Abstract
EMAGE (http://genex.hgu.mrc.ac.uk/Emage/database) is a freely available, curated database of gene expression patterns generated by in situ techniques in the developing mouse embryo. It is unique in that it contains standardized spatial representations of the sites of gene expression for each gene, denoted against a set of virtual reference embryo models. As such, the data can be interrogated in a novel and abstract manner by using space to define a query. Accompanying the spatial representations of gene expression patterns are text descriptions of the sites of expression, which also allows searching of the data by more conventional text-based methods.
INTRODUCTION
With the completion of whole genome sequencing of various model organisms, the challenge facing modern biology now is to determine the biological roles and interactions that each gene and their products play. Central to addressing this problem is an understanding of the sites of expression (at both the transcript and protein level) throughout development as well as in the adult organism. Techniques for determining sites of expression in situ such as immunohistochemistry and in situ hybridization offer direct visualization of the accumulation of gene products at cellular resolution, and as such, are central to the elucidation of gene function in complex tissues.
Traditionally, data of this type has been archived using the conventional method of publication in a journal of at least one photograph of the expression results with an accompanying brief description of the sites of expression as determined by the authors. Whilst this method does allow for the distribution of the information, data published in this way is often impossible to retrieve from the literature unless the gene or sites of expression in question are mentioned in the literature citation, i.e. in the title, abstract, keywords or MeSH (Medical Subject Headings, http://www.nlm.nih.gov/mesh/meshhome.html) terms of the paper. In the case of gene expression patterns, this is usually not included in the citation unless the paper is specifically written to describe the expression pattern itself. When this information is included in the citation, it is still often difficult to retrieve owing to the use of non-standardized language by authors to describe the sites of gene expression. In addition, patterns are often not completely described by the authors. This may arise from a lack of anatomical knowledge that is required to identify and name all of the structures that express the gene, or from the complexities of a text-based description required to describe a complex expression pattern.
To help address these problems, we have developed EMAGE, a database in which gene expression patterns detected using in situ techniques are described uniquely by using a combination of both text (i.e. standardized words are used to list the anatomical parts that express a gene) and space (i.e. standardized spatial representations are used to show the sites of expression). A proportion of the data in EMAGE has been published previously in the literature and our approach extends the amount of information that can be extracted from the original data images as well as providing a new access gateway and novel analysis possibilities to these data. This approach also allows access to expression data for users who are not, or are only superficially, familiar with the anatomy of the mouse embryo.
DATABASE STRUCTURE
Concept
The framework that houses all data contained in the EMAGE database is the EMAP Digital Atlas of Mouse Development (1). This interactive atlas contains two parts—a hierarchically organized ontology of anatomical terms for all Theiler stages (2,3) of mouse development [which is also employed by our colleagues at the GXD database to index expression data from multiple experimental sources (4)], and a set of virtual 3D mouse embryo models for most post-implantation stages of Theiler stages of development. In a proportion of the 3D embryo models, 3D anatomical domains have been delineated within them and these correspond directly to terms in the ontology (1). See Table 1 for a summary of the current status of this framework for each Theiler Stage and also online Supplementary Data for a document which gives a detailed description of the concepts of the database framework and the rules and definitions of the annotation.
Table 1.
Current status of the framework housing EMAGE data
| Theiler stage | Anatomy ontology | 3D embryo model | 3D anatomy domains in model |
|---|---|---|---|
| 01 | + | − | n/a |
| 02 | + | − | n/a |
| 03 | + | − | n/a |
| 04 | + | − | n/a |
| 05 | + | − | n/a |
| 06 | + | − | n/a |
| 07 | + | + | + |
| 08 | + | + | + |
| 09 | + | + | + |
| 10 | + | + | + |
| 11 | + | + | + |
| 12 | + | + | + |
| 13 | + | + | + |
| 14 | + | + | + |
| 15 | + | + | − |
| 16 | + | + | − |
| 17 | + | + | − |
| 18 | + | + | − |
| 19 | + | + | − |
| 20 | + | + | + |
| 21 | + | − | n/a |
| 22 | + | − | n/a |
| 23 | + | − | n/a |
| 24 | + | − | n/a |
| 25 | + | − | n/a |
| 26 | + | − | n/a |
For all embryonic Theiler stages (TS01-26) an ontology of anatomical terms have been developed. A 3D embryo model is present to spatially house EMAGE data at TS07-20 inclusive, and of these, 3D anatomical regions have been defined within the TS07-14 and TS20 embryo models. The work of refining the anatomical ontology, building embryo models and delineating anatomy is ongoing.
Software
The database software has client–server architecture. The application-domain information is stored in an ObjectStore (http://www.progress.com) database. The database includes C++ server and Java Swing (http://www.sun.com) client software. The C++ server accesses the database and runs on a Sun Solaris server. The Java client software is delivered by Java Web Start (http://www.sun.com) or directly through a web browser and communicates with the C++ server via Corba (http://www.orbacus.com).
CONTENTS OF THE EMAGE DATABASE
Every EMAGE entry contains at least one original data photograph showing expression as detected with either one nucleic acid probe or antiserum. Also contained in the entry, is an accompanying spatial and/or text annotation, which describes the sites of expression as seen in the assay image(s). Further information on the contents of an entry can be found in the ‘Data Entry’ section.
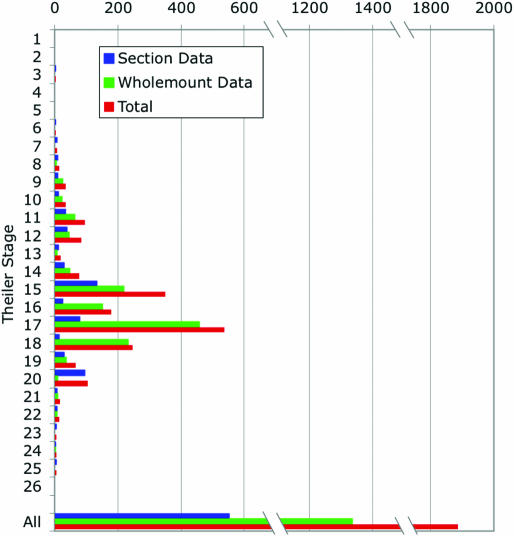
As of July 2005, there were 1905 entries covering 704 genes and 22 Theiler stages of development. Of these entries, 1345 correspond to data from wholemount and 560 from sections of in situ/immunohistochemistry experiments (for a further breakdown see Figure 1). Of all entries, 10% correspond to direct submissions from individual laboratories, 44% to data that has been published previously in the literature and 46% are submissions from screening consortia.
Figure 1.
Contents of the public EMAGE database as of July 2005. The numbers of wholemount (green), section (blue) and total (red) EMAGE entries per Theiler Stage in the public EMAGE database is shown.
DATA QUERYING IN EMAGE
Querying via the Java interface
Downloading and starting the Java interface
The EMAGE Java interface can be downloaded from the EMAGE homepage (http://genex.hgu.mrc.ac.uk/Emage/database) and will run on any platform (Windows, MacOSX and UNIX/Linux) with Java v1.4.2+ installed. Platform-specific instructions are given on the website. Following installation, the Java interface can be launched from the EMAGE homepage by clicking the ‘START’ icon. Every time the interface is subsequently launched when the computer is connected to the internet, the latest version of the software is installed. The data in EMAGE can also be accessed in HTML, without downloading the Java interface (see below).
Using the Java interface to browse EMAGE data
When the Java interface launches, a ‘Browse’ window is automatically loaded. This gives an indication of the number of public entries in the database at that time. The list can be sorted by gene or Theiler stage and is presented as a hierarchically organized tree. Branches of the tree can be opened by clicking on the toggles. Double/right clicking on a term in the tree (i.e. gene symbol, Theiler stage, EMAGE ID) will load thumbnail images of the original data in the adjacent panel. Individual EMAGE entries can be opened by double/right clicking on the thumbnail images (for a movie of this type of search in action see online Supplementary Data).
Performing spatial searches in the EMAGE Java interface
By using the menu option Central Database > Search By Any, a user can formulate the query: ‘What (genes) are (detected/possibly detected/not detected) in the following (region) at (Theiler Stage X)?’.
Two types of spatial expression data are held in the database—those with original data from sectioned samples and those with original data as images of wholemount stained embryos. These two distinct data types are treated separately in the database and are queried independently of each other.
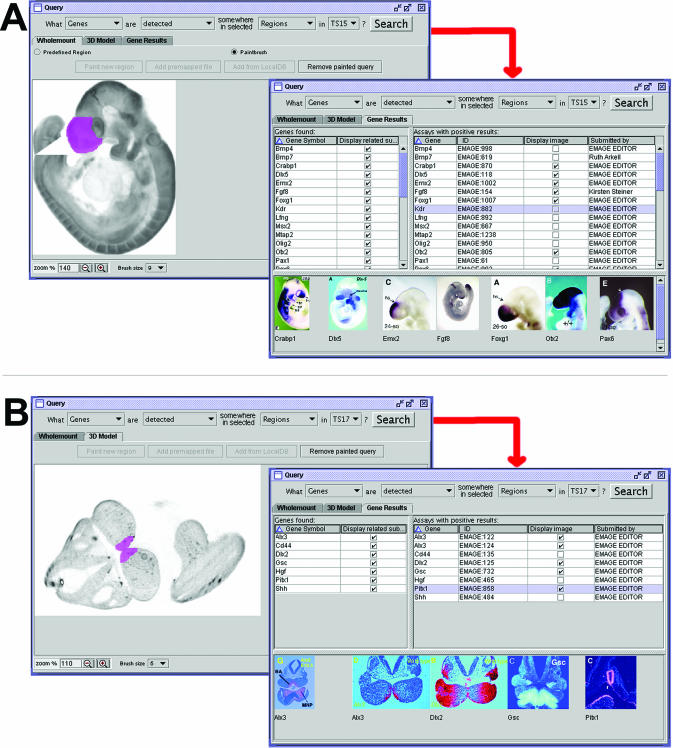
The user defines their query area by painting an arbitrary region, either onto a lateral (left or right) wholemount view or into the 3D space of one of the EMAP 3D virtual embryo models. This allows either wholemount or 3D (section) data to be retrieved that spatially intersects with the query domain. See Figure 2 for examples and also online Supplementary Data for movies of these types of searches being performed.
Figure 2.
Spatial queries formulated using the EMAGE Java interface. Examples are shown of the query type ‘What genes are expressed in this region, at this stage?’ for (A) ‘wholemount’ data and (B) 3D data. Shown in the left hand panel in each case are arbitrary regions that have been defined using a simple paint tool (shown in magenta). Following searching of the central EMAGE database, a list of genes expressed somewhere in the specified region are returned, and the user can choose to display thumbnails of original data (shown in the right panels). Further details associated with an entry [such as probe, specimen, submitter information, links, further original data images (full-size) and the annotation] can be obtained by double clicking on the thumbnail image (for movies of these and other types of searches see online Supplementary Data).
In addition, the following query type can be formulated: ‘What (regions) (express/possibly express/do not express) the following (genes) at (Stage X)?’ The user defines their gene of interest and subsequently a list of 2D and/or 3D regions are returned with accompanying original data images. The 2D (wholemount) regions are displayed directly in the Java interface. Returned 3D regions can be downloaded (in woolz image format, http://genex.hgu.mrc.ac.uk/Software/woolz/) from the Java interface and subsequently visualized using accompanying MAPaint software (http://genex.hgu.mrc.ac.uk/MouseAtlasCD/Software.html). MAPaint requires a UNIX operating environment (Linux, MacOSX, Solaris and so on), is free and can be obtained by emailing a request to ma-cdrom@hgu.mrc.ac.uk. For movies of these types of searches being performed see online Supplementary Data.
Performing text-based searches in the EMAGE Java interface
Following selection of the menu option Central Database > Search By Any, a user can also formulate the query: ‘What (genes) are (detected/possibly detected/not detected) in the following (named anatomical components) at (Theiler Stage X)?’. The user is presented with the ontology tree of anatomical terms for the Theiler stage chosen and can browse through the list or query it by text to find instances of the term of interest. Common synonyms are included in the anatomy nomenclature database and will also be returned from text queries of the ontology (e.g. searching the list for ‘mesencephalon’ at TS15 will return the synonym ‘future midbrain’). Appropriate terms are selected by the user by clicking on them and they will change colour to magenta denoting the term is selected for search. Following the search, a list of genes are returned with accompanying original data images (for a movie showing a search of this type see Supplementary Data).
In addition, the following query type can be formulated: ‘What (named anatomical components) (express/possibly express/do not express) the following (genes) at (Stage X)?’. The ontology tree of anatomical terms for the Theiler stage chosen is returned with relevant terms highlighted (in magenta). Because of the hierarchical nature of the ontology tree, all terms including and higher than those directly annotated are returned from this query type when querying for structures expressing the gene, and all terms including and lower than those directly annotated are returned when querying for structures with no expression of the gene detected. For a movie showing a search of this type see online Supplementary Data.
Querying EMAGE via HTML
EMAGE data is also accessible via HTML. This allows searching of the central EMAGE database by anatomical structure at a particular Theiler stage. The HTML search is accessed from the homepage of EMAGE (http://genex.hgu.mrc.ac.uk/Emage/database/) by clicking on the ‘SEARCH’ icon under the ‘WEB SEARCH EMAGE’ link. The user is presented with a Java Applet interface that allows selection of a Theiler stage and subsequent browsing of the ontology of anatomical terms. Selection of an ontology term allows searching of EMAGE to find genes expressed within that structure. The results are presented as a list and by following links, the user is able to peruse the content of individual EMAGE entries to view the original data images and associated annotation.
EMAGE data can also be reached from an Ensembl (5) mouse gene report via a distributed annotation system [DAS, (6)] server. Under these circumstances, EMAGE is listed amongst the DAS resources in the Ensembl entry and when selected, results are presented in a tabular format displaying the gene symbol as well as the Theiler stages and the total number of tissues in which expression is annotated in EMAGE for the gene. Selection of any of these fields allows the user to browse relevant lists and through a series of links ultimately peruse the content of individual EMAGE entries.
DATA SOURCING AND ENTRY IN EMAGE
Data sourcing
Data in EMAGE comes from a variety of sources. Submissions can be sent directly from individual labs/screening consortia. In these circumstances, the EMAGE Java interface can be used to create a local, private database and then send individual or multiple entries to EMAGE Editorial Staff for curation. Alternatively, appropriate images or specimens can be sent to EMAGE Editorial Staff for data mapping and entry, either by email (to ma-edit@hgu.mrc.ac.uk) or by post (to EMAGE Editorial Office, MRC Human Genetics Unit, Western General Hospital, Crewe Road, Edinburgh, EH4 2XU, UK).
A proportion of the data in EMAGE has been published previously in the literature and incorporation of this data is performed in collaboration with the GXD database (MGI) (4). GXD curators scan the literature and annotate the EMAP text anatomy ontology (7) to indicate the sites of gene expression according to each author's text description. GXD and EMAGE have joint global copyright agreements with the Company of Biologists Ltd and Elsevier B.V. to reproduce images that have been published in the journals Development, Developmental Biology, Gene Expression Patterns and Mechanisms of Development. At EMAGE, we scan through available images associated with GXD entries and spatially map data from suitable images. This process extracts more information from the experiment about the sites of gene expression than is commonly achieved using a text annotation according to the author's brief original description alone.
Data entry
Entries can be made for in situ hybridization, immunohistochemistry or transgenic reporter experiments that reflect the endogenous pattern of gene expression. Information included in an EMAGE entry includes: (i) Probe/antibody information—including a unique probe identifier, Mouse Gene Nomenclature Committee approved gene name and symbol (with associated MGI gene ID), nucleic acid sequence of the probe or amino acid sequence of the target epitope of the antisera (when known) and the labelling and visualization methods used. (ii) Information about the Specimen—including the Theiler stage, alternate staging method and value (e.g. dpc, somite number), strain, sex, fixation/embedding treatment and notes useful for interpretation of the data. At least one original data image is always included. (iii) Submitter/author contact details. (iv) Links to relevant data in other databases—e.g. PubMed and MGI/GXD. (v) Annotation—if the annotation is spatial, this includes a digital representation of the sites and levels of gene expression in the data embryo in spatially equivalent regions of the appropriately stage-matched virtual embryo model.
Briefly, spatial annotation involves the ‘warping’ and subsequent extraction of signal of a digital representation of the original data image into the standard spatial context of one of the EMAP digital embryo models. This is achieved using the accompanying MAPaint program (for further information on the spatial mapping process see http://genex.hgu.mrc.ac.uk/MouseAtlasCD/html/guides/sect070104.htm and http://genex.hgu.mrc.ac.uk/MouseAtlasCD/html/guides/wm070104.htm).
The annotation can also include a simple text annotation to the EMAP anatomy ontology which may also include information on levels and patterns (e.g. graded, restricted and so on) of expression in the original data image.
Levels are denoted broadly to reflect regions of strongest, moderate and weakest expression in the data image, as well as regions of possible expression, and regions where no expression is detected in the assay. The colour convention employed is red for ‘strongest’, yellow for ‘moderate’, blue for ‘weakest’, green for ‘possible’ and cyan for ‘not detected’.
In cases where spatial annotation has been performed into the 3D space of an embryo model with 3D anatomical domains defined within it, the expression domain will intersect with regions of known anatomy. Under these circumstances, the spatial annotation is therefore accompanied by an automatically generated text annotation to the corresponding terms in the anatomical ontology.
FUTURE DIRECTIONS
EMAGE will continue to acquire and spatially map gene expression patterns in the developing mouse embryo. This includes data from the literature in conjunction with our colleagues at the GXD database (4), as well as data acquired from large-scale expression screening consortia as well as that from individual labs. We are currently developing tools to allow direct spatial comparisons of multiple complex patterns and hierarchical clustering of these to segregate any number of patterns into groups showing spatial similarities. We are also working towards incorporation of full 3D datasets into EMAGE that are derived from stained whole mount embryos imaged using Optical Projection Tomography (8).
USER SUPPORT
There is dedicated User Support for EMAGE. Please refer issues to ma-edit@hgu.mrc.ac.uk.
We also provide Hands-On User Courses on a regular basis. For details see http://genex.hgu.mrc.ac.uk/Emage/courses/course.html.
CITING EMAGE
To reference the EMAGE database, please cite this article. For referring to specific data entries in the database, please list the EMAGE:ID and also mention that the data was retrieved from the EMAGE database, MRC Human Genetics Unit, Edinburgh, UK (http://genex.hgu.mrc.ac.uk).
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
Supplementary Material
Acknowledgments
EMAGE is supported by the Medical Research Council. We would like to thank Dr Martin Ringwald and our colleagues at the GXD database (MGI, Jackson Laboratory, Maine USA) for input and also The Company of Biologists Ltd and Elsevier B.V. for copyright agreements allowing the reproduction of images from Development, Developmental Biology, Mechanisms of Development and Gene Expression Patterns. Funding to pay the Open Access publication charges for this article was provided by the Medical Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Baldock R.A., Bard J.B., Burger A., Burton N., Christiansen J., Feng G., Hill B., Houghton D., Kaufman M., Rao J., et al. EMAP and EMAGE: a framework for understanding spatially organized data. Neuroinformatics. 2003;1:309–325. doi: 10.1385/NI:1:4:309. [DOI] [PubMed] [Google Scholar]
- 2.Kaufman M. The Atlas of Mouse Development. London: Academic Press; 1992. [Google Scholar]
- 3.Theiler K. The House Mouse: Atlas of Embryonic Development. NY: Springer-Verlag; 1989. [Google Scholar]
- 4.Hill D.P., Begley D.A., Finger J.H., Hayamizu T.F., McCright I.J., Smith C.M., Beal J.S., Corbani L.E., Blake J.A., Eppig J.T., et al. The mouse Gene Expression Database (GXD): updates and enhancements. Nucleic Acids Res. 2004;32:D568–D571. doi: 10.1093/nar/gkh069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hubbard T., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowell R.D., Jokerst R.M., Day A., Eddy S.R., Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bard J.L., Kaufman M.H., Dubreuil C., Brune R.M., Burger A., Baldock R.A., Davidson D.R. An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech. Dev. 1998;74:111–120. doi: 10.1016/s0925-4773(98)00069-0. [DOI] [PubMed] [Google Scholar]
- 8.Sharpe J., Ahlgren U., Perry P., Hill B., Ross A., Hecksher-Sorensen J., Baldock R., Davidson D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.