb ORF. This polar effect was confirmed by an assay of β-galactosidase activity in the strain harboring pFS30, a plasmid that carries only the last 89 bp (that cannot be translated) of mxiEb upstream from mxiD-lacZ (Fig. 1). This strain contained 380 U of β-galactosidase activity, i.e., 3.3 times more than the strain harboring pFS25 (carrying the 610-bp mxiEb ORF that cannot be translated).
The strain harboring pFS23 (wild-type slippage site) contained 285 U of β-galactosidase activity. Since transcriptional slippage results in the production of one-third of the mxiE mRNA population in which mxiEb is in the mxiEa reading frame (9), the β-galactosidase activity obtained in strain harboring pFS23 is consistent with that calculated by using the slippage efficiency (one-third) and β-galactosidase activities assayed with strains harboring pFS24 (mxiEb always translated; 592 U) and pFS25 (mxiEb not translated; 115 U), as follows: (1/3 × 592) + (2/3 × 115) = 274 U. Moreover, the β-galactosidase activity assayed in the strain harboring pFS23 (wild-type slippage site) was half of that assayed in the strain harboring pFS24 (mxiEb always translated). This indicates that encoding of MxiE by two ORFs that are placed in the same reading frame by transcriptional slippage, as opposed to encoding of MxiE by a single ORF, leads to a twofold reduction in MxiD expression.
In conclusion, transcriptional slippage in mxiE controls production of MxiE, the transcription activator regulated by TTSA activity, and influences both transcription and translation of mxiD, encoding a TTSA component. Compared to a situation in which mxiEa and mxiEb would be in the same reading frame, encoding of MxiE by transcriptional slippage decreases production of MxiE and MxiD by three- and twofold, respectively. Paradoxically, the “error” made by the RNA polymerase over the mxiE slippage site, i.e., the incorporation of one nontemplated nucleotide, positively controls elongation of the mRNA downstream from the slippage site.
REFERENCES
- 1.Buchrieser, C., P. Glaser, C. Rusniok, H. Nedjari, H. d'Hauteville, F. Kunst, P. Sansonetti, and C. Parsot. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760-771. [DOI] [PubMed] [Google Scholar]
- 2.Larsen, B., N. M. Wills, C. Nelson, J. F. Atkins, and R. F. Gesteland. 2000. Nonlinearity in genetic decoding: homologous DNA replicase genes use alternatives of transcriptional slippage or translational frameshifting. Proc. Natl. Acad. Sci. USA 97:1683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mavris, M., A. L. Page, R. Tournebize, B. Demers, P. Sansonetti, and C. Parsot. 2002. Regulation of transcription by the activity of the Shigella flexneri type III secretion apparatus. Mol. Microbiol. 43:1543-1553. [DOI] [PubMed] [Google Scholar]
- 4.Newton, W. A., J. R. Beckwith, D. Zipser, and S. Brenner. 1965. Nonsense mutants and polarity in the lac operon of Escherichia coli. J. Mol. Biol. 14:290-296. [DOI] [PubMed] [Google Scholar]
- 5.Nudler, E., A. Mustaev, E. Lukhtanov, and A. Goldfarb. 1997. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell 89:33-41. [DOI] [PubMed] [Google Scholar]
- 6.Oppenheim, D. S., and C. Yanofsky. 1980. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics 95:785-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parsot, C. 2005. Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol. Lett. 252:11-18. [DOI] [PubMed] [Google Scholar]
- 8.Parsot, C., E. Ageron, C. Penno, M. Mavris, K. Jamoussi, H. d'Hauteville, P. Sansonetti, and B. Demers. 2005. A secreted anti-activator, OspD1, and its chaperone, Spa15, are involved in the control of transcription by the type IIII secretion apparatus activity in Shigella flexneri. Mol. Microbiol. 56:1627-1635. [DOI] [PubMed] [Google Scholar]
- 9.Penno, C., P. Sansonetti, and C. Parsot. 2005. Frameshifting by transcriptional slippage is involved in production of MxiE, the transcription activator regulated by the activity of the type III secretion apparatus in Shigella flexneri. Mol. Microbiol. 56:204-214. [DOI] [PubMed] [Google Scholar]
- 10.Wagner, L. A., R. B. Weiss, R. Driscoll, D. S. Dunn, and R. F. Gesteland. 1990. Transcriptional slippage occurs during elongation at runs of adenine or thymine in Escherichia coli. Nucleic Acids Res. 18:3529-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]