Abstract
The Nucleolar Proteome Database (NOPdb) archives data on >700 proteins that were identified by multiple mass spectrometry (MS) analyses from highly purified preparations of human nucleoli, the most prominent nuclear organelle. Each protein entry is annotated with information about its corresponding gene, its domain structures and relevant protein homologues across species, as well as documenting its MS identification history including all the peptides sequenced by tandem MS/MS. Moreover, data showing the quantitative changes in the relative levels of ∼500 nucleolar proteins are compared at different timepoints upon transcriptional inhibition. Correlating changes in protein abundance at multiple timepoints, highlighted by visualization means in the NOPdb, provides clues regarding the potential interactions and relationships between nucleolar proteins and thereby suggests putative functions for factors within the 30% of the proteome which comprises novel/uncharacterized proteins. The NOPdb (http://www.lamondlab.com/NOPdb) is searchable by either gene names, nucleotide or protein sequences, Gene Ontology terms or motifs, or by limiting the range for isoelectric points and/or molecular weights and links to other databases (e.g. LocusLink, OMIM and PubMed).
INTRODUCTION
The nucleolus is the most prominent structure within the eukaryotic nucleus and is known for its role in ribosomal RNA (rRNA) transcription, processing and the subsequent assembly of processed rRNA with ribosomal proteins to form ribosomal subunits (1–3). Recent studies suggested that the mammalian nucleolus may also play roles in tumourigenesis (4), viral replication (5) and cellular stress responses (6). However, the pathway and the identities of the molecular machineries involved in these mechanisms within this nuclear organelle remained largely unknown. Due to its inherent high density, nucleoli from cultured human cells can be isolated readily from sonicated nuclear extracts (7). Taking advantage of this, we and others have previously employed mass spectrometry (MS) techniques to identify the protein components from highly purified nucleolar preparations (8–10). Furthermore, fluorescent protein-tagging experiments and photobleaching analyses have vividly demonstrated the dynamic nature of the nucleolar proteome, where proteins only accumulate in the nucleolus either under specific metabolic conditions, or at specific cell cycle stages (11). Recently, we have extended our MS analyses to measure the dynamic behaviour of the nucleolar proteome by quantitating the relative level of individual nucleolar components upon transcriptional inhibition using a method known as stable isotope labelling with amino acids in cell culture (SILAC) (12).
DATABASE ACCESS AND CONTENT
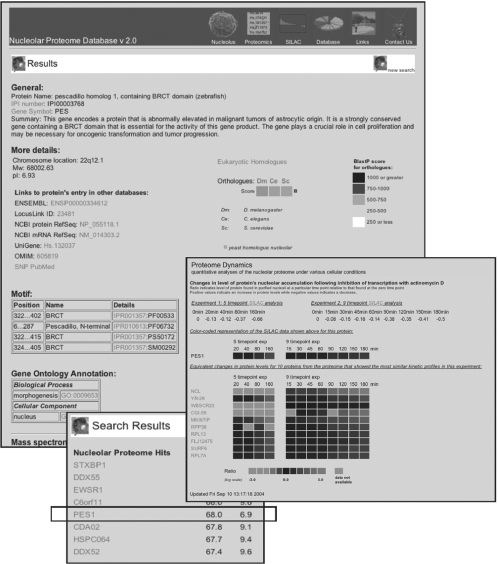
To facilitate the analysis of these quantitative proteomic data, we have established the Nucleolar Proteome Database (NOPdb), a database aiming to archive all the human nucleolar proteins identified by MS analyses so far (13). The current version 2.0 of the database is available at http://www.lamondlab.com/NOPdb/ and is searchable by gene name/symbol, protein sequence, motif (14–16), Gene Ontology (GO) terms (17) or by setting the range of the predicted isoelectric point and/or molecular weight (Figure 1). To date, NOPdb archives 728 human nucleolar proteins (covering ∼2.5% of the predicted human proteome) verified by multiple MS analyses and documents the quantitative changes in protein levels for 498 of these proteins at multiple timepoints after transcription is inhibited by treating cells with Actinomycin D.
Figure 1.
Snapshots of the NOPdb (http://www.lamondlab.com/NOPdb/). The database was searched against molecular weights between 65 and 70 kDa and here we show an overview page for the PES1 protein (pescadillo) documenting its motif distribution, its GO annotations, its identification history by multiple MS analysis and its quantitative data from SILAC analyses. Proteins of similar kinetic profiles based on correlation coefficient are identified for future investigation. The kinetic profiles are ranked according to the Pearson's correlation coefficients for the log value of the peak intensities of multiple peptides at a particular timepoint normalized to the respective peak intensities at zero timepoints.
The NOPdb provides (i) information on gene sequences and chromosomal localization, (ii) information on primary protein sequence (including protein sequence, predicted isoelectric point and molecular weight and motif structure) and (iii) information about putative nucleolar protein homologues in fruitfly, nematode and yeast, and also their localization data in these species, if available (18,19). A dedicated section for MS data has included the identification history of these nucleolar proteins in multiple MS analyses, peptide sequences deduced by tandem MS and the details of the MS experiments. Functions of these proteins are described using GO terms and detailed comments manually curated in the Entrez Gene database (20). In addition, the NOPdb also acts as a gateway to other databases, including NCBI LocusLink (20), OMIM (21), PubMed (9), UniGene (20) and ENSEMBL (22).
ACCESS TO PROTEOME DYNAMICS
A general problem experienced in proteome analyses is the abundance of novel/uncharacterized proteins (∼30% in the case of the nucleolus) where limited information is available regarding their function (9,13). Therefore, the availability of quantitative information allows for the first time the ability to annotate/classify the proteome according to the changes in individual protein levels at multiple timepoints upon drug treatment. Analogous to the gene expression profiles generated for microarray data (23), we used SILAC data to generate a unique kinetic profile over time for each protein, where the relative abundance of each protein is compared with its respective level at the initial timepoint. Unlike microarray data, the quantitative measurements are made at the post-transcriptional level. The changes in the levels of protein in the nucleolus after drug treatment likely reflect their respective functional roles. Moreover, proteins with similar kinetic profiles based on Pearson's correlation coefficients can be identified, through the visualization means in the NOPdb, where available. This information makes direct predictions that can subsequently be tested both in vivo and in vitro.
PERSPECTIVES
Future versions of the NOPdb will include additional kinetic profiles for each protein, based on their responses to both different drug treatments and other metabolic and cell cycle variations. Clustering of such data may offer useful information for predicting the potential functions of these novel proteins (24). Apart from shedding light to the functions of novel proteins, clustered protein groups can be served as refined sets for motif search. Bioinformatic tools will also be developed to provide means to interact with the related microarray data deposited in the public domain. Comparison of these profiles with gene expression profiles from parallel microarray data may yield fresh understanding of the post-transcriptional regulation of the corresponding genes. Current analyses on the primary sequences deposited in the NOPdb determined a number of properties of the nucleolar proteome in terms of the distribution of amino acid/short peptide composition (13), domain structure and GO terms (Supplementary Tables 1 and 2), which are statistically different from the profiles of proteins accumulated within other cellular structures or organelles. In summary, the NOPdb provides a useful resource for the scientific community to explore the plurifunctionality of nucleolus, where further surprises are probably still in store.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
Supplementary Material
Acknowledgments
A.K.L.L. was supported by a Croucher Foundation Scholarship. A.I.L. is a Wellcome Trust Principal Research Fellow. The Human Frontier Science Program is acknowledged for a research grant entitled ‘Functional organization of the cell nucleus investigated through proteomics and molecular dynamics’. The work in the Lamond laboratory is supported by the Wellcome Trust and work in the Mann laboratory is funded by a Danish National Research Foundation grant to the Centre for Experimental Bioinformatics. Funding to pay the Open Access publication charges for this article was provided by Joint Information Systems Committee of the UK.
Conflict of interest statement. None declared.
REFERENCES
- 1.Leary D.J., Huang S. Regulation of ribosome biogenesis within the nucleolus. FEBS Lett. 2001;509:145–150. doi: 10.1016/s0014-5793(01)03143-x. [DOI] [PubMed] [Google Scholar]
- 2.Tschochner H., Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 3.Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruggero D., Pandolfi P.P. Does the ribosome translate cancer? Nature Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 5.Hiscox J.A. The nucleolus—a gateway to viral infection? Arch. Virol. 2002;147:1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olson M.O. Sensing cellular stress: another new function for the nucleolus? Sci. STKE. 2004;2004:pe10. doi: 10.1126/stke.2242004pe10. [DOI] [PubMed] [Google Scholar]
- 7.Busch H., Muramatsu M., Adams H., Steele W.J., Liau M.C., Smetana K. Isolation of nucleoli. Exp. Cell Res. 1963;24(Suppl 9):150–163. [PubMed] [Google Scholar]
- 8.Scherl A., Coute Y., Deon C., Calle A., Kindbeiter K., Sanchez J.C., Greco A., Hochstrasser D., Diaz J.J. Functional proteomic analysis of human nucleolus. Mol. Biol. Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersen J.S., Lyon C.E., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 10.Andersen J.S., Lam Y.W., Leung A.K., Ong S.E., Lyon C.E., Lamond A.I., Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 11.Leung A.K., Lamond A.I. The dynamics of the nucleolus. Crit. Rev. Eukaryot. Gene Expr. 2003;13:39–54. doi: 10.1615/critreveukaryotgeneexpr.v13.i1.40. [DOI] [PubMed] [Google Scholar]
- 12.Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Leung A.K., Andersen J.S., Mann M., Lamond A.I. Bioinformatic analysis of the nucleolus. Biochem. J. 2003;376:553–569. doi: 10.1042/BJ20031169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Barrell D., Bateman A., Binns D., Biswas M., Bradley P., Bork P., et al. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 2003;31:315–318. doi: 10.1093/nar/gkg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 19.Mewes H.W., Amid C., Arnold R., Frishman D., Guldener U., Mannhaupt G., Munsterkotter M., Pagel P., Strack N., Stumpflen V., et al. MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 2004;32:D41–D44. doi: 10.1093/nar/gkh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler D.L., Church D.M., Edgar R., Federhen S., Helmberg W., Madden T.L., Pontius J.U., Schuler G.D., Schriml L.M., Sequeira E., et al. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 2004;32:D35–D40. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamosh A., Scott A.F., Amberger J., Bocchini C., Valle D., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birney E., Andrews D., Bevan P., Caccamo M., Cameron G., Chen Y., Clarke L., Coates G., Cox T., Cuff J., et al. Ensembl 2004. Nucleic Acids Res. 2004;32:D468–D470. doi: 10.1093/nar/gkh038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spellman P.T., Sherlock G., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.