Abstract
In recent years, the Munich Information Center for Protein Sequences (MIPS) yeast protein–protein interaction (PPI) dataset has been used in numerous analyses of protein networks and has been called a gold standard because of its quality and comprehensiveness [H. Yu, N. M. Luscombe, H. X. Lu, X. Zhu, Y. Xia, J. D. Han, N. Bertin, S. Chung, M. Vidal and M. Gerstein (2004) Genome Res., 14, 1107–1118]. MPact and the yeast protein localization catalog provide information related to the proximity of proteins in yeast. Beside the integration of high-throughput data, information about experimental evidence for PPIs in the literature was compiled by experts adding up to 4300 distinct PPIs connecting 1500 proteins in yeast. As the interaction data is a complementary part of CYGD, interactive mapping of data on other integrated data types such as the functional classification catalog [A. Ruepp, A. Zollner, D. Maier, K. Albermann, J. Hani, M. Mokrejs, I. Tetko, U. Güldener, G. Mannhaupt, M. Münsterkötter and H. W. Mewes (2004) Nucleic Acids Res., 32, 5539–5545] is possible. A survey of signaling proteins and comparison with pathway data from KEGG demonstrates that based on these manually annotated data only an extensive overview of the complexity of this functional network can be obtained in yeast. The implementation of a web-based PPI-analysis tool allows analysis and visualization of protein interaction networks and facilitates integration of our curated data with high-throughput datasets. The complete dataset as well as user-defined sub-networks can be retrieved easily in the standardized PSI-MI format. The resource can be accessed through http://mips.gsf.de/genre/proj/mpact.
INTRODUCTION
The analysis of numerous genomes over the past decade contributed substantially to a comprehensive understanding of the complex biological processes in living cells since the ‘parts list’ of a genome lacks any information on the action of genes in context provided by the cellular environment. Several types of interaction networks such as metabolic pathways, regulatory modules or signaling cascades, which require coordinated action of many different proteins can be distinguished. The most exhaustively studied model for functional interactions in eukaryotes is the yeast Saccharomyces cerevisiae. In addition to the impressive number of individual experiments that uncover protein–protein interactions (PPIs) in yeast, data generated by several high-throughput techniques are available. Especially, large-scale yeast-two-hybrid analysis added valuable information to the understanding of the protein network in yeast (1,2). However, a major disadvantage of most high-throughput approaches is their significant rate of false-positive interactions. The overlap between the two large but independent yeast-two-hybrid data sets has been found to be remarkably low which gave rise to the question of how these data should be weighted. Since no straightforward benchmark standards of truth are available, manually curated data in the MPact dataset are accepted as a trusted standard (3,4).
Not only providing a sound reference for the evaluation of experimental results, MPact was used intensively for the validation of bioinformatics methods for predicting functional associations from experimental data. It was shown that genes with similar expression profiles are more likely to encode interacting proteins, thus describing a subset of functional modules, named ‘party hubs’, in contrast to ‘date hubs’ which consists of interacting proteins not synchronized by co-regulation (5–9). Extracting information from scientific literature and subsequent processing for systematic storage is a time consuming and expensive task. Accordingly, only few databases of manually compiled PPIs exist. CYGD (10), DIP (11), BIND (12), MINT (13) and HPRD (14) are important resources of this kind.
As applications mapping experimental data to PPIs ask not only for dynamic and interactive ways of retrieval, a new section of the Munich Information Center for Protein Sequences (MIPS) Genome Research Environment (GenRE) (http://mips.gsf.de/genre/proj/genre) was developed. This section, initially designed as a generic and versatile data structure for interaction data, was used to structure interaction data on mammalian proteomes (15). Using MPact, mapping of interaction data to other secondary information such as the functional classification catalog (FunCat) or mapping of interaction data to related proteomes is feasible (16). For instance the latter revealed that the likelihood of having an ortholog in other ascomycota species correlates with the number of interacting partners which show a clear preference to be pairwise conserved as a pair (17).
We describe MPact, a manually annotated protein interaction database in yeast as a reference for the experimental and theoretical work to elucidate the characteristics of cellular protein interaction networks (3,5,7,9). The power of the manually curated data set is illustrated by the network of proteins involved in signal transduction as an example. In addition, we describe a web-based tool, that allows scientists to analyze user-defined PPI-networks enabling investigation of protein subsets of interest. The resource can be accessed through http://mips.gsf.de/genre/proj/mpact.
METHODS
Software development
The MIPS interaction information resource is divided into several physically separated independent databases. This approach was chosen to fulfill different requirements of diverse protein interaction projects at MIPS like the MPPI resource (15).
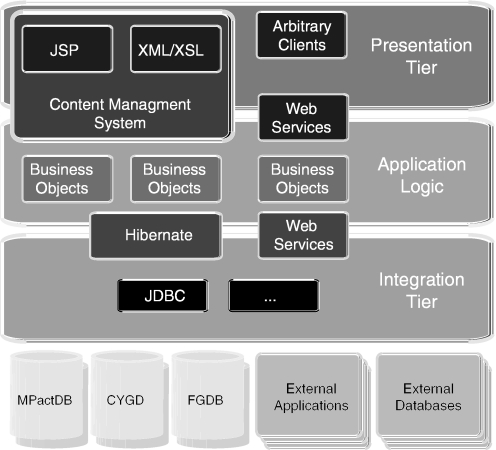
To avoid redundancy and possible inconsistencies, we focus on interaction relevant information and retrieve additional information about the interaction partners from related databases. Therefore, we decided to implement the resource with a component oriented approach. The MIPS GenRE (http://mips.gsf.de/genre/proj/genre) concept is built on linked but distributed components following the J2EE (http://java.sun.com/j2ee/) specification. The design principles of GenRE allow for seamless integration of different data sources and their representation as domain objects. The advantage of GenRE is its modularity that can be part of integrated distributed environments by introducing a multi-tier architecture with separated layers (Figure 1).
Figure 1.
The GenRE n-tier architecture. JSP—JavaServer Pages; XML—Extensible Markup Language; XSL—Extensible Stylesheet Language; JDBC—Java DataBase Connectivity; CYGD—Comprehensive Yeast Genome Database; and FGDB—Fusarium graminearum Genome DataBase.
The core classes comply with a light-weight object-oriented data model able to map the minimal information about protein interactions (http://mips.gsf.de/genre/proj/mpact/info/about.html), in accordance with the PSI-MI standard for exchange of protein interaction data (18). PSI-MI specifies minimal requirements for the description of molecular interactions like confidence levels and information necessary for protein identification. Additionally, it provides controlled vocabularies for experiment types or the role of the interactor in the experiment (e.g. bait and prey). The classes are mapped within the integration layer using Hibernate, an object/relational mapping technology (http://hibernate.org/) and do not access the databases directly. Retrieval of the data is performed by data access objects using the Hibernate persistence mechanism. Supplementary information about the interaction partners, such as functional annotation or localization, is accessed with similar components already available in GenRE.
On top of the core classes we developed components located in the application tier for further processing. Data is wrapped into a generic XML format allowing HTML generation by XSL style sheet transformation for the presentation layer. The generic XML format contains all the interaction information, including protein and gene annotation from in-house databases. Furthermore, the relevant subset of this information can be compiled into PSI-MI XML documents.
We restrict the access not only to internal applications but offering the same functionality also for web-wide external access. Therefore we also developed a HOBIT service layer (http://hobit.sf.net) based on the web service technology to share MPact in a programming language independent and web-wide way with the public domain. The MPact web service is accessible at http://mips.gsf.de/proj/hobitws/services/PsimiService?wsdl.
RESULTS
Data collection and retrieval
Although attempts have been published for natural language analysis and text-mining techniques (19,20), automatic extraction of information from scientific articles is still in its infancy and does not compete yet with high-quality manual annotation. While many journals require authors to deposit sequence information for new proteins and genes in one of the publicly available sequence databases, other knowledge such as protein interactions, regulation, signaling, cellular location or function is rarely submitted to an appropriate database or in an appropriate notation and hence are effectively lost for systematic approaches. The MIPS group has acquired long-standing experience in protein and genome annotation contributing to the protein database PIR-International as well as several genomes such as yeast and Arabidopsis thaliana. (21–25). In order to annotate PPIs, relevant articles are selected from PubMed using text-mining tools and processed by a human expert. The collection of yeast PPI data was started in the context of the original effort to annotate the S.cerevisiae genome for the Comprehensive Yeast Genome Database (CYGD) (25). As a consequence, our protein interaction data is well integrated with other CYGD data such as description and the localization data or functional classification of proteins, using the FunCat annotation scheme (26). Since then, newly discovered PPIs have been added continuously. Moreover, CYGD continues integrating data from high-throughput experiments.
Key information in the annotation of PPIs are the identification of the interacting partners, the kind of experiment as well as the original source of information (PubMed ID). For a standardized specification of experiments an evidence catalog exists. This is in line with the requirements for PSI-MI compliant annotation (see below). Based on this information it is possible to filter the data according to interaction type (physical/genetic) or to restrict the analysis to a certain type of experiment. As large-scale experiments have their unique strengths and weaknesses, and produce a significant fraction of false positives, it is important to distinguish this data from individual and manually extracted interactions described in the literature (3). We clearly make a distinction in our data using the ‘high-throughput’ tag (htp), indicating that these interactions should be filtered first while browsing the data or performing in-depth analysis.
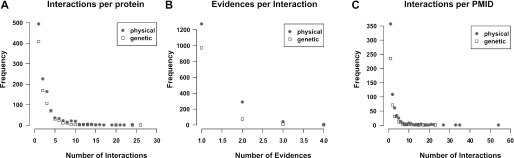
The reliability for any individual interaction described in the database increases by the number of annotated evidences. In MPact, the manually extracted data have on average 2.6 interactions per protein and are annotated with 1.2 evidences per interaction; 2.5 interactions are published per reference (Figure 2). In contrast to the lower quality of high-throughput data sets, highly reliable co-immunoprecipitation—together with affinity chromatography—experiments are the major source for the extracted data (Table 1).
Figure 2.
(A–C)Statistical analysis of the manually extracted data.
Table 1.
Distribution of evidences of the manually extracted data
| Evidence | % | No. |
|---|---|---|
| Co-immunoprecipitation | 43.7 | 777 |
| Two-hybrid | 40.0 | 712 |
| Affinity chromatography | 10.7 | 191 |
| Gel retardation | 1.9 | 33 |
| Centrifugation | 1.5 | 26 |
| Crosslinking | 1.1 | 19 |
| In vitro reconstitution | 0.7 | 12 |
| Overlay assay | 0.4 | 8 |
Browsing through the MPact protein interaction space
The database can be accessed through http://mips.gsf.de/genre/proj/mpact. Further details concerning the implementation are described in the method section.
Several types of predefined queries are available. ‘Query by Protein’ offers simple queries by searching for interactions of individual proteins by their systematic name, gene name or aliases. Queries are not limited to single proteins; alternatively selections using attributes such as functional categories based on the MIPS FunCat (26), cellular localization and EC number are possible.
Complex confinements of the search space are possible using ‘Query by Interaction’. Several filters can be applied; searches between two distinct individual proteins or lists of proteins can be performed. The result set delivers all interactions with at least one partner from each list. As in the ‘Query by Protein’ form, combinations of attributes are available. To consider the different strengths of certain interaction detection methods the user can choose to display only interactions derived from a specific method based on the PSI-MI controlled vocabulary. To distinguish the manually extracted data as described above, high-throughput experiments can be excluded. Since MPact contains both physical and genetic interactions we provide separate exclusion for these types. Finally, interactions described in a certain reference (PubMed ID) can be selected.
Search results are presented as tables, depending on the selection of the short or long output option different levels of detail are displayed. Short description of the interaction participants are linked to the corresponding entries of the CYGD database. For convenient navigation through the interaction network a link to the direct interactions of a specific participant is available. The long format additionally provides details such as the type of experimental evidence, PubMed references and a description of the interaction. MPact offers the possibility of extracting this result set in the standardized PSI-MI format.
Complementary to the tabular format, visualization of the interaction graph or its selected subgraphs is offered. Edges of the interaction graph are colored according to the number of evidences supporting the respective interaction. Additional information from CYGD including the functional annotation of the interacting proteins is included. Visualizations may be downloaded in PDF format for offline use and to allow enlargement of interesting regions even for very large graphs. As an example for the visualization and analysis of a complex cellular protein interaction network we focus on signal transduction processes.
Survey of the MPact dataset on signal transduction processes
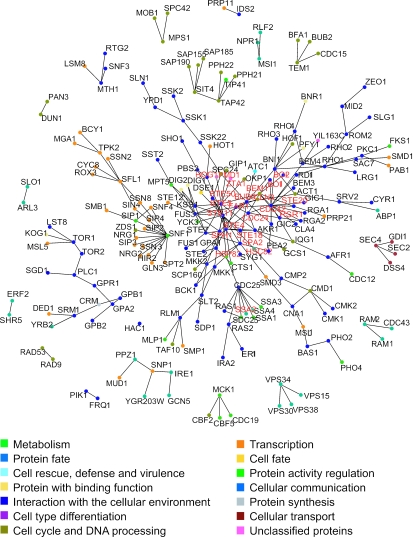
Protein interactions involved in signaling pathways provide a suitable example to illustrate the information collected and structured by MPact. Figure 3 shows all proteins of S.cerevisiae that have been annotated as signal transduction proteins and their physical interactions. Our dataset contains evidence of physical interactions with at least one partner for 190 (204 physical and genetic) out of a total of 231 signaling proteins. The vast majority of signal transduction proteins are connected through one large network including 51 members. While the majority of proteins in the graph are connected to only one or two binding partners a few nodes exhibit connections with a large number of other proteins thus serving as signaling hubs in the interaction network. This characteristic feature of scale-free networks has been shown to be applicable to most known biological interactions (27). The complete network as well as the signal transduction network follows a power law distribution indicating a scale-free behavior. The overall picture in Figure 3 shows that signal transduction in yeast is a highly complex network in which regulatory proteins do not necessarily interact directly but are linked through different players.
Figure 3.
Interaction network of signal transduction proteins restricted to physical interactions, high-throughput interactions not included. Node colors indicate the most prevalent functional category assigned to all proteins of a sub-network.
To get a notion of the completeness of our interaction collection, we compared our dataset with signal transduction pathways documented in the KEGG pathway database (28). In KEGG, signal transduction in yeast is shown for the MAPK signaling pathway, two-component system, second messenger pathway and phosphatidylinositol signaling system.
The MAPK pathways are highly conserved signaling units present in all eukaryotes, where they play essential roles in the response to environmental signals and hormones, growth factors and cytokines. They control cell growth, morphogenesis, proliferation and stress response (29). Figure 3 shows that members of the MAPK pathway appear in the centre of the large network, which agrees with the pivotal role of the MAPK pathway in information transfer processes. In KEGG, 54 proteins are displayed in the MAPK pathway of S.cerevisiae. Of these proteins 40 were annotated in CYGD as involved in signal transduction. Proteins that were assigned to signal transduction in KEGG but not in CYGD were found to be linked only peripherally to signal transduction processes. It is a general problem in the functional assignment of proteins, how to distinguish between core proteins of a biological process and others that are only associated with it. In KEGG, a total of 40 protein–protein relations are represented by single arrows or lines, which could theoretically also be found as PPI in MPact. In fact, our dataset includes 27 PPIs which are also part of the KEGG dataset. Differences between KEGG and our manually annotated dataset can originate for different reasons.
Annotation of our dataset is not comprehensive but represents only a fraction of the published PPIs. Accordingly, many interactions are indicated as high-throughput data but have not yet been published as individual experiments. For example, four PPIs from the MAPK pathway in KEGG can be found in mass-spectrometry analysis of protein complexes and large-scale two-hybrid experiments, respectively.
Interaction between proteins in signal transduction does not necessarily depend on physical interaction but may occur indirectly via regulation on the level of transcription.
The two-component system of yeast in KEGG consists of three proteins and their interactions. These are redundant in the MAPK pathway and completely represented in our dataset. The second messenger pathway and phosphatidylinositol signaling system in KEGG are represented by 17 and 9 proteins, respectively; 14 and 6 of those are found in our dataset.
Although physical interaction is not an obligatory condition in many signal transduction processes a comparison with three important signaling cascades taken from the KEGG database revealed good coverage of the respective pathways by our physical interaction data.
CONCLUSIONS
A comprehensive resource on yeast protein interaction data was set up as a reference for comparative genomics and setting a standard for other organisms such as human (15). To access the data a convenient data structure as well as a public interface is available allowing user-defined analysis of sub-networks and data retrieval in the standardized PSI-MI format. The data resource is interlinked with the CYGD database enabling in-depth mapping and analysis employing functional classification or localization data. As the resource is continuously updated its value for the community will steadily increase in future.
Acknowledgments
We thank Louise Riley for critical reading of the manuscript, Gisela Fobo, Barbara Brauner, Goar Frishman, Corinna Montrone and Irmtraud Dunger for excellent annotation. This work was supported by a grant of the German Federal Ministry of Education and Research (BMBF) within the BFAM framework (031U112C/212C), the European Commission (QLRI-CT 1999-01333) and the Impuls- und Vernetzungsfonds der Helmholtz-Gemeinschaft Deutscher Forschungszentren eV. Funding to pay the Open Access publication charges for this article was provided by the GSF National Research Center for Environment and Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 2.Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Mering C., Krause R., Snel B., Cornell M., Oliver S.G., Fields S., Bork P. Comparative assessment of large-scale data sets of protein–protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 4.Jansen R., Yu H., Greenbaum D., Kluger Y., Krogan N.J., Chung S., Emili A., Snyder M., Greenblatt J.F., Gerstein M. A Bayesian networks approach for predicting protein–protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 5.Ge H., Liu Z.H., Church G.M., Vidal M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nature Genet. 2001;29:482–486. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- 6.Teichmann S.A., Babu M.M. Conservation of gene co-regulation in prokaryotes and eukaryotes. Trends Biotechnol. 2002;20:407–410. doi: 10.1016/s0167-7799(02)02032-2. [DOI] [PubMed] [Google Scholar]
- 7.Tornow S., Mewes H.W. Functional modules by relating protein interaction networks and gene expression. Nucleic Acids Res. 2003;31:6283–6289. doi: 10.1093/nar/gkg838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spirin V., Mirny L.A. Protein complexes and functional modules in molecular networks. Proc. Natl Acad. Sci. USA. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han J.D.J., Bertin N., Hao T., Goldberg D.S., Berriz G.F., Zhang L.V., Dupuy D., Walhout A.J.M., Cusick M.E., Roth F.P., et al. Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 10.Güldener U., Münsterkötter M., Kastenmüller G., Strack N., Van Helden J., Lemer C., Richelles J., Wodak S.J., Garcia-Martinez J., Perez-Ortin J.E., et al. CYGD: the Comprehensive Yeast Genome Database. Nucleic Acids Res. 2005;33:D364–D368. doi: 10.1093/nar/gki053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. The Database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bader G.D., Betel D., Hogue C.W.V. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 14.Peri S., Navarro J.D., Kristiansen T.Z., Amanchy R., Surendranath V., Muthusamy B., Gandhi T.K.B., Chandrika K.N., Deshpande N., Suresh S., et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:D497–D501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pagel P., Kovac S., Oesterheld M., Brauner B., Dunger-Kaltenbach I., Frishman G., Montrone C., Mark P., Stümpflen V., Mewes H.W., et al. The MIPS mammalian protein–protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 16.Yu H.Y., Luscombe N.M., Lu H.X., Zhu X.W., Xia Y., Han J.D.J., Bertin N., Chung S., Vidal M., Gerstein M. Annotation transfer between genomes: protein–protein interologs and protein–DNA regulogs. Genome Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagel P., Mewes H.W., Frishman D. Conservation of protein–protein interactions—lessons from ascomycota. Trends Genet. 2004;20:72–76. doi: 10.1016/j.tig.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Hermjakob H., Montecchi-Palazzi L., Bader G., Wojcik R., Salwinski L., Ceol A., Moore S., Orchard S., Sarkans U., von Mering C., et al. The HUPOPSI's molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 19.Muller H.M., Kenny E.E., Sternberg P.W. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:1984–1998. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krallinger M., Erhardt R.A.A., Valencia A. Text mining approaches in molecular biology and biomedicine. Drug Discov. Today. 2005;10:439–445. doi: 10.1016/S1359-6446(05)03376-3. [DOI] [PubMed] [Google Scholar]
- 21.Wu C.H., Huang H.Z., Arminski L., Castro-Alvear J., Chen Y.X., Hu Z.Z., Ledley R.S., Lewis K.C., Mewes H.W., Orcutt B.C., et al. The protein information resource: an integrated public resource of functional annotation of proteins. Nucleic Acids Res. 2002;30:35–37. doi: 10.1093/nar/30.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruepp A., Graml W., Santos-Martinez M.L., Koretle K.K., Volker C., Mewes H.W., Frishman D., Stocker S., Lupas A.N., Baumeister W. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature. 2000;407:508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- 23.Salanoubat M., Lemcke K., Rieger M., Ansorge W., Unseld M., Fartmann B., Valle G., Blocker H., Perez-Alonso M., Obermaier B., et al. Sequence and analysis of chromosome 3 of the plant Arabidopsis thaliana. Nature. 2000;408:820–822. doi: 10.1038/35048706. [DOI] [PubMed] [Google Scholar]
- 24.Galagan J.E., Calvo S.E., Borkovich K.A., Selker E.U., Read N.D., Jaffe D., FitzHugh W., Ma L.J., Smirnov S., Purcell S., et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 25.Mewes H.W., Albermann K., Bahr M., Frishman D., Gleissner A., Hani J., Heumann K., Kleine K., Maierl A., Oliver S.G., et al. Overview of the yeast genome. Nature. 1997;387:7–8. doi: 10.1038/42755. [DOI] [PubMed] [Google Scholar]
- 26.Ruepp A., Zollner A., Maier D., Albermann K., Hani J., Mokrejs M., Tetko I., Güldener U., Mannhaupt G., Münsterkötter M., et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barabasi A.L., Oltvai Z.N. Network biology: understanding the cell's functional organization. Nature Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 28.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]