Abstract
The snoRNA-LBME-db is a dedicated database containing human C/D box and H/ACA box small nucleolar RNAs (snoRNAs), and small Cajal body-specific RNAs (scaRNAs). C/D box and H/ACA box snoRNAs are part of ribonucleoparticles that guide 2′-O-ribose methylation and pseudouridilation, respectively, of selected residues of 28S, 18S or 5.8S rRNAs or of the spliceosomal U6 RNA. Similarly, scaRNAs guide modifications of the spliceosomal RNAs transcribed by RNA polymerase II (U1, U2, U4, U5 and U12) and are often composed of both C/D box and H/ACA box domains. However, some snoRNAs do not function as modification guide RNAs, but rather as RNA chaperones during the maturation of pre-rRNA. The database was built by a compilation of the literature, and comprises human sno/scaRNAs that were experimentally verified, as well as the human orthologs of snoRNAs that were cloned in other vertebrate species, and some snoRNAs that are predicted by bioinformatics search in loci submitted to genomic imprinting, but have not all been experimentally verified. For each entry, the database identifies the modified nucleotide(s) in the target RNA(s), indicates the corresponding predicted base pairing, gives a few pertinent references and provides a link to the position of the sno/scaRNA on the UCSC Genome Browser. The ‘Find guide RNA’ function allows one to find the sno/scaRNAs predicted to guide the modification of a particular nucleotide in the rRNA and spliceosomal RNA sequences. The ‘Browse’ function allows one to download the sequences of selected sno/scaRNAs in the FASTA format. The database is available online at http://www-snorna.biotoul.fr/. It can also be accessed from the human UCSC Genome Browser via the sno/miRNA track.
INTRODUCTION
Small nucleolar RNAs (snoRNAs) are 60–150 nt long non-coding RNAs (but up to 608 nt for yeast snR30) that guide modifications of selected nucleotides in rRNAs or spliceosomal RNAs (1–3). From a structural basis, snoRNAs fall into two groups (4). The C/D box snoRNAs serve as guides for the 2′-O-ribose methylation of rRNAs or snRNAs and are characterized by the conserved boxes C (RUGAUGA) and D (CUGA) and often divergent copies of these boxes, C′ and D′. They possess 10–21 nt long antisense elements located upstream of the D and/or D′ boxes, so that the nucleotide of the target RNA paired with the fifth nucleotide upstream of the D/D′ box is 2′-O-ribose methylated (5,6). The C/D box snoRNAs are part of sno ribonucleoparticles (snoRNPs) that also comprise the NOP1/fibrillarin methyltransferase enzyme, and the NOP56 (NOL5A), NOP58 and NHP2L1 (Snu13, 15.5 kDa) proteins (7,8).
The H/ACA box snoRNAs are guides for the isomerization of uridine residues into pseudouridine, and are characterized by two imperfect hairpin structures separated by a single-stranded hinge containing the H box (ANANNA), and followed by a short tail containing the ACA (sometimes AUA, and AAA for yeast snR44) motif located 3 nt before the snoRNA 3′ end. Two short antisense sequences located in an internal loop of the 5′ and/or 3′ hairpin can base pair upstream and downstream of the targeted, unpaired uridine that is located 14 or 15 bp upstream of the H and/or ACA box (9,10). H/ACA box snoRNAs are associated with four proteins including dyskerin (the pseudouridine synthase), NOLA1 (GAR1), NOLA2 (NHP2) and NOLA3 (NOP10).
A new facet of modification guide RNAs appeared with the discovery of U85, a 330 nt long RNA composed of an H/ACA box domain embedded in a C/D box domain. Accordingly, U85 guides both the pseudouridylation of U46 and the 2′-O-ribose methylation of C46 of the U5 spliceosomal RNA (11). Subsequently, other composite guide RNAs were cloned and found to share the two characteristics of, first, guiding modification of snRNAs transcribed by RNA polymerase II (U1, U2, U4, U5 and U12, but not U6) and, second, accumulating in the Cajal bodies (12). Such guide RNAs were thus named small Cajal body-specific RNAs (scaRNAs). In addition to composite C/D-H/ACA RNAs, scaRNAs can be composed of two H/ACA domains, or of one or two C/D box domains.
Rather than guiding RNA modifications, a small number of C/D box snoRNAs are suspected to act as RNA chaperone during various steps of pre-rRNA processing, again by a base pairing mechanism. Moreover, a growing number of both C/D box and H/ACA box snoRNAs do not display antisense sequences compatible with a modification guiding function for rRNAs or snRNAs. These so-called ‘orphan’ snoRNAs might guide the modification of other classes of RNAs. In particular, the C/D box snoRNA HBII-52 possesses a perfect 18 nt long complementarity to the 3′-untranslated region of the serotonin receptor 5HT-2c mRNA. Interestingly, the base predicted to be 2′-O-ribose methylated is subjected to A/I RNA editing.
The vast majority of human sno/scaRNAs are encoded in introns of so-called host genes, in the sense orientation, and are produced by exonucleolytic processing of the debranched intron after splicing. Although many host genes are related to ribosome biogenesis and nucleolar function, snoRNA host genes are involved in a variety of biological function. A growing number of host genes appear to be devoid of protein coding potential. In addition, a small number of sno/scaRNAs are produced from independent RNA polymerase II transcriptional units. Those include the C/D box snoRNAs U3, U8 and U13, as well as the recently characterized methylation guides mgU2-25/61, mgU2-19/30 and mgU12-22/U4-8 (13).
AIMS AND CONTENT OF snoRNA-LBME-db
The snoRNA-LBME-db (http://www-snorna.biotoul.fr/) was created to collect the available information on human snoRNAs and scaRNAs, including their predicted target nucleotide(s), and the potential base-pairing interactions with their target RNA(s). This was done by an exhaustive analysis of the pertinent literature, rather than automatic extraction from other databases. Indeed, the sequence of many snoRNAs have not been deposited in databases. Even so, the database entries give in general no information on the modified nucleotide and base-pairing interactions with target RNAs. Moreover, we included in the database the probable orthologs of several murine sno/scaRNAs sequenced by Huttenhofer et al. (14) in their extensive RNOmics approach, although their existence in human has not been always experimentally established. Special cases are the HBII-52 and HBII-85 C/D box snoRNA clusters. Their members (47 and 27 for the HBII-52 and HBII-85 families, respectively) were characterized by a bioinformatics search in introns of the large, non-coding SNURF-SNRNP-UBE3A antisense transcript from the imprinted locus involved in the Prader-Willi/Angelman syndrome. Two other such families of snoRNAs, named 14q(I) (9 members) and 14q(II) (31 members), were characterized at another imprinted locus on chromosome 14. Owing to sequence similarities, it could not be assessed by northern blot if each individual member of these four families are expressed. However, all of them are included in the database.
Several snoRNAs are often clustered in different introns of the same host gene. The examination of conserved sequences in additional introns of these genes can lead to the discovery of new snoRNA candidates. This is the case for the C/D box snoRNA mgh18S-121, which was discovered in an intron of the RPL23A gene, that hosts the C/D box snoRNAs U42A and U42B. This new snoRNA is predicited to guide the 2′-O-ribose methylation of U121 of 18S rRNA. As this nucleoside was experimentally shown to be methylated, mgh18S-121 was included in the database, although its expression was not assessed.
With these caveats, all sno/scaRNAs from the database have been experimentally demonstrated in human, or for some of them, in the mouse.
To facilitate the identification of sno/scaRNAs genomic localization and their host gene, each entry of the database possesses a link to its position in the human UCSC Genome Browser (15). Moreover, the database can be accessed from the sno/miRNA track of the UCSC Genome Browser, which combines human snoRNAs and scaRNAs from the snoRNA-LBME-db and human miRNAs from the miRBase section of Rfam (16).
DATABASE DESIGN
The snoRNA-LBME-db home page
The home page gives basic information on sno/scaRNA biology, including:
the protein composition of the snoRNPs (with links to the corresponding entries in the GeneLinx and GeneCards databases);
the predicted secondary structure of C/D box and H/ACA box snoRNAs and composite scaRNAs, and base-pairing interactions with target RNAs;
information on the RNA component of telomerase, hTR, a special case of H/ACA box RNA, with appropriate links to the OMIM database;
short information on the expression of sno/scaRNAs from their host gene or from an independent transcriptional unit;
basic information on the relationship between snoRNAs and genomic imprinting;
selected reviews in the sno/scaRNA field.
Most importantly, the home page defines the GenBank accession numbers of the rRNA sequences that were used for numbering the modified nucleotides. The various laboratories in the field used indeed different reference sequences, making it difficult sometimes to compare their data. The human 18S rRNA sequence is included in the four GenBank entries X03205, K03432, M10098 and U13369 (nt 3657–5527), which differ by few nucleotides. In particular, the sequence from U13369 contains two extra nucleotides (1641 and 1649) absent from the three other sequences, as well from the mouse (X00686) and pig (AY265350 and NR_002170) 18S rRNA sequences. For a sake of constancy, we adopted the human X03205 sequence, which is also used by the Small Subunit rRNA modification Database (17).
The human 5.8S rRNA sequence from GenBank entry J01866 contains two extra nucleotides (nt 51 and 52) compared with the sequence from U13369 (nt 6623–6779). Although these 2 nt are also found in some 5.8S rRNA sequences from mouse (J01871), rat (J01881) and Xenopus borealis (K01375), the shorter form agrees with many sequences from different organisms. Therefore, we selected the 5.8S rRNA sequence from GenBank entry U13369 as the sequence of reference for our database.
For the human 28S rRNA sequence, we used nt 7935–12 969 of the U13369 pre-rRNA sequence, rather than the 10 nt shorter M11167 GenBank entry, as the former was more similar to sequences from other mammalian species.
The search page
The search page allows one to find a sno/scaRNA by typing its name from the literature (e.g. U85, ACA17, mgU12-22/U4-8, Z32 and E2). The RNAs can be also searched by incomplete names, such as mg and HBII-. When a snoRNA has been given alternative names by different laboratories (such as E1 and U17A, U39 and U55), the user is directed to a common entry, so that the database is non-redundant. The only exception concerns the scaRNAs mgU12-22/U4-8 and U91, as the latter corresponds to the 3′ half of the former. As these two RNAs were cloned separately, and appear to have distinct subnuclear localizations, they constitute two separate entries. The human orthologs of mouse snoRNAs cloned by Huttenhoffer et al. (14) were named by changing MBI- and MBII- to HBI- and HBII-, respectively.
RNAs can also be searched by length, GenBank accession number or target RNA. Typing 28S in the ‘Target RNA’ window will give a list of 28S rRNA modification guides. Typing ‘no’ gives the list of orphan snoRNAs without documented or predicted target RNA.
The sno/scaRNA pages
The database contains a total of 361 entries: 257 C/D box snoRNAs [149 by counting only one example of each of the 14q(I) (9 members), 14q(II) (31 members), HBII-85 (27 members) and HBII-52 (47 members) clusters], 86 H/ACA snoRNAs and 18 scaRNAs. Among those, 114 have no documented target RNA. The number of these so-called orphan snoRNAs reduces to 40 when counting 1 per cluster.
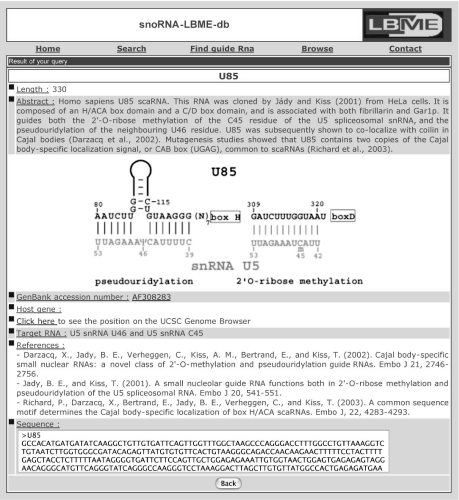
As an example, the page corresponding to U85 scaRNA is reproduced on Figure 1.
Figure 1.
Sample of snoRNA-LBME-db entry. Hyperlinks are provided with GenBank, the UCSC Genome Browser and PubMed.
The individual pages for snoRNAs or scaRNAs give the following information:
the most common name from the literature.
the length of the RNA.
an abstract that gives information on the manner the RNA was characterized, its predicted function (RNA modification guide, RNA chaperone and orphan RNA), with pertinent references. Other sno/scaRNAs with the same host gene are listed.
a drawing that indicates the predicted base pairing with the target RNA(s). This is unique among other vertebrate non-coding RNA databases.
the GenBank accession number, with a hyperlink to GenBank.
information on the host gene. Generally, genes are designated by the acronym used by the UCSC Genome Browser.
a hyperlink to the UCSC Genome Browser that will direct to the position of the sno/scaRNA gene in the human genome.
information on the target RNA(s) and nucleotide(s).
pertinent references with hyperlinks to PubMed.
the sequence in the FASTA format.
The ‘Find guide RNA’ pages
Individual pages for the 28S, 18S and 5.8S rRNAs, and the U1, U2, U4, U5, U6 and U12 snRNAs allow one to find sno/scaRNAs, if any, which are predicted to guide the modification of a particular nucleotide in these ARNs. One can thus see, for example, that pseudouridylation of 28S rRNA U4523 is predicted to be guided by both H/ACA box snoRNAs ACA7 and HBI-6. Pointing a nucleotide for a few seconds will display its numbering in the target RNA sequence. Clicking on the sno/scaRNA name will direct to the corresponding entry.
The Browse page
This page allows one to select sno/scaRNAs and get their sequence in the FASTA format. Pressing the ‘Toggle all’ allows to retrieve all sequences from the database. Clicking on the sno/scaRNA name will direct to the corresponding page.
Accessing the snoRNA-LBME-db from the UCSC Genome Browser
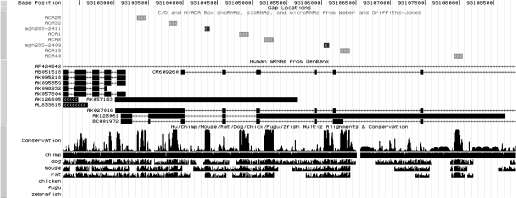
The snoRNAs and scaRNAs from the snoRNA-LBME-db were aligned against the human genome using BLAT (18) on the public USCS Genome Browser server. In a few cases, no exact match was found for the published sequence. In these cases, which probably correspond to sequencing errors, we adopted the best BLAT hit, which differed from the published sno/scaRNA sequence by 1–3 nt. The BLAT results were used to write a track named sno/miRNA that combines the sno/scaRNAs from the snoRNA-LBME-db, and miRNAs from miRBase (16). The C/D box snoRNAs, H/ACA snoRNAs, scaRNAs and miRNAs are indicated by different colors (Figure 2). Clicking on the name of a particular RNA directs one to the corresponding entry in the pertinent database, snoRNA-LBME-db or miRBase.
Figure 2.
Sample of snoRNA annotation on the human UCSC Genome Browser. C/D and H/ACA box snoRNAs are colored dark gray and light gray, respectively; and scaRNAs, white (not shown). In the case of the cluster shown, the examination of the Hu/Chimp/Mouse/Rat/Dog/Chick/Fugu/Zfish Multiz Alignment & Conservation track reveals that sequences corresponding to snoRNAs are more conserved than those of the exons of the putative host gene.
The sno/miRNA track allows one to visualize the position of a sno/scaRNA or a miRNA relative to other annotations [e.g. RefSeq genes, mRNAs and expressed sequence tags (ESTs)]. Moreover, the use of the sno/miRNA table in the ‘Genes and Gene Prediction Tracks’ allows one to answer many questions, such as the identity of sno/miRNAs that intersect with an EST in the entire genome or at a particular chromosome interval.
Future developments
The present nomenclature for sno/scaRNAs does not follow a general logic. Various names were given by different laboratories, such as En, Un, ACAn, Zn, HBII-n, HBI-n, 14q(I-n), snRn and mgU6-n (where n is an integer). Most of such names do not give any hint on the corresponding structural and functional family of the snoRNA. A name like mgU6-53 indicates that the snoRNA is predicted to guide the 2′-O-ribose methylation of nucleoside 53 of the U6 snRNA. Although logical, this nomenclature becomes complicated when a snoRNA (e.g. mgU12-22/U4-8) guides modifications in different RNAs and appears difficult to extend to scaRNAs (e.g. U85), which guide both a methylation and a pseudouridylation. We thus hope that the setting up of this database will provide the scientific community with an opportunity to adopt an unified nomenclature for sno/scaRNAs. In that case, newly discovered sno/scaRNAs might be submitted to the snoRNA-LBME-db to be attributed a consensus name.
This database is for the moment restricted to human. It would be useful to extend it to mouse by both collecting the published sequences and searching for mouse orthologs of human sno/scaRNAs. Moreover, we plan to build tables linking the human modification sites and snoRNAs with their homologs in other species like Saccharomyces cerevisiae.
Acknowledgments
We thank our colleagues at the Laboratoire de Biologie Moléculaire Eucaryote (Toulouse, France) for their help in setting up this database, and in particular Dr Jean-Pierre Bachellerie for collecting snoRNA sequences. We thank the UCSC Genome Browser team, in particular Jim Kent, Dona Karolchik, Fan Hsu and Hiram Clawson, for their help in the building of the human sno/miRNA track. We thank Dr Emmanuel Käs and Dr Tamas Kiss (Toulouse, France) for critically reading this manuscript. This work was supported by grants from CNRS and La Ligue Nationale contre le Cancer to Tamas Kiss. Funding to pay the Open Access publication charges for this article was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bachellerie J.P., Cavaille J., Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 2.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 4.Henras A.K., Dez C., Henry Y. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 2004;14:335–343. doi: 10.1016/j.sbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Cavaille J., Nicoloso M., Bachellerie J.P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 6.Kiss-Laszlo Z., Henry Y., Bachellerie J.P., Caizergues-Ferrer M., Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 7.Filipowicz W., Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 8.Weinstein L.B., Steitz J.A. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 9.Ganot P., Bortolin M.L., Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 10.Ni J., Tien A.L., Fournier M.J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 11.Jady B.E., Kiss T. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 2001;20:541–551. doi: 10.1093/emboj/20.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darzacq X., Jady B.E., Verheggen C., Kiss A.M., Bertrand E., Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tycowski K.T., Aab A., Steitz J.A. Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr. Biol. 2004;14:1985–1995. doi: 10.1016/j.cub.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Huttenhofer A., Kiefmann M., Meier-Ewert S., O'Brien J., Lehrach H., Bachellerie J.P., Brosius J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCloskey J.A., Rozenski J. The Small Subunit rRNA Modification Database. Nucleic Acids Res. 2005;33:D135–138. doi: 10.1093/nar/gki015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]