Abstract
Fission yeast Pot1 (protection of telomeres) is a single-stranded telomeric DNA binding protein with a critical role in ensuring chromosome stability. A putative human homolog (hPot1) was previously identified, based on moderate sequence similarity with fission yeast Pot1 and telomere end-binding proteins from ciliated protozoa. Using indirect immunofluorescence, we show here that epitope-tagged hPot1 localizes to telomeres in interphase nuclei of human cells, consistent with a direct role in telomere end protection. The hPOT1 gene contains 22 exons, most of which are present in all cDNAs examined. However, four exons are subject to exon skipping in some transcripts, giving rise to five splice variants. Four of these are ubiquitously expressed, whereas the fifth appears to be specific to leukocytes. The resultant proteins vary significantly in their ability to form complexes with single-stranded telomeric DNA as judged by electrophoretic mobility shift assays. In addition to these splice variants, the Pot1 family is expanded by the identification of six more genes from diverse species. Pot1-like proteins have now been found in plants, animals, yeasts, and microsporidia.
Telomeres, the protein-DNA complexes at eukaryotic chromosome ends, protect the termini from end-to-end fusions and nucleolytic degradation. In most cells the DNA portion of telomeres consists of short repeats, which are rich in guanines on the strand running 5′ to 3′ towards the chromosome end. This strand is synthesized by the ribonucleoprotein complex telomerase. The catalytic subunit, telomerase reverse transcriptase (TERT), uses part of an RNA subunit as a template for DNA synthesis (14, 21). Telomeres range in length from 50 bp in hypotrichous ciliated protozoa to 150 kb in some strains of mice, with the length of each telomere usually fluctuating around a species-specific mean value. In all organisms where telomeres have been examined more closely, a 3′ overhang of the G-rich strand is present at the termini (1, 18, 22, 23, 37, 39).
Numerous proteins have been identified that bind to the double- or single-stranded portion of telomeres and play roles in end protection and telomere length regulation (reviewed in reference 3). In Oxytricha nova, a hypotrichous ciliate, a telomere end-binding protein (TEBP), consisting of α and β subunits, binds specifically to the single-stranded DNA overhangs at the ends of telomeres (8, 12, 13). The crystal structure of this ternary complex revealed the presence of four oligonucleotide/oligosaccharide binding folds (OB-folds), three of which are involved in DNA binding and one of which is involved in protein-protein interactions (16). Related proteins have been found in other ciliated protozoa including Stylonychia mytilis (10) and Euplotes crassus (36); the latter contains two proteins related to the α subunit but apparently contains no β subunit.
In Saccharomyces cerevisiae, the single-stranded overhang is bound by Cdc13, a protein with a pivotal role in protecting the chromosome end (11, 30) and in recruiting telomerase to the telomere (7) or activating it at the telomere (34). Single-stranded telomeric DNA binding proteins are not the only solution to end protection, however. Telomeres from humans, trypanosomes, and ciliate germ line nuclei have all been reported to form T-loop structures in which the single-stranded overhang is inserted into the upstream double-stranded region (15, 26, 27). In vitro, T-loop formation is facilitated by the telomere repeat binding factor Trf2 (15). Consistent with a role of Trf2 in end protection, a dominant-negative version of the protein causes a loss of G-strand overhangs and end-to-end fusions (35).
Recently a single-stranded telomeric DNA binding protein has been identified in Schizosaccharomyces pombe (1). The protein shares weak sequence similarity with the N-terminal DNA binding domain of TEBPs from ciliated protozoa and was named Pot1 (protection of telomeres) based on the rapid loss of telomeric DNA that occurs following deletion of the gene. Whereas most cells lacking Pot1 die due to sequence loss and end-to-end chromosome fusions, a few survivors emerge that have circularized all three chromosomes, thereby escaping the need for chromosome end maintenance. A human sequence homolog of the S. pombe protein has also been described (1). Purified fission yeast and human Pot1 (hPot1) proteins bind specifically to the G-rich strand of their own telomeric DNA but not to the complementary C-rich strand or double-stranded telomeric DNA, consistent with a role in binding to the 3′ overhang at the ends of telomeres in vivo (1).
Here we show by indirect immunofluorescence that the hPot1 protein resides at the telomeres in vivo as judged by colocalization with the telomere repeat binding factor Trf2 and hRap1. Together with previous results, this finding strongly suggests that hPot1 is a bona fide telomere end-binding protein and a functional homolog of S. pombe Pot1 (SpPot1). Based on sequence similarity, six new members of the Pot1/TEBP family are identified from diverse organisms including mouse and mustard weed. We have further analyzed the human and mouse POT1 gene structure and isolated five splice variants of hPOT1, for which tissue-specific expression and distinct DNA-binding properties are presented.
MATERIALS AND METHODS
Cloning and plasmids.
The plasmid pPB330 was constructed by inserting full-length hPOT1 cDNA into the plasmid pcDNA/FRT/V5-His (Invitrogen) such that the protein is expressed in-frame with C-terminal V5 and His6 tags. Plasmid pPB341 is a derivative of pIND V5-His C (Invitrogen) containing full-length hPOT1 cDNA under the control of a ponasterone A-inducible promoter.
Human POT1 splice variants were amplified by PCR from human cDNA using primers TCTACAGAATCAATGTCTTTGGTTCCAGC and GATTACATCT TCTGCAACTGTGGTGTCA. PCR products were gel purified, cloned into the plasmid pCR2.1 TOPO (Invitrogen), and analyzed by automated sequencing.
Cell culture, transfection, and immunofluorescence.
EcR293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, l-glutamine (2 mM), and Zeocin (0.4 mg/ml). HeLaI.2.11 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, l-glutamine (2 mM), and 1× nonessential amino acids (Invitrogen).
For analysis by immunofluorescence, cells were seeded onto coverslips in tissue culture dishes. After 16 h, cells were transfected with the plasmid pPB341 (EcR293) or the plasmid pPB330 (HeLaI.2.11) using lipofectamin 2000 (Invitrogen). The medium was replaced after 4 h, and after 24 h expression of V5-tagged hPot1 was induced in EcR293 cells by addition of ponasterone A to a final concentration of 3 μM.
Forty-eight hours posttransfection, cells were washed twice in phosphate-buffered saline (PBS) and fixed for 10 min in 4% formaldehyde. Cell membranes were permeabilized with 0.1% Triton X-100 in PBS for 10 min. After a brief rinse in PBS, cells were incubated for 1 h in 1% bovine serum albumin in PBS followed by 1 h in a solution containing PBS, 1% bovine serum albumin, and primary antibodies. V5-tagged hPot1 was detected using a monoclonal mouse anti-V5 antibody (Invitrogen) at a concentration of 1 μg/ml. Polyclonal antibodies against hRap1 (#666) were a generous gift from Titia de Lange and were used at a 1:1,000 dilution. Polyclonal antibodies against Trf2 were purchased from Santa Cruz Biotechnology and used at a concentration of 0.2 μg/ml. After four 10-min washes in PBS, samples were incubated for 1 h with secondary antibodies Alexa Fluor 488 goat antimouse and Alexa Fluor 594 goat antirabbit (Molecular Probes) at a concentration of 2 μg/ml. Prior to being mounted on slides, the coverslips were washed three times in PBS containing 1% bovine serum albumin and twice in PBS. DNA was visualized by adding 4′,6′-diamidino-2-phenylindole (0.2 ng/ml) to the final wash. Images were acquired using a Zeiss Axioplan 2 microscope equipped with a charge-coupled device camera. Images were merged and analyzed using Slidebook software (Intelligent Imaging).
Expression analysis.
Expression levels of hPOT1 in different tissues were assessed using first-strand cDNA panels (Clontech) normalized to the expression of four housekeeping genes. Transcripts were amplified using AdvanTaq Plus polymerase (Clontech) and the following pairs of oligonucleotides: GGGCAAAGCAGAAGTGGACGGAGCATC and ATTGACAGATAACATCTGAATGCTGATTGGCTGTC (exons 9 to 12); TTCAGATGTTATCTGTCAATCAGAACCTG and ATGTATTGTTCCTTGTATAAGAAATGGTGC (exons 12 to 16); CAGCACCATTTCTTATACAAGGAACAATAC and GATTACATCTTCTGCAACTGTGGTGTCA (exons 16 to 20); ACACCCCTGAATCAACTTAAGGGTGGT and ATTGACAGATAACATCTGAATGCTGATTGGCTGTC (exons 6 to 12). Template DNA was denatured at 94°C for 30 s, and hPOT1 fragments were amplified using the following cycling parameters: 94°C for 5 s, 72°C for 120 s (5 cycles); 94°C for 5 s, 70°C for 120 s (5 cycles); and 94°C for 5 s, 68°C for 120 s (26 cycles). Samples were taken after a total of 28, 30, 32, and 34 cycles. As a control, G3PDH was amplified under the same conditions for 22, 24, and 26 cycles with the primers TGAAGGTCGGAGTCAACGGATTTGGT and CATGTGGGCCATGAGGTCCACCAC. Reaction products were subjected to electrophoresis on 1.1% Tris-borate EDTA agarose gels run for 2.5 h followed by staining with Vistra Green (1 μg/ml; Molecular Probes) for 1 h. The relative abundance of PCR products was determined using a Typhoon PhosphorImager and ImageQuant software. For each PCR product the rate of accumulation was monitored to ensure that comparisons were made within the linear range for all samples. Each experiment also included a no-cDNA control to ensure the absence of contamination.
In vitro translation and DNA binding.
Human POT1 splice variants 1, 2, 3, and 5 were subcloned into a T7 expression vector for in vitro protein expression using the TNT coupled reticulocyte lysate kit (Promega). In a 50-μl reaction mixture, 3 μg of plasmid DNA was incubated with 4 μl of [35S]methionine (1,000 Ci/mmol; 10 mCi/ml) and 25 μl of rabbit reticulocyte lysate under the conditions recommended by the manufacturer. A fraction of each reaction was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and relative amounts of produced protein were quantified using a PhosphorImager.
DNA binding assays (10-μl reaction mixtures) contained 2 μl of translation reaction product, the 5′ 32P-labeled telomeric oligonucleotide GGTTAGGGTTAGGGTTAGGG, 25 mM HEPES-NaOH (pH 7.5), 100 mM NaCl, 1 mM EDTA, and 5% glycerol. Herring sperm DNA (50 μg/ml) and the oligonucleotide TTAATTAACCCGGGGATCCGGCTTGATCAACGAATGATCC (2.5 μM) were added as nonspecific competitors. Where appropriate, RNA oligonucleotide (UUAGGG)5 was added to a 2.5 μM final concentration. Reactions were incubated for 10 min at room temperature, and protein-DNA complexes were analyzed by electrophoresis at 4°C on a 4 to 20% polyacrylamide Tris-borate EDTA gel run at 150 V for 80 min.
RESULTS
Immunolocalization of hPot1.
Previous work has shown that purified hPot1 protein binds specifically to the G-rich strand of telomeric DNA (1), suggesting that in vivo the protein may bind to the single-stranded extension of the G-rich strand at the ends of chromosomes. To examine the subcellular localization of hPot1, human embryonic kidney EcR293 cells were transiently transfected with a plasmid encoding V5-epitope-tagged hPot1. Detection of hPot1 by indirect immunofluorescence revealed a punctate nuclear staining pattern in interphase nuclei (Fig. 1A and D). No staining was observed in cells transfected with an empty control vector or a plasmid encoding hPot1 lacking the V5-epitope tag (data not shown).
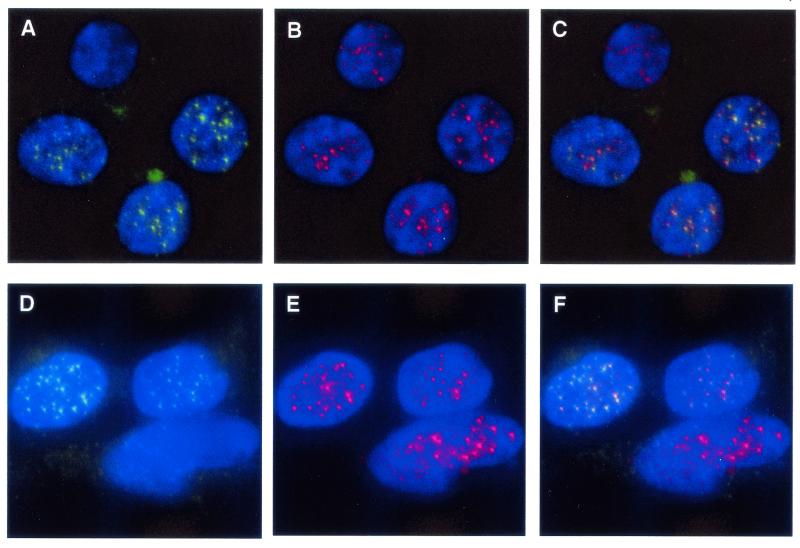
FIG. 1.
Human Pot1 protein colocalizes with hRap1 in EcR293 cells. Actively growing cells expressing V5-epitope-tagged hPot1 were fixed in formaldehyde and stained for Pot1 with a monoclonal V5 antibody and for hRap1 with a polyclonal rabbit antibody. Nuclei were visualized by 4′,6′-diamidino-2-phenylindole staining of chromosomal DNA. (A and D) Nuclear hPot1 foci (green). Due to the nature of transient transfections, not all cells express V5-tagged Pot1. (B and E) hRap1 staining (red). (C) Merged images from panels A and B; colocalization of V5-hPot1 and hRap1 is indicated by yellow color. (F) Merged images from panels D and E.
The hRap1 protein has been shown to localize to telomeres via its interaction with Trf2 (20). To determine whether the nuclear hPot1 foci coincide with hRap1 staining and hence the telomeres, hRap1 was visualized using a polyclonal antibody raised against the protein (Fig. 1B and E). An overlay of the two images revealed a strong correlation in the staining patterns, indicated by the yellow color (Fig. 1C and F). Nuclei which did not express V5-tagged hPot1, presumably because they were not transfected, showed only the hRap1 staining and no yellow foci (top nucleus in Fig. 1A to C and bottom nucleus in Fig. 1D to F).
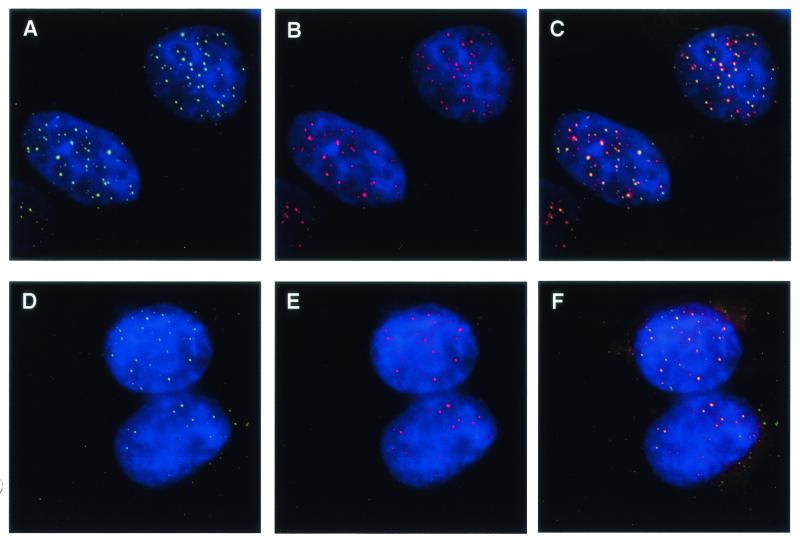
HeLaI.2.11 cells are a subclone of HeLaI (31) containing exceptionally long telomeres of 25 to 40 kb (35). Due to the increased number of binding sites per telomere, these cells have proven particularly useful in the immunolocalization of telomeric proteins (32, 33, 35). After transfection of these HeLa cells with a plasmid encoding V5-epitope-tagged hPot1, distinct nuclear foci were observed in interphase nuclei by indirect immunofluorescence (Fig. 2A and D). Staining of the same cells for hRap1 (Fig. 2B) or Trf2 (Fig. 2E) also resulted in a punctate nuclear pattern as previously reported (2, 4, 20). In each case the majority of hPot1 foci coincided with hRap1 (Fig. 2C) or Trf2 (Fig. 2F). We therefore conclude that hPot1 is present at the telomeres in interphase nuclei of EcR293 and HeLaI.2.11 cells.
FIG. 2.
Colocalization of hPot1 with hRap1 and Trf2 in HeLaI.2.11 cells. Actively growing cells expressing V5-epitope-tagged hPot1 were fixed in formaldehyde and stained for Pot1 with a monoclonal V5 antibody and for hRap1 or Trf2 with polyclonal rabbit antibodies. Nuclei were visualized by DAPI staining of chromosomal DNA. (A and D) Nuclear hPot1 foci (green). (B) hRap1 staining (red). (C) Merged images form panels A and B; colocalization of V5-hPot1 and hRap1 is indicated by yellow color. (E) hTrf2 staining (red). (F) Merged images from panels D and E.
Evolutionary conservation of Pot1 proteins.
The fission yeast and human Pot1 proteins were identified owing to a weak sequence similarity with the N-terminal regions of telomere end-binding proteins from ciliated protozoa (1). Based on the assumption that this family of proteins might be more widely conserved, we used PSI-BLAST to search for members in other organisms. A cDNA from the green monkey Macaca fascicularis was found to be 98% identical (99% similar) to human Pot1 over the entire length of the open reading frames. A mouse homolog was identified based on a sequence identity of 75% (84% similarity).
Unlike the case for the human, monkey, and mouse sequences, the similarity of human and fission yeast Pot1 with the ciliate telomere end-binding proteins is limited to approximately the N-terminal fifth of the proteins (1). Using this N-terminal region of hPot1 in a PSI-BLAST search, we identified several other putative family members (E value of <10−28). Notably, there appear to be two proteins in Arabidopsis thaliana that fall into this category (sequence accession number NP_196249, E = 8 × 10−54 after two rounds; and sequence accession number NP_178592, E = 10−28 after three rounds). Because the NP_196249 protein is more closely related to the Pot1 proteins from other organisms, this protein was named AtPot1, whereas the NP_178592 protein is referred to as AtPot2. Other candidate family members were identified in the yeast Neurospora crassa (accession number CAD11392; E = 4 × 10−33 after four rounds) and in the microsporidium Encephalitozoon cuniculi (accession number CAD26625) (E = 10−34 after four rounds). An alignment of the N-terminal regions of Pot1 proteins revealed the presence of a few key residues that are conserved between all sequences (Fig. 3).
FIG. 3.
Alignment of N-terminal regions of Pot1-like proteins from Homo sapiens (Hs) (sequence accession number AK001935), M. fascicularis (Mf) (BAB62219), Mus musculus (Mm) (AAH16121), A. thaliana (At1, NP_196249; At2, NP_178592), E. cuniculi (Ec) (CAD26625), S. pombe (Sp) (NP_594453), and N. crassa (Nc) (CAD11392). Protein sequences were aligned in ClustalW using the default values. Amino acids identical or similar in more than four sequences are shown as black letters shaded in gray, amino acids identical or similar in more than six sequences are shown as white letters shaded in gray, and those identical or similar in all sequences are shaded black. The first and last amino acid positions are indicated for each sequence.
Genomic sequence and gene structure.
Using sequence data from BAC clones RP11-304A23 (accession number AC096665), RP11-563L14 (AC110791), and RP5-907C10 (AC004925), we assembled a genomic sequence of approximately 120 kb spanning the entire hPOT1 gene. The gene is located on chromosome 7. The positions of 22 exons were mapped based on cDNA sequences and consensus splice sites (Fig. 4A). The assembled DNA sequence can be viewed at http://petunia.colorado.edu/pot1/hpot1_gdna.html.
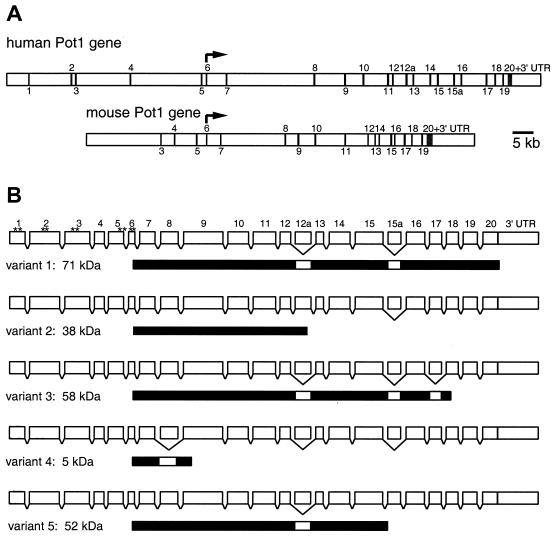
FIG. 4.
Gene structure and splice variants of hPOT1. (A) Structural organization of human and mouse POT1 genes. Exons are indicated by vertical bars and were numbered after the 20 exons found in the hPOT1 cDNA clone hCT24115 were mapped onto the genomic sequence. The 5′ ends of other hPOT1 cDNA clones are located in exons 2 (accession number AK022580) and 5 (AK001935 and BC002923). Exons specific to splice variants 2 and 5 are numbered 12a and 15a, respectively. Translation of human and mouse POT1 starts in exon 6 and ends in exon 20. (B) Structure of the hPOT1 splice variants. Exons are shown as white boxes (length proportional to exon size); introns are generally long and are not shown to scale. Skipping of an exon is indicated by a bracket below the box. The protein products are shown under each splice variant as black bars. For splice variant 1 the positions of the first 10 AUGs are indicated by asterisks.
The genomic sequence of mouse POT1 (MmPOT1) was extracted from the Celera database using the MmPot1 protein sequence and BLAST. Based on cDNAs BC016121 and NM_133931 as well as our own sequencing data, we mapped 18 exons onto the genomic DNA (Fig. 4A). The exon containing the start codon was given the number 6 to facilitate comparison with the human sequence. The exon size and the position of splice sites are highly conserved between human and mouse for exons 6 to 20. The mouse POT1 gene spans approximately 70 kb on chromosome 6.
In eukaryotes, protein synthesis usually begins with the first AUG codon from the 5′ end of an mRNA. Surprisingly, the AUG we had previously predicted as the start of the 634-amino-acid hPot1 protein maps to the 6th exon in the human sequence and is the 10th AUG from the 5′ end of the most complete cDNA available to us (hCT24115; Celera). If translation was initiated at any of the first nine AUGs (Fig. 4B), the resulting peptides would be between 4 and 71 amino acids in length, because each AUG is followed by an in-frame stop codon. Barring complex translational frame-shifting, the bulk of the hPot1 protein would be produced only if the 10th AUG were used. Neither the 10th nor any of the preceding AUGs is embedded within a Kozak consensus sequence for initiation of translation (19).
The existence of several exons and AUGs upstream of the POT1 start codon is conserved in mouse and Macaca (data not shown). In the MfPOT1 cDNA (sequence accession number AB066545), all exon sizes are conserved with those of human sequence, and the DNA sequences corresponding to exons 2 to 6 are 97% identical with the human sequence (data not shown). For the most complete cDNA isolated from mouse, the 5′ untranslated region consists of three exons.
Expression of hPOT1 splice variants.
Human POT1 was first cloned from ovary cDNA by amplifying the predicted open reading frame using primers corresponding to sequences in exons 6 and 20. Sequencing of the cloned PCR products revealed the presence of four alternatively spliced forms of hPOT1 (Fig. 4B). Splice variant 1 is the form previously described by members of our group (1) and is identical to several cDNA sequences deposited in the public database (e.g., NM_015450, AK001935, AK001230, AK022580, and BC002923). In variant 2, use of an alternative 5′ splice site adds exon 12a between exons 12 and 13. Although the isolated cDNAs included exons 13 to 20, a stop codon in exon 12a is expected to terminate translation, resulting in a 38-kDa protein. Variants 3 and 4 originate by exon skipping of exon 17 and exon 8, respectively. The protein produced from a message lacking exon 17 should be 58 kDa in size, since splicing of exon 16 to exon 18 results in a change of open reading frame and termination within exon 18. Similarly, skipping of exon 8 should result in a truncated protein of only 5 kDa.
A fifth splice form was identified when hPOT1 was amplified and cloned from cDNA isolated from peripheral blood leukocytes. This form includes an additional exon 15a between exons 15 and 16 (Fig. 4B). Exon 15a harbors a stop codon in-frame with the hPOT1 open reading frame, resulting in the production of a 52-kDa protein.
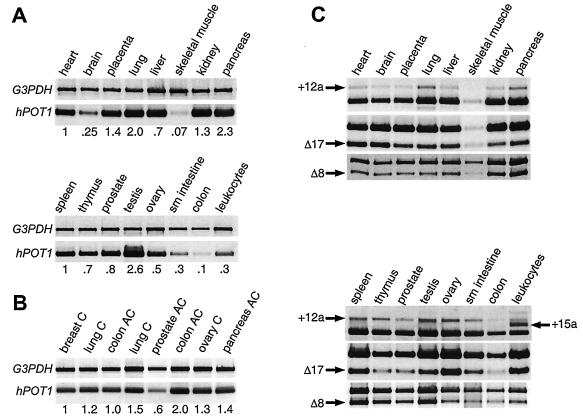
Multitissue cDNA panels were used to systematically survey for the presence of splice variants in different tissues by PCR. This method allows unambiguous identification of different splice forms and is more sensitive than Northern analysis. A number of primer pairs were synthesized, each covering a region of the sequence. Because no variation had been observed between exons 9 and 12, primers spanning this region were used to compare the total levels of all hPOT1 transcripts between different tissues. Differences in the amount of cDNA used from each tissue were accounted for by normalizing the hPOT1 PCR amplification products against a ∼1-kb fragment of the housekeeping gene for glyceraldehyde-3-phosphate dehydrogenase (G3PDH), which was amplified in parallel. To compare the relative amounts of hPOT1 mRNA within each panel, the values obtained for the heart and spleen were set to 1. Although hPOT1 expression was observed in all tissues, it was highest in testis and low in skeletal muscle and colon (Fig. 5A). Human Pot1 expression was also detected in eight samples of cDNA derived from a variety of carcinomas and adenocarcinomas (Fig. 5B). The ubiquitous presence of hPOT1 mRNA suggests that the protein plays a role in the maintenance of telomeres in all cells, unlike TERT, which is predominantly found in the germ line and in tumors.
FIG. 5.
Expression analysis of hPOT1 splice variants. (A) Exons 9 to 12 of hPOT1 were amplified from multitissue cDNA panels, and relative expression levels compared to those for heart (top panel) and spleen (bottom panel) are given. G3PDH was amplified in parallel and used for normalization. (B) Same as panel A except that cDNA samples isolated from tumors were used: breast carcinoma gi-101, lung carcinoma gi-117, colon adenocarcinoma cx-1, lung carcinoma lx-1, prostatic adenocarcinoma pc-3, colon adenocarcinoma gi-112, ovarian carcinoma gi-102, and pancreatic adenocarcinoma gi-103. Breast carcinoma gi-101 was used for normalization. (C) The following regions of hPOT1 mRNA were amplified to detect the presence of different splice forms: exons 12 to 16; products containing exon 12a (variant 2) or exon 15a (variant 5) are indicated by arrows. Exons 16 to 20; product lacking exon 17 (variant 3) is indicated. Exons 6 to 12; product lacking exon 8 (variant 4) is indicated. The unmarked band in each lane corresponds to transcripts that are not variant in this region.
The presence or absence of splice variant 2 in different tissues was assayed using a primer pair spanning exons 12 to 16. PCR from all tissues produced two bands, and sequencing confirmed that inclusion of exon 12a was responsible for the production of the upper band (Fig. 5C and data not shown). Due to the low transcript levels in colon and skeletal muscle, the presence of splice variant 2 was confirmed after additional rounds of amplification (data not shown). When cDNA derived from peripheral blood leukocytes was used in the PCR spanning exons 12 to 16, a new PCR product was detected. Sequencing revealed that this additional product included exon 15a (splice variant 5; Fig. 4B).
Primer pairs spanning exons 16 to 20 and exons 6 to 12 were used to detect splice variants 3 and 4, respectively. Skipping of exon 17 (variant 3) and exon 8 (variant 4) was apparent in all tissues (Fig. 5C). We conclude that splice variants 1 to 4 coexist in all tissues examined, whereas splice variant 5 was detected only in peripheral blood leukocytes.
DNA binding activity of proteins encoded by splice variants.
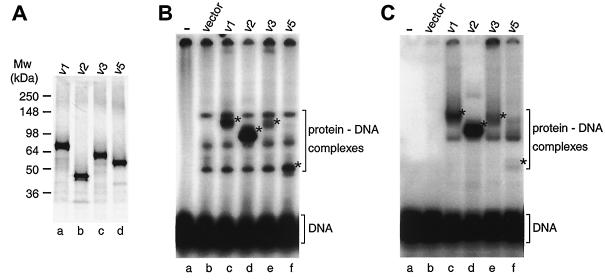
Sequence alignments and structural comparison of hPot1 with ciliate telomere end-binding proteins suggested that the N-terminal region of hPot1 is responsible for DNA binding (1, 29). As this region is present in splice forms 1, 2, 3, and 5, we wanted to test whether the encoded proteins would interact with telomeric DNA. The four variants were expressed in rabbit reticulocyte lysate in the presence of [35S]methionine and gave rise to proteins of the expected relative sizes (Fig. 6A).
FIG. 6.
Binding of hPot1 splice variant proteins to telomeric DNA. (A) sodium dodecyl sulfate-polyacrylamide gel showing in vitro-translated 35S-labeled splice forms 1, 2, 3, and 5. These variants contain 12, 6, 6, and 6 methionine residues, respectively, which accounts for their different signal strengths. Quantification revealed that actual expression levels are within 1.5-fold of each other. (B) Electrophoretic mobility shift assay of a 32P-labeled telomeric oligonucleotide in the presence of hPot1 variants. Protein-DNA complexes specific to hPot1 variants are indicated by asterisks. The three bands seen above the free DNA in lane b represent proteins from the reticulocyte extract binding to the telomeric DNA oligonucleotide and persist in lanes c to f. (C) As with panel B, except that RNA oligonucleotide (UUAGGG)5 was added as competitor.
When incubated with telomeric DNA, a distinct shift was observed for each splice variant (Fig. 6B). Intriguingly, the amount of protein-DNA complex formed with variant 2 was sixfold greater than for variant 1, suggesting a higher affinity of the former splice form for telomeric DNA (lanes c and d). Moreover, variants 3 and 5 produced only weak bands corresponding to hPot1-DNA complexes (lanes e and f). When different concentrations of the proteins were incubated together, competing for binding to the telomeric DNA, the apparent binding affinity of variant 2 was 8 to 9 times greater than that of variant 1, ∼20 times greater than that of variant 3, and ∼30 times greater than that of variant 5 (data not shown). Variant 5 formed a complex of much greater electrophoretic mobility than expected from its protein molecular weight relative to the other variants. This could represent a different stoichiometry of binding or a more compact conformation of the protein-DNA complex.
Addition of an excess of unlabeled RNA oligonucleotide of telomeric sequence eliminated most of the background binding of proteins from the reticulocyte lysate to the telomeric DNA oligonucleotide (compare lanes b in Fig. 6B and C). The relative differences in amounts of DNA-Pot1 variant complexes and their electrophoretic mobilities persisted in the presence of the RNA competitor (Fig. 6C) as well as at two different protein concentrations (data not shown). In summary, the four splice variants of hPot1 appear to have quite different telomeric DNA binding properties and may therefore play distinct roles in telomere maintenance.
DISCUSSION
Using indirect immunofluorescence, we show here that epitope-tagged hPot1 localizes to distinct nuclear foci in EcR293 and HeLa cells. The hPot1 staining largely coincides with the position of telomeres as evidenced by dual labeling for other telomeric proteins, namely hRap1 and Trf2. Together with the sequence similarity between human and S. pombe Pot1 proteins and biochemical analysis of single-stranded telomeric DNA binding (1), these results strongly suggest that hPot1 is indeed a telomeric protein.
Database searches revealed new homologs of the human and fission yeast Pot1 proteins in diverse species including animals, plants, fungi, and microsporidia. This wide distribution of Pot1/TEBP family members leads us to speculate that related telomere end-binding proteins are likely to be present in most eukaryotes. We have sequenced four new splice variants of human POT1 and mapped the exons of the human and mouse proteins onto the respective genomic sequences. Amplification of mRNA demonstrated that four splice variants are expressed in a wide variety of tissues, whereas the fifth splice variant appears to be specific to peripheral blood leukocytes. Notably, four of the five Pot1 protein variants have distinct DNA-binding properties, indicating that they may fulfill different functions within the cell.
A growing family of single-stranded telomeric DNA binding proteins.
Until recently, telomere end-binding proteins, as first described for O. nova (12, 13), were thought to be specific to ciliated protozoa. This perception was largely based on the inability to identify related genes from other species based on sequence homology and the fact that the structure of a T-loop seemed incompatible with the presence of a DNA end-binding protein. The exponential increase in available sequence data finally led to the identification of the fission yeast and human Pot1 proteins (1), followed by putative homologs in macaque monkeys, mice, mustard weed, N. crassa, and E. cuniculi.
The difficulty in identifying members of this family is due to the small amount of primary sequence conservation that is necessary to maintain the structure of an OB-fold (28). This point was nicely illustrated by a recent report on the nuclear magnetic resonance structure of the budding yeast Cdc13 protein (24). Although no sequence similarity was apparent between Cdc13 and TEBPs or Pot1, the Cdc13 DNA-binding domain conforms to the OB-fold and is structurally similar to the N-terminal OB-fold of O. nova TEBP. This exciting finding suggests that the OB-fold may be a universally conserved structure for binding of single-stranded telomeric DNA. There are also some functional similarities between SpPot1 and Cdc13, since deletion of either protein leads to a rapid loss of telomeric DNA (1, 11). It is currently unclear, however, whether there is an ancestral relationship between Cdc13 and Pot1/TEBP or whether convergent evolution may explain the structural similarity. An important question to answer in this context is whether Pot1 proteins play the same pivotal role as Cdc13 in recruiting telomerase as well as an Stn1-containing protective complex (30).
Splice variants with distinct biochemical properties.
The observation that the hPOT1 gene may encode at least five distinct proteins adds to the growing list of examples demonstrating that much of the complexity of higher organisms arises from alternative splicing (25). Alternative splicing has also been reported for the human TERT gene (17, 38), and telomerase activity appears to be regulated, at least in part, via alternative splicing, resulting in the production of inactive or dominant-negative forms of hTERT (5, 6, 40).
It is tempting to speculate that the hPot1 variants will have distinct functions in vivo, since they interact differently with telomeric DNA. Most interestingly, splice variant 2 protein, which lacks almost half the sequence of variant 1, appeared to have the highest affinity for DNA. Increased DNA binding affinity following truncation of the C terminus has previously been reported for SpPot1 (1) and O. nova TEBP (9). The human protein, however, provides the first example where the shorter version could be produced in vivo via alternative splicing. It will be of interest to determine whether different splice forms associate with the telomere during specific stages of the cell cycle, or whether they may interact with the telomere in different conformations—such as within the structure of a T-loop or with the open conformation. In addition to modulating the affinity for telomeric DNA, the different C termini may also be involved in mediating contacts with additional proteins.
Acknowledgments
We thank J. Richard McIntosh for access to the fluorescence microscope, Titia de Lange for antibodies and HeLaI.2.11 cells, Karen Goodrich for oligonucleotide synthesis, Deborah Wuttke and Diana Baumann for critical reading of the manuscript, and the members of the Cech laboratory for their comments and suggestions. The mouse sequence analysis was generated with the Celera Discovery System and Celera Genomics associated databases.
This work was supported by the Howard Hughes Medical Institute.
REFERENCES
- 1.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. [DOI] [PubMed] [Google Scholar]
- 2.Bilaud, T., C. Brun, K. Ancelin, C. E. Koering, T. Laroche, and E. Gilson. 1997. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17:236-239. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 4.Broccoli, D., A. Smogorzewska, L. Chong, and T. de Lange. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17:231-235. [DOI] [PubMed] [Google Scholar]
- 5.Cerezo, A., H. Kalthoff, M. Schuermann, B. Schafer, and P. Boukamp. 2002. Dual regulation of telomerase activity through c-Myc-dependent inhibition and alternative splicing of hTERT. J. Cell Sci. 115:1305-1312. [DOI] [PubMed] [Google Scholar]
- 6.Colgin, L. M., C. Wilkinson, A. Englezou, A. Kilian, M. O. Robinson, and R. R. Reddel. 2000. The hTERTalpha splice variant is a dominant negative inhibitor of telomerase activity. Neoplasia 2:426-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 8.Fang, G., and T. R. Cech. 1993. Oxytricha telomere-binding protein: DNA-dependent dimerization of the alpha and beta subunits. Proc. Natl. Acad. Sci. USA 90:6056-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang, G., J. T. Gray, and T. R. Cech. 1993. Oxytricha telomere-binding protein: separable DNA-binding and dimerization domains of the alpha-subunit. Genes Dev. 7:870-882. [DOI] [PubMed] [Google Scholar]
- 10.Fang, G., and T. R. Cech. 1991. Molecular cloning of telomere-binding protein genes from Stylonychia mytilis. Nucleic Acids Res. 19:5515-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottschling, D. E., and V. A. Zakian. 1986. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell 47:195-205. [DOI] [PubMed] [Google Scholar]
- 13.Gray, J. T., D. W. Celander, C. M. Price, and T. R. Cech. 1991. Cloning and expression of genes for the Oxytricha telomere-binding protein: specific subunit interactions in the telomeric complex. Cell 67:807-814. [DOI] [PubMed] [Google Scholar]
- 14.Greider, C. W., and E. H. Blackburn. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331-337. [DOI] [PubMed] [Google Scholar]
- 15.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 16.Horvath, M. P., V. L. Schweiker, J. M. Bevilacqua, J. A. Ruggles, and S. C. Schultz. 1998. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell 95:963-974. [DOI] [PubMed] [Google Scholar]
- 17.Kilian, A., D. D. Bowtell, H. E. Abud, G. R. Hime, D. J. Venter, P. K. Keese, E. L. Duncan, R. R. Reddel, and R. A. Jefferson. 1997. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet. 6:2011-2019. [DOI] [PubMed] [Google Scholar]
- 18.Klobutcher, L. A., M. T. Swanton, P. Donini, and D. M. Prescott. 1981. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc. Natl. Acad. Sci. USA 78:3015-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak, M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, B., S. Oestreich, and T. de Lange. 2000. Identification of human Rap1: implications for telomere evolution. Cell 101:471-483. [DOI] [PubMed] [Google Scholar]
- 21.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 22.Makarov, V. L., Y. Hirose, and J. P. Langmore. 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88:657-666. [DOI] [PubMed] [Google Scholar]
- 23.McElligott, R., and R. J. Wellinger. 1997. The terminal DNA structure of mammalian chromosomes. EMBO J. 16:3705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitton-Fry, R. M., E. M. Anderson, T. R. Hughes, V. Lundblad, and D. S. Wuttke. 2002. Conserved structure for single-stranded telomeric DNA recognition. Science 296:145-147. [DOI] [PubMed] [Google Scholar]
- 25.Modrek, B., and C. Lee. 2002. A genomic view of alternative splicing. Nat. Genet. 30:13-19. [DOI] [PubMed] [Google Scholar]
- 26.Munoz-Jordan, J. L., G. A. Cross, T. de Lange, and J. D. Griffith. 2001. t-loops at trypanosome telomeres. EMBO J. 20:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murti, K. G., and D. M. Prescott. 1999. Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc. Natl. Acad. Sci. USA 96:14436-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murzin, A. G. 1993. OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 12:861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peersen, O. B., J. A. Ruggles, and S. C. Schultz. 2002. Dimeric structure of the Oxytricha nova telomere end-binding protein alpha-subunit bound to ssDNA. Nat. Struct. Biol. 9:182-187. [DOI] [PubMed] [Google Scholar]
- 30.Pennock, A., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 31.Saltman, D., R. Morgan, M. L. Cleary, and T. de Lange. 1993. Telomeric structure in cells with chromosome end associations. Chromosoma 102:121-128. [DOI] [PubMed] [Google Scholar]
- 32.Smith, S., and T. de Lange. 2000. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10:1299-1302. [DOI] [PubMed] [Google Scholar]
- 33.Smogorzewska, A., B. van Steensel, A. Bianchi, S. Oelmann, M. R. Schaefer, G. Schnapp, and T. de Lange. 2000. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taggart, A., S. Teng, and V. Zakian. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297:1023-1026. [DOI] [PubMed] [Google Scholar]
- 35.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 36.Wang, W., R. Skopp, M. Scofield, and C. Price. 1992. Euplotes crassus has genes encoding telomere-binding proteins and telomere-binding protein homologs. Nucleic Acids Res. 20:6621-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wellinger, R. J., A. J. Wolf, and V. A. Zakian. 1993. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72:51-60. [DOI] [PubMed] [Google Scholar]
- 38.Wick, M., D. Zubov, and G. Hagen. 1999. Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT). Gene 232:97-106. [DOI] [PubMed] [Google Scholar]
- 39.Wright, W. E., V. M. Tesmer, K. E. Huffman, S. D. Levene, and J. W. Shay. 1997. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 11:2801-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi, X., D. M. White, D. L. Aisner, J. A. Baur, W. E. Wright, and J. W. Shay. 2000. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia 2:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]