Abstract
Microarray studies of the Escherichia coli response to nitric oxide and nitrosative stress have suggested that additional transcriptional regulators of this response remain to be characterized. We identify here the product of the yjeB gene as a negative regulator of the transcription of the ytfE, hmpA and ygbA genes, all of which are known to be upregulated by nitrosative stress. Transcriptional fusions to the promoters of these genes were expressed constitutively in a yjeB mutant, indicating that all three are targets for repression by YjeB. An inverted repeat sequence that overlaps the −10 element of all three promoters is proposed to be a binding site for the YjeB protein. A similar inverted repeat sequence was identified in the tehA promoter, which is also known to be sensitive to nitrosative stress. The ytfE, hmpA, ygbA, and tehA promoters all caused derepression of a ytfE-lacZ transcriptional fusion when present in the cell in multiple copies, presumably by a repressor titration effect, suggesting the presence of functional YjeB binding sites in these promoters. However, YjeB regulation of tehA was weak, as judged by the activity of a tehA-lacZ fusion, perhaps because YjeB repression of tehA is masked by other regulatory mechanisms. Promoters regulated by YjeB could be derepressed by iron limitation, which is consistent with an iron requirement for YjeB activity. The YjeB protein is a member of the Rrf2 family of transcriptional repressors and shares three conserved cysteine residues with its closest relatives. We propose a regulatory model in which the YjeB repressor is directly sensitive to nitrosative stress. On the basis of similarity to the nitrite-responsive repressor NsrR from Nitrosomonas europaea, we propose that the yjeB gene of E. coli be renamed nsrR.
Pathogenic strains of enteric bacteria are exposed to high concentrations of nitric oxide (NO) made by the inducible NO synthase of host phagocytic cells, such as macrophages (11). Species (such as Escherichia coli) that utilize nitrate and nitrite as electron acceptors for anaerobic respiration probably also generate low levels of NO as a consequence of the reduction of nitrite (4, 19). Since NO is reactive and toxic, bacteria may require enzyme systems that protect against NO, whether the source is endogenous or exogenous (32). In the case of E. coli, flavohemoglobin, flavorubredoxin, and the cytochrome c nitrite reductase, Nrf, are enzymes that utilize NO as a substrate and have established roles in mediating NO resistance (15, 31, 34). Several regulatory proteins have been shown to be sensitive to NO in vitro or in vivo, or both, and so may mediate adaptive responses to NO exposure. These include SoxR, OxyR, FNR, MetR, and Fur (5, 6, 10, 12, 17), though in each case the principal function of the regulator is to sense another signal (superoxide, hydrogen peroxide, oxygen, homocysteine, and iron, respectively). The only regulator known to serve exclusively as an NO sensor in E. coli is NorR, which activates transcription of the norVW genes encoding the flavorubredoxin and associated flavoprotein that together reduce NO to nitrous oxide (7, 13, 14, 16, 18).
Three studies have made use of microarrays to evaluate the response of the E. coli transcriptome to exposure to NO or other reagents that cause nitrosative stress (12, 21, 27). These experiments differed in their choice of medium (rich or defined), growth conditions (aerobic or anaerobic, batch or continuous culture) and the reagents used to impose nitrosative stress (S-nitrosoglutathione, acidified nitrite, or aqueous NO), which may in part account for the different gene sets identified in each case. Only three transcription units were found to be upregulated in all three studies: norVW, hmpA, and nrdH. The norVW genes are activated in response to NO by NorR (7, 14, 42). Regulation of the hmpA gene has been extensively studied, and it has been shown that hmpA transcription is subject to repression mediated by MetR and FNR. It has been proposed that upregulation of hmpA by NO involves inactivation of MetR by nitrosation of its corepressor, homocysteine (25), and inactivation of FNR by reaction of NO with the protein-bound [Fe-S] cluster (5). The third gene identified in all three array studies was nrdH, the first gene of an operon encoding components of a ribonucleotide reductase, the expression of which has not been studied.
Microarray analysis of cells grown in rich medium and exposed to either S-nitrosoglutathione (GSNO) or acidified nitrite identified a number of genes that remained inducible in strains lacking the NO-responsive regulators mentioned above (27). Thus, it was proposed that the E. coli genome encodes another NO-responsive regulatory protein (27). One of the potential targets for the unidentified regulator was ytfE, which can also be strongly induced by aqueous NO in anaerobic cultures (21). The ytfE homologue of Salmonella enterica serovar Typhimurium (designated nipC) was identified in a screen for promoters inducible by acidified nitrite (23). Homologues of ytfE are also NO inducible in the denitrifying organisms Ralstonia eutropha and Pseudomonas stutzeri, which make NO as an intermediate of denitrification (29, 44). Despite its widespread nature and conserved expression pattern, the function of the ytfE gene product remains unknown. Mutation of the ytfE gene of E. coli causes an increased sensitivity to NO through a mechanism that has not been defined (21). In the present study, we identify the product of the yjeB gene as the regulator of ytfE transcription. We identify a sequence in the ytfE promoter that we propose to be required for regulation by YjeB and show that at least two other promoters that share this sequence (hmpA and ygbA) are targets for regulation by YjeB.
MATERIALS AND METHODS
Bacterial strains and plasmids and culture conditions.
Bacterial strains and plasmids used in the present study are listed in Table 1. The rich medium for routine culturing of E. coli was Lennox (L) broth (tryptone 10 g liter−1; yeast extract 5 g liter−1; NaCl 5 g liter−1). Cultures for preparation of electrocompetent cells were grown in SOB medium, and the cells were prepared according to standard protocols (37). Cultures for β-galactosidase assays were grown either in L broth, or in a mineral salts medium (40), supplemented with glucose (0.2 and 0.5% [wt/vol], for aerobic and anaerobic cultures, respectively), Casamino Acids (0.5 g liter−1), vitamin B1 (0.01 mg liter−1), and other supplements as indicated. Aerobic cultures were grown in 20 ml of medium in 250-ml flasks, which were shaken at 250 rpm. Anaerobic cultures were grown standing in filled bottles. For β-galactosidase assays, cultures were treated with NO sources when in early log phase (optical density at 650 nm of 0.15 to 0.3) and were assayed 90 to 120 min later while still in log phase. Treatment with NO sources caused partial or no growth inhibition, depending on growth conditions and the reagent used. Cultures for iron starvation experiments were grown aerobically in L broth as described previously (33). NO was prepared as an aqueous solution by reaction of sodium nitrite with sulfuric acid (33); GSNO and N-(2-aminoethyl)-N-(2-hydroxy-2-nitrosohydrazino)-1,2-ethylenediamine (Spermine NONOate) were purchased from Calbiochem.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| BW25113 | lacIqrrnBT14 ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | Barry Wanner |
| MC1000 | araD139 Δ(ara-leu) Δ(codB-lacI) galK16 galE15 relA1 rpsL spoT1 | E. coli Genetic Stock Center |
| EC100D pir-116 | mcrA Δ(mrr hsdRMS mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu) galU galK rpsL nupG pir-116 | Epicentre |
| NovaBlue Singles | endA1 hsdR17(rK12− mK12+) supE44 thi-1 recA1 gyrA96 relA1 lac F′ [proA+B+lacIqZΔM15::Tn10] | Novagen |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac F′ [proA+B+lacIqZΔM15::Tn10] | Stratagene |
| TP1000 | araD Δ(argF-lac)U169 rpsL relA flbB ptsF devC rbsR mobAB::kan | Tracy Palmer |
| JOEY19 | MC1000 (λRS45 ytfE-lacZ) | This work |
| JOEY24 | MC1000 (λRS45 ytfE-lacZ) yjeB::EZ-Tn5TM <R6Kγori/KAN-2> | This work |
| JOEY28 | MC1000 (λRS45 ytfE-lacZ) yjeB::EZ-Tn5TM <R6Kγori/KAN-2> | This work |
| JOEY30 | MC1000 (λRS45 ytfE-lacZ) yjeB::EZ-Tn5TM <R6Kγori/KAN-2> | This work |
| JOEY59 | MC1000 (λRS45 ytfE-lacZ) yjeB::kan | This work |
| JOEY60 | MC1000 yjeB::kan | This work |
| JOEY61 | MC1000 (λRS45 ytfE-lacZ) ΔyjeB | This work |
| JOEY62 | MC1000 ΔyjeB | This work |
| JOEY72 | MC1000 (λRS45 hmpA-lacZ) | This work |
| JOEY73 | MC1000 (λRS45 ygbA-lacZ) | This work |
| JOEY75 | MC1000 (λRS45 tehA-lacZ) | This work |
| JOEY76 | MC1000 ΔyjeB (λRS45 hmpA-lacZ) | This work |
| JOEY77 | MC1000 ΔyjeB (λRS45 ygbA-lacZ) | This work |
| JOEY79 | MC1000 ΔyjeB (λRS45 tehA-lacZ) | This work |
| JOEY83 | MC1000 (λRS45 yjeB-lacZ) | This work |
| JOEY84 | MC1000 ΔyjeB (λRS45 yjeB-lacZ) | This work |
| Plasmids | ||
| pRS415 | lacZ fusion vector | Valley Stewart |
| pSTBlue-1 | Cloning vector | Novagen |
| pKD4 | Barry Wanner | |
| pCP20 | Barry Wanner | |
| pGIT1 | 205-bp ytfE promoter fragment in pSTBlue-1 | This work |
| pGIT2 | 354-bp hmpA promoter fragment in pSTBlue-1 | This work |
| pGIT3 | 292-bp ygbA promoter fragment in pSTBlue-1 | This work |
| pGIT5 | 232-bp tehA promoter fragment in pSTBlue-1 | This work |
| pGIT8 | 205-bp ytfE promoter fragment with ΔA deletion in pSTBlue-1 | This work |
| pGIT9 | yjeB and its 5′ noncoding region cloned in pSTBlue-1 | This work |
Construction of reporter fusions.
The noncoding region upstream of ytfE was amplified by PCR using primers ytfEP1 and ytfEP3, with a BamHI site incorporated at the 5′ end of the gene-proximal primer, to facilitate subsequent cloning reactions (all primer sequences are available from the authors on request). The PCR product was cloned into pSTBlue-1 by using the Perfectly Blunt Cloning Kit (Novagen), and the DNA sequence of the insert was confirmed. The ytfE promoter was then cloned into pRS415 on an EcoRI-BamHI fragment (using the EcoRI site in pSTBlue-1) and was transferred to λRS45 by homologous recombination in MC1000 (39). The resulting phage was used to lysogenize MC1000, generating strain JOEY19. Mono-lysogens were initially distinguished from di-lysogens by β-galactosidase assays and were subsequently confirmed by PCR (35). Reporter fusions to the noncoding regions upstream of hmpA, ygbA, tehA, and yjeB were made by the same strategy.
Transposon mutagenesis and cloning of insertions.
Strain JOEY19 was subjected to transposon mutagenesis using the EZ-Tn5<R6Kγori/KAN-2>Tnp Transposome kit (Epicenter) according to the manufacturer's instructions. Briefly, the Transposome complex was electroporated into JOEY19, and then dilutions of the cell suspension were plated on to L agar, supplemented with kanamycin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Approximately 10,000 colonies were screened, from which 17 with a blue-colony phenotype were selected for further characterization. β-Galactosidase activity was assayed in overnight cultures of these 17 strains; 14 gave activities only ∼2-fold higher than the JOEY19 control, and 3 (named JOEY24, JOEY28, and JOEY30) gave extremely high activities. Genomic DNA was prepared from these three strains (using the Wizard Genomic DNA purification kit from Promega), digested with EcoRI, and then immediately ligated. Ligation reactions were electroporated into strain EC100D pir-116 and kanamycin-resistant transformants selected. Plasmid DNAs were purified from these strains and were sequenced with primers KAN-2 FP-1 and R6KAN-2 RP-1 (Epicenter). Sequence data were used to interrogate the E. coli MG1655 genome sequence (http://genolist.pasteur.fr/Colibri/) to identify the sites of transposon insertion. Transposon EZ-Tn5 generates a 9-bp duplication at its insertion site, and the three insertions mapped to coordinates 4404305 to 4404313, 4404500 to 4404508, and 4404536 to 4404544 (in strains JOEY24, JOEY28, and JOEY30, respectively) in the E. coli genome.
Other genetic methods.
The coding region of the yjeB gene was replaced with a kanamycin resistance cassette using a the λred recombinase method with primers yjeBP1 and yjeBP2 and pKD4 as the template (8). The mutation was transferred to strains MC1000 and JOEY19 by P1 transduction. To convert the insertion mutation to an unmarked deletion, the strains were transformed with pCP20, and kanamycin/ampicillin-sensitive transformants were identified after colony purification at 43°C (8). The structures of the insertion and deletion mutants were confirmed at each step by PCR. For complementation tests, the yjeB gene and 5′ noncoding region were amplified with the primers yjeB5′ and yjeB3′, and the PCR product was cloned into pSTBlue-1.
Site-directed mutagenesis was done with the QuikChange II kit (Stratagene) according to the manufacturer's instructions, with pGIT1 DNA as the template. The products of mutagenesis were sequenced prior to further analysis.
RNA methods.
Cultures for RNA isolation were grown anaerobically and exposed to nitrite for 30 min prior to mRNA isolation. RNA was purified from growing cultures with QIAGEN RNeasy minikits according to the manufacturer's instructions. Transcript start sites were determined by 5′-RACE (5′-Full RACE Core Set; Takara) according to the manufacturer's directions. Primers for RACE were designed according to the manufacturers' guidelines.
RESULTS
Regulation of the ytfE gene.
In microarray experiments, transcription of ytfE was activated by treatment of aerobic cultures grown in rich medium with GSNO or acidified nitrite and of anaerobic cultures grown in minimal medium with aqueous NO (21, 27). However, in continuous cultures grown in minimal medium, ytfE expression did not respond to GSNO (12). We have studied here the activity of a ytfE-lacZ transcriptional fusion that is integrated into the chromosome of strain JOEY19. Expression of the reporter fusion was measured in cultures grown under both aerobic and anaerobic conditions and exposed to physiological and chemical sources of NO (Table 2). The basal level of activity of the ytfE promoter in cultures grown aerobically or anaerobically in glucose minimal medium was low (<50 U). The addition of 0.02 mM aqueous NO to anaerobic cultures caused an ∼16-fold increase in ytfE promoter activity (Table 2), in agreement with data from microarray experiments (21). Treatment with 0.1 mM GSNO caused an ∼5-fold increase in ytfE promoter activity in aerobic cultures, and an ∼14-fold increase in anaerobic cultures (Table 2). The relatively small effect of GSNO in aerobic cultures is surprising, in view of the 38-fold increase in ytfE expression that was observed by microarray analysis of cultures grown aerobically in rich medium and exposed to 0.1 mM GSNO (27). Therefore, we assayed ytfE-lacZ expression in cultures grown aerobically in L broth and exposed to 0.1 mM GSNO for 1 h in log phase. Under these conditions, ytfE promoter activity (664 ± 42 U) was much higher than in cultures grown in minimal medium and showed an induction ratio similar to that seen in the microarray analysis. One possible explanation for the medium effect is that growth in tryptone-based rich medium induces the expression of oligopeptide permeases that can transport GSNO into the cell (9). This consideration may not apply in anaerobic cultures (in which ytfE expression is more sensitive to GSNO), where the extracellular NO that is released by the slow homolytic decomposition of GSNO is more stable to auto-oxidation than in the presence of oxygen. Treatment with Spermine NONOate (which decomposes with a half-life of approximately 40 min at neutral pH and 37°C, releasing one molar equivalent of NO) caused a large increase in ytfE promoter activity under both aerobic and anaerobic conditions (Table 2). Presumably, the continuous provision of NO by the decomposition of Spermine NONOate provides a steady-state concentration of NO that is sufficient to overcome the tendency of NO to auto-oxidize in the presence of oxygen; thus, the NONOate is an efficient inducer of ytfE in aerobic cultures.
TABLE 2.
Activity of the ytfE promoter activity under different growth conditions
| Supplementa (concn [mM]) | Mean β-galactosidase activityb ± SD
|
|
|---|---|---|
| Aerobic | Anaerobic | |
| None | 27 ± 1.2 | 34 ± 2.5 |
| Aqueous NO (0.02) | NDc | 551 ± 50 |
| GSNO (0.1) | 133 ± 18 | 468 ± 138 |
| Spermine NONOate (0.05) | 943 ± 53 | 575 ± 172 |
| Nitrate (50) | 40 ± 1.2 | 458 ± 86 |
| Nitrite (5) | 166 ± 8.7 | 1,202 ± 195 |
Cultures were grown aerobically and anaerobically in minimal medium with glucose as the carbon and energy source. Culture media were supplemented as indicated; nitrate was present throughout growth, and all other supplements were added to growing cultures 90 to 120 min prior to sampling for β-galactosidase assays.
For each condition, β-galactosidase activity was assayed in duplicate in at least three independently grown cultures. Units of activity are as defined by Miller (26).
ND, not done.
The addition of either nitrate or nitrite to anaerobic cultures also caused activation of ytfE-lacZ expression. Elimination of nitrate respiration by introduction of a mobAB mutation (which prevents biosynthesis of the molybdenum cofactor required by nitrate reductase) into JOEY19 eliminated activation of ytfE by nitrate (data not shown). This shows that activation of ytfE by nitrate requires nitrate reduction and indicates that ytfE is not a member of the nitrate/nitrite-responsive NarXL/NarQP regulon. The most likely explanation for the effects of nitrate and nitrite on ytfE transcription is that the promoter is activated by the low levels of NO that are made by E. coli as a by-product of nitrate and nitrite respiration (4, 20). The expression of ytfE is not upregulated by hydrogen peroxide or by superoxide (30, 46), so it seems that the ytfE promoter is sensitive specifically to reagents that cause nitrosative stress.
The yjeB gene encodes a repressor of ytfE expression.
The ytfE-lacZ fusion is upregulated by multiple copies of the ytfE promoter, a finding suggestive of negative regulation (see below). To identify the gene encoding the repressor of ytfE, JOEY19 was subjected to transposon mutagenesis, and mutants in which the ytfE promoter was derepressed were identified by screening on media containing X-Gal. Three transposon insertion mutations gave rise to high and constitutive ytfE promoter activities (Fig. 1); the three insertions mapped to different positions within the coding region of yjeB. Precise replacement of the yjeB gene by a kanamycin resistance cartridge (8) also caused derepression of the ytfE promoter (Fig. 1), confirming that constitutive ytfE promoter activity in the transposon insertion mutants is caused by the insertions and not by adventitious second-site mutations. The kanamycin resistance cartridge was removed by site-specific recombination, leaving an unmarked deletion of yjeB (8). The ytfE promoter was also derepressed by this deletion of yjeB (Fig. 1). The yjeB gene is the promoter proximal gene of a transcription unit that also includes rnr, encoding RNase R, and yifH and yifI, the products of which may also have roles in RNA metabolism (3). The yjeB gene is predicted to encode a transcriptional repressor of the Rrf2 family (http://pfam.wustl.edu/index.html), and no transposon insertions that caused derepression of ytfE were recovered in rnr, yifH, or yifI. Thus, it is likely that derepression of ytfE is a direct consequence of mutations in the coding region of yjeB. Accordingly, the yjeB mutant phenotype could be complemented in trans by the yjeB gene cloned on a plasmid (Fig. 1), confirming that polar effects of yjeB mutations do not contribute to derepression of the ytfE promoter. The complemented strain showed a smaller (though still significant) response to nitrite (data not shown) and Spermine NONOate (Fig. 1). We suggest that the presence of multiple copies of the yjeB gene causes an enhanced repression of the ytfE promoter, such that it is less sensitive to treatments that cause a larger derepression in the wild-type background.
FIG. 1.
Activity of the ytfE promoter in yjeB mutant backgrounds. Cultures were grown aerobically and were treated with 0.05 mM Spermine NONOate (striped bars) for 90 min in log phase or were not treated (open bars) and then were assayed for β-galactosidase. The data are shown for only one of the transposon insertion mutants (in JOEY30); the two other mutants behaved in an indistinguishable fashion. The β-galactosidase activity was assayed in duplicate in three independently grown cultures; standard deviations are shown. Units of activity are as defined by Miller (26).
Other targets for YjeB regulation.
Transformation of JOEY19 with pGIT1, which contains the ytfE promoter fragment cloned in a high-copy-number vector, gave rise to high β-galactosidase activities from the ytfE-lacZ fusion (Table 3). This result is most easily explained by a repressor titration effect (41) and is consistent with negative regulation of the ytfE promoter by YjeB. The ytfE promoter region contains a prominent inverted repeat sequence, (AAGATGcATTTaAAATaCATCTT), which is not the recognition sequence for any known regulatory protein. We determined the start site of the ytfE mRNA, which identifies the −10 motif of the ytfE promoter as the TAAAAT sequence close to the center of the inverted repeat (Fig. 2). Deletion of the AT base pair at the center of dyad symmetry of this putative YjeB binding site eliminated activity in the repressor titration assay (Table 3). This result is consistent with the hypothesis that the inverted repeat is the binding site for the YjeB repressor. We were unable to recover Lac+ fusion phages containing this promoter, presumably because the deletion (which is within the −10 sequence) eliminates ytfE promoter activity.
TABLE 3.
Derepression of the ytfE promoter by repressor titration
| Plasmid | Promoter | Inverted repeat sequencea | Mean β-galactosidase activityb ± SD |
|---|---|---|---|
| None | None | 73 ± 5 | |
| pSTBlue-1 | None | 51 ± 1 | |
| pGIT1 | ytfE | AAGATGCATTTAAAATACATCTT | 2,738 ± 426 |
| pGIT8 | ytfE ΔA deletion | AAGATGCATTT-AAATACATCTT | 80 ± 6 |
| pGIT2 | hmpA | AAGATGCATTTGAGATACATCAA | 2,832 ± 291 |
| pGIT4 | ygbA | AAGATGTAATATAAATACATCTT | 1,426 ± 329 |
| pGIT5 | tehA | AAAATGCATTTCAAATATACTTT | 1,477 ± 92 |
Inverted repeat sequences similar to that in the ytfE promoter region are shown; underlined nucleotides match those at equivalent positions in ytfE.
Cultures were grown aerobically in L broth for 16 h. β-Galactosidase activity was assayed in duplicate in three independently grown cultures. Units of activity are as defined by Miller (26).
FIG. 2.
Organization of the ytfE, hmpA, ygbA, and tehA promoters. Sequences that match the inverted repeat that is the proposed YjeB binding site in ytfE (AAGATG-ATTT-AAAT-CATCTT) are underlined. Promoter elements (−10 sequences) and start codons are shown in boldface, and transcription start sites are indicated (+1). The mRNA start site and promoter sequences for hmpA have been reported previously (24). Start sites for ytfE, ygbA, and tehA were determined in the present study.
The cis-acting sequence hypothesized to be required for YjeB regulation of ytfE was used to search the E. coli genome (http://genolist.pasteur.fr/Colibri/), and similar sequences were found in the promoter regions of the hmpA, tehA, and ygbA genes (Table 3 and Fig. 2). All three of these genes can be activated either by GSNO or by aqueous NO in microarray experiments (21, 27). Transcription from the ygbA promoter was shown to be sensitive to low concentrations of GSNO, and this gene was suggested to belong to a yet-to-be discovered regulon (27). The noncoding regions from upstream of the three genes were cloned and tested in the repressor titration assay. The ygbA and tehA promoter fragments caused a 20- to 25-fold derepression of the ytfE-lacZ fusion (Table 3), which is consistent with the presence of YjeB binding sites. The hmpA promoter fragment caused an ∼39-fold derepression of the ytfE promoter, similar to the level of derepression seen with the ytfE promoter itself (Table 3).
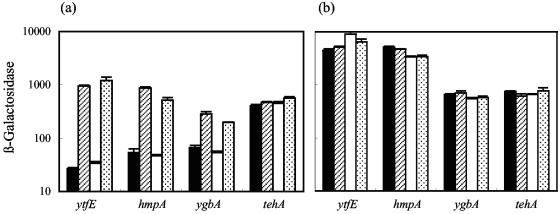
To investigate the regulation of hmpA, tehA, and ygbA, each promoter was fused to lacZ to generate a chromosomal transcriptional fusion. Assays of β-galactosidase from these fusions were used to measure promoter activities under different growth conditions, and in the strain deleted for yjeB (Fig. 3). The hmpA and ygbA promoter fusions showed very similar patterns of activity to the ytfE-lacZ fusion. Both had low activities in aerobic and anaerobic cultures and could be upregulated by treatment of aerobic cultures with Spermine NONOate and of anaerobic cultures with nitrite (Fig. 3). In a yjeB mutant background, the hmpA and ygbA promoters were constitutively active. Thus, the hmpA and ygbA promoters are targets for repression mediated by YjeB, which is consistent with the presence of potential YjeB binding sites in these promoters, as revealed by the repressor titration assay. The tehA promoter contains a putative YjeB binding site (Table 3); yet assays of the tehA-lacZ fusion suggested that YjeB has only a small (though reproducible) effect on tehA promoter activity. The tehA promoter had a high activity in untreated cultures, which increased <2-fold in response to NO sources or to the yjeB mutation (Fig. 3). It is possible that growth conditions that reveal maximal YjeB regulation of the tehA promoter have not yet been identified.
FIG. 3.
Activities of the ytfE, hmpA, ygbA, and tehA promoters in yjeB+ (a) and ΔyjeB (b) strains. Cultures were grown aerobically (solid and striped bars) and anaerobically (open and stippled bars) and were treated with 0.05 mM Spermine NONOate (striped bars) or 5 mM nitrite (stippled bars) 90 to 120 min prior to assay for β-galactosidase. Units of activity are as defined by Miller (26).
The transcription start sites of the tehA and ygbA promoters were determined by 5′-RACE, using mRNA isolated from cultures grown anaerobically and treated with nitrite. The start site of the hmpA mRNA has been determined previously (24). In all three cases, and as is the case for ytfE, the likely YjeB binding site overlaps or partially overlaps the −10 element of the promoter (Fig. 2), suggesting that YjeB represses transcription by interfering directly with transcription initiation by RNA polymerase.
We were unable to detect a possible YjeB binding site in the yjeB promoter, and assays of a yjeB-lacZ fusion showed that yjeB expression is not subject to autoregulation. Furthermore, expression of the yjeB-lacZ fusion is not regulated by sources of NO (data not shown). Therefore, the NO responsiveness of targets for YjeB regulation does not involve regulation of expression of the yjeB gene.
Iron regulation of YjeB-repressed promoters.
It has been shown previously that the hmpA promoter switches on in response to iron limitation, imposed by the addition of 0.1 to 0.4 mM 2′2′-dipyridyl to cultures growing in rich medium (33). A strong response to iron starvation was observed in aerobic cultures (33), excluding inactivation of FNR as the sole mechanism involved. We speculated that stimulation of hmpA by iron chelation might reflect an iron requirement for efficient repression by YjeB. We therefore assayed YjeB-regulated promoters for their response to iron limitation, using growth conditions similar to those used previously with the hmpA promoter (33). In agreement with the previous results, we found that the hmpA promoter was significantly upregulated by treatment of aerobic cultures with 0.4 mM 2′2′-dipyridyl (Table 4). The other promoters known to be strongly regulated by YjeB (ytfE and ygbA) were similarly responsive to iron limitation (Table 4). Thus, the response of the hmpA promoter to iron limitation appears to be a common feature of YjeB-repressed promoters, and we propose that YjeB is, either directly or indirectly, inactivated by iron limitation. The tehA promoter was not derepressed by iron limitation (Table 4), which is consistent with the weak regulation of this promoter by YjeB.
TABLE 4.
YjeB-regulated promoters are derepressed by iron limitation
| Promoter | Mean β-galactosidase activitya ± SD
|
|
|---|---|---|
| − Dipyridyl | + 0.4 mM dipyridyl | |
| ytfE | 81 ± 3.1 | 1,387 ± 164 |
| hmpA | 143 ± 23 | 1,301 ± 57 |
| ygbA | 162 ± 6 | 744 ± 32 |
| tehA | 2,084 ± 102 | 1,172 ± 74 |
DISCUSSION
We have identified the product of the yjeB gene as a regulator of three promoters (ytfE, hmpA, and ygbA) that are stimulated by sources of NO. We assume that YjeB-regulated genes have a physiological role in protecting the cell against NO. This is undoubtedly the case for the flavohemoglobin, which has an established role in mediating NO resistance (13, 32). Mutation of ytfE causes an enhanced sensitivity to NO, and the YtfE protein is predicted to contain iron, though it is of unknown function (21). The biochemical function of the product of the ygbA gene is not known. The YjeB protein is a member of the Rrf2 family of transcriptional repressors, named for a regulator of the hmc operon of Desulfovibrio vulgaris (22). The best-characterized member of the Rrf2 family is the IscR protein of E. coli, which contains a [2Fe-2S] cluster, and regulates the transcription of isc genes, the products of which have roles in the biogenesis of [Fe-S] clusters (38). IscR-mediated repression can be reversed by iron starvation (28), an effect similar to that reported here for YjeB. Another Rrf2 family member is NsrR of Nitrosomonas europaea, which represses the transcription of the nirK gene encoding a copper nitrite reductase (1). NsrR-mediated repression is reversed in vivo by nitrite, suggesting that NsrR activity is sensitive to nitrite or a product of nitrite metabolism (1). In Rhodobacter capsulatus E1F1, a homologue of the N. europaea nsrR gene is in a cluster of genes required for nitrate assimilation, including the hcp gene encoding a hydroxylamine reductase that protects against reactive intermediates or by-products of nitrate metabolism (2). The RirA protein is an Rrf2-type regulator from Rhizobium leguminosarum, which is a repressor of genes involved in iron assimilation (45). All of these proteins, including YjeB, share three conserved cysteines, although with somewhat variable spacing. Since these are the only cysteine residues in IscR, it is likely, although not proven, that they provide three of the ligands to the [2Fe-2S] cluster (38). It has been suggested that the NsrR-type proteins also contain an [Fe-S] cluster that is liganded by the three conserved cysteine residues and can be inactivated by either nitrite or hydroxylamine (38). On the basis of these properties of the Rrf2 family members, and our genetic data, we propose that YjeB is an NO-sensitive repressor of its target promoters. Although it is not possible at this stage to completely exclude indirect effects, it seems likely that YjeB is a direct repressor of the transcription of its targets (see below). We also cannot be certain that NO is the principal or sole inducer of YjeB-regulated genes, although all of the available evidence points to this being the case. On the basis of similarity to the other Rrf2 family members, we suggest that YjeB contains an NO-labile [Fe-S] cluster and that iron starvation causes the synthesis of an inactive apo form of the protein.
The tehA promoter is activated threefold by aqueous NO in anaerobic cultures (21) and contains an inverted repeat resembling the putative YjeB binding site (Table 3) overlapping the −10 sequence (Fig. 2). However, the promoter is apparently subject to a very weak regulation by YjeB (Fig. 3). We were unable to detect any additional transcription initiation site for tehA using RNA isolated from cells grown under different conditions, and assays of the tehA-lacZ fusion under different growth conditions did not reveal additional patterns of regulation. Nevertheless, it is possible that we have yet to identify the growth conditions that are required for maximal YjeB-mediated repression of tehA. Alternatively, the YjeB binding site in the tehA promoter may be weak, and the activity in the repressor titration assay could be high as a simple consequence of a copy number effect. The putative YjeB binding sites in the strongly regulated ytfE, ygbA, and hmpA promoters are characterized by a conserved inverted repeat motif, GATG-N11-CATC (Table 3 and Fig. 2). The putative site in the tehA promoter diverges from this pattern (Table 3), so the quality of the binding site may explain the lack of, or weak, regulation by YjeB. The products of the tehA and tehB genes are required for resistance to tellurite and a range of other toxic compounds (43), and their role, if any, in the response to NO remains to be determined.
The regulation of the hmpA promoter by YjeB is, presumably, superimposed on the previously described regulatory mechanisms involving FNR (5) and MetR (25). Another study has, however, reported that NO induction of hmpA in aerobic cultures is independent of MetR (27). If this is the case, then there must be a mechanism for NO induction of hmpA in aerobic cultures that involves neither MetR nor FNR (which is inactive in the presence of oxygen) and that mechanism is presumably the one described here involving YjeB. FNR binds to the hmpA promoter in vitro (5) and the FNR binding site (TTGAG----ATCAA) overlaps the presumed YjeB binding site (Fig. 2). It will be interesting to elucidate the mechanistic relationship between FNR- and YjeB-mediated repression of the hmpA promoter.
A recent analysis using comparative genomics has predicted a consensus YjeB binding site similar to the one described here and has also predicted that ytfE, ygbA, and hmpA belong to the YjeB regulon in E. coli (36). Our experimental data agree with these predictions, and the bioinformatics further predicts that YjeB interacts directly with the sequence proposed here to be its binding site (36). It has been suggested that YjeB is an orthologue of the nitrite-responsive repressor NsrR from N. europaea (1, 36). NsrR is not responsive to nitroprusside (a source of NO+) but shows an enhanced response to nitrite at low pH (1). The data are consistent with the possibility that the signal that inactivates NsrR is not nitrite per se but rather the NO formed by nitrite reductase catalyzed nitrite reduction or formed abiotically from nitrite under acid conditions. The related NsrR protein of R. capsulatus E1F1 may also respond to a reactive nitrogen species (2). On the basis of these similarities, it is appropriate that the yjeB gene of E. coli be renamed nsrR, and it will be of interest to determine the ligand specificities of the NsrR proteins. Bioinformatic analysis suggests that NsrR orthologues mediate responses to NO in a wide range of gram-negative and gram-positive bacteria (36).
Acknowledgments
We are grateful to Barry Wanner, Mary Berlyn, Tracy Palmer, and Valley Stewart for providing strains and plasmids; to Ray Dixon, Rainer Cramm, Andy Johnston, and Valley Stewart for helpful discussions; to Ray Dixon for comments on the manuscript; and to Dmitry Rodionov for sharing data prior to publication. We thank Yanling Wang and Meeyoung Park for their contributions to the early stages of this project.
This study was supported by grant MCB-0517174 from the National Science Foundation.
REFERENCES
- 1.Beaumont, H. J. E., S. I. Lens, W. N. M. Reijnders, H. V. Westerhoff, and R. J. M. van Spanning. 2004. Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol. Microbiol. 54:148-158. [DOI] [PubMed] [Google Scholar]
- 2.Cabello, P., C. Pino, M. F. Olmo-Mira, F. Castillo, M. D. Roldan, and C. Moreno-Vivian. 2004. Hydroxylamine assimilation by Rhodobacter capsulatus E1F1: requirement of the hcp gene (hybrid cluster protein) located in the nitrate assimilation nas gene region for hydroxylamine reduction. J. Biol. Chem. 279:45485-45494. [DOI] [PubMed] [Google Scholar]
- 3.Cairrao, F., A. Cruz, H. Mori, and C. M. Arraiano. 2003. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 50:1349-1360. [DOI] [PubMed] [Google Scholar]
- 4.Corker, H., and R. K. Poole. 2003. Nitric oxide formation by Escherichia coli: dependence on nitrite reductase, the NO-sensing regulator Fnr, and flavohemoglobin Hmp. J. Biol. Chem. 278:31584-31592. [DOI] [PubMed] [Google Scholar]
- 5.Cruz-Ramos, H., J. Crack, G. Wu, M. N. Hughes, C. Scott, A. J. Thomson, J. Green, and R. K. Poole. 2002. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 21:3235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Autréaux, B., D. Touati, B. Bersch, J. M. Latour, and I. Michaud-Soret. 2002. Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc. Natl. Acad. Sci. USA 99:16619-16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Autréaux, B., N. P. Tucker, R. Dixon, and S. Spiro. 2005. A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature 437:769-772. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Groote, M. A., D. Granger, Y. Xu, G. Campbell, R. Prince, and F. C. Fang. 1995. Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc. Natl. Acad. Sci. USA 92:6399-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding, H., and B. Demple. 2000. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc. Natl. Acad. Sci. USA 97:5146-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang, F. C. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2:820-832. [DOI] [PubMed] [Google Scholar]
- 12.Flatley, J., J. Barrett, S. T. Pullan, M. N. Hughes, J. Green, and R. K. Poole. 2005. Transcriptional responses of Escherichia coli to S-Nitrosoglutathione under defined chemostat conditions reveal major changes in methionine biosynthesis. J. Biol. Chem. 280:10065-10072. [DOI] [PubMed] [Google Scholar]
- 13.Gardner, A. M., and P. R. Gardner. 2002. Flavohemoglobin detoxifies nitric oxide in aerobic, but not anaerobic, Escherichia coli. Evidence for a novel inducible anaerobic nitric oxide-scavenging activity. J. Biol. Chem. 277:8166-8171. [DOI] [PubMed] [Google Scholar]
- 14.Gardner, A. M., C. R. Gessner, and P. R. Gardner. 2003. Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli. Role of NorR and σ54 in the nitric oxide stress response. J. Biol. Chem. 278:10081-10086. [DOI] [PubMed] [Google Scholar]
- 15.Gardner, A. M., R. A. Helmick, and P. R. Gardner. 2002. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 277:8172-8177. [DOI] [PubMed] [Google Scholar]
- 16.Gomes, C. M., J. B. Vicente, A. Wasserfallen, and M. Teixeira. 2000. Spectroscopic studies and characterization of a novel electron-transfer chain from Escherichia coli involving a flavorubredoxin and its flavoprotein reductase partner. Biochemistry 39:16230-16237. [DOI] [PubMed] [Google Scholar]
- 17.Hausladen, A., C. T. Privalle, T. Keng, J. DeAngelo, and J. S. Stamler. 1996. Nitrosative stress: activation of the transcription factor OxyR. Cell 86:719-729. [DOI] [PubMed] [Google Scholar]
- 18.Hutchings, M. I., N. Mandhana, and S. Spiro. 2002. The NorR protein of Escherichia coli activates expression of the flavorubredoxin gene norV in response to reactive nitrogen species. J. Bacteriol. 184:4640-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji, X. B., and T. C. Hollocher. 1989. Nitrate reductase of Escherichia coli as a NO-producing nitrite reductase. Biochem. Arch. 5:61-66. [Google Scholar]
- 20.Ji, X. B., and T. C. Hollocher. 1988. Reduction of nitrite to nitric oxide by enteric bacteria. Biochem. Biophys. Res. Commun. 157:106-108. [DOI] [PubMed] [Google Scholar]
- 21.Justino, M. C., J. B. Vicente, M. Teixeira, and L. M. Saraiva. 2005. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 280:2636-2643. [DOI] [PubMed] [Google Scholar]
- 22.Keon, R. G., R. Fu, and G. Voordouw. 1997. Deletion of two downstream genes alters expression of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough. Arch. Microbiol. 167:376-383. [DOI] [PubMed] [Google Scholar]
- 23.Kim, C. C., D. Monack, and S. Falkow. 2003. Modulation of virulence by two acidified nitrite-responsive loci of Salmonella enterica serovar Typhimurium. Infect. Immun. 71:3196-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Membrillo-Hernandez, J., G. M. Cook, and R. K. Poole. 1997. Roles of RpoS (σS), IHF and ppGpp in the expression of the hmp gene encoding the flavohemoglobin (Hmp) of Escherichia coli K-12. Mol. Gen. Genet. 254:599-603. [DOI] [PubMed] [Google Scholar]
- 25.Membrillo-Hernandez, J., M. D. Coopamah, A. Channa, M. N. Hughes, and R. K. Poole. 1998. A novel mechanism for upregulation of the Escherichia coli K-12 hmp (flavohaemoglobin) gene by the ‘NO releaser’, S-nitrosoglutathione: nitrosation of homocysteine and modulation of MetR binding to the glyA-hmp intergenic region. Mol. Microbiol. 29:1101-1112. [DOI] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Mukhopadhyay, P., M. Zheng, L. A. Bedzyk, R. A. LaRossa, and G. Storz. 2004. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl. Acad. Sci. USA 101:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Outten, F. W., O. Djaman, and G. Storz. 2004. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52:861-872. [DOI] [PubMed] [Google Scholar]
- 29.Pohlmann, A., R. Cramm, K. Schmelz, and B. Friedrich. 2000. A novel NO-responding regulator controls the reduction of nitric oxide in Ralstonia eutropha. Mol. Microbiol. 38:626-638. [DOI] [PubMed] [Google Scholar]
- 30.Pomposiello, P. J., M. H. J. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poock, S. R., E. R. Leach, J. W. Moir, J. A. Cole, and D. J. Richardson. 2002. Respiratory detoxification of nitric oxide by the cytochrome c nitrite reductase of Escherichia coli. J. Biol. Chem. 277:23664-23669. [DOI] [PubMed] [Google Scholar]
- 32.Poole, R. K. 2005. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem. Soc. Trans. 33:176-180. [DOI] [PubMed] [Google Scholar]
- 33.Poole, R. K., M. F. Anjum, J. Membrillo-Hernandez, S. O. Kim, M. N. Hughes, and V. Stewart. 1996. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J. Bacteriol. 178:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poole, R. K., and M. N. Hughes. 2000. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36:775-783. [DOI] [PubMed] [Google Scholar]
- 35.Powell, B. S., M. P. Rivas, D. L. Court, Y. Nakamura, and C. L. Turnbough, Jr. 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodionov, D. A., I. L. Dubchak, A. P. Arkin, E. J. Alm, and M. S. Gelfand. 2005. Dissimilatory metabolism of nitrogen oxides in Bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol., e55. [DOI] [PMC free article] [PubMed]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schwartz, C. J., J. L. Giel, T. Patschkowski, C. Luther, F. J. Ruzicka, H. Beinert, and P. J. Kiley. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 98:14895-14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 40.Spencer, M. E., and J. R. Guest. 1973. Isolation and properties of fumarate reductase mutants of Escherichia coli. J. Bacteriol. 114:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 42.Tucker, N. P., B. D'Autréaux, D. J. Studholme, S. Spiro, and R. Dixon. 2004. DNA-binding activity of the Escherichia coli nitric oxide sensor NorR suggests a conserved target sequence in diverse Proteobacteria. J. Bacteriol. 186:6656-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner, R. J., D. E. Taylor, and J. H. Weiner. 1997. Expression of Escherichia coli TehA gives resistance to antiseptics and disinfectants similar to that conferred by multidrug resistance efflux pumps. Antimicrob. Agents Chemother. 41:440-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vollack, K. U., and W. G. Zumft. 2001. Nitric oxide signaling and transcriptional control of denitrification genes in Pseudomonas stutzeri. J. Bacteriol. 183:2516-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeoman, K. H., A. R. J. Curson, J. D. Todd, G. Sawers, and A. W. B. Johnston. 2004. Evidence that the Rhizobium regulatory protein RirA binds to cis-acting iron-responsive operators (IROs) at promoters of some Fe-regulated genes. Microbiology 150:4065-4074. [DOI] [PubMed] [Google Scholar]
- 46.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]