Abstract
Chromosome translocation and gene fusion are frequent events in the human genome and are often the cause of many types of tumor. ChimerDB is the database of fusion sequences encompassing bioinformatics analysis of mRNA and expressed sequence tag (EST) sequences in the GenBank, manual collection of literature data and integration with other known database such as OMIM. Our bioinformatics analysis identifies the fusion transcripts that have non-overlapping alignments at multiple genomic loci. Fusion events at exon–exon borders are selected to filter out the cloning artifacts in cDNA library preparation. The result is classified into two groups—genuine chromosome translocation and fusion between neighboring genes owing to intergenic splicing. We also integrated manually collected literature and OMIM data for chromosome translocation as an aid to assess the validity of each fusion event. The database is available at http://genome.ewha.ac.kr/ChimerDB/ for human, mouse and rat genomes.
INTRODUCTION
Chromosomal aberrations are frequently observed in many hematologic and solid tumors (1,2). Various large-scale and high-throughput techniques, such as chromosome banding (1,3), comparative genomic hybridization (CGH) (4) and fluorescence in situ hybridization (FISH) (5), are being used in modern cancer cytogenetics to detect structural and copy number changes in chromosomes. The most common type of mutation among the known cancer genes is chromosomal translocation (6). It can deregulate the gene expression by disrupting the promoter region of the gene or by joining the gene with enhancer elements like immunoglobulin or T-cell receptor genes (7). Alternatively, fusion of two coding regions creates a chimeric gene that encodes a fusion protein that interferes with the normal regulating pathways (2,8).
The most famous example is the fusion protein BCR–ABL, the target protein of the drug gleevec treating chronic myeloid leukemia (CML) (8,9). CML is associated in most cases with a chromosomal translocation between chromosomes 9 and 22 that creates the Philadelphia chromosome. The BCR gene in chr22 is fused with the gene ABL in chr9, so called the t(9;22)(q34;q11) translocation. The tyrosine kinase activity of ABL is constantly activated by the BCR gene (GTPase activator) in the fusion protein, resulting in the rapid cellular mitosis and inability of the cell to perform apoptosis. Gleevec inhibits the tyrosine kinase ability of the BCR–ABL fusion protein. Successful development of gleevec opened an era of targeted molecular therapy.
Chimeric sequences can be generated from other mechanisms too. Two adjacent, independent genes may be co-transcribed and the intergenic region is spliced out so that the resulting fused transcript possesses exons from both genes (10). This phenomenon, termed as cotranscription and intergenic splicing (CoTIS), can lead to fusion protein or altered promoter region for the downstream gene in the same way as chromosome translocation. Furthermore, trans-splicing can join two independently transcribed mRNA sequences at canonical exon–exon borders. Even though several cases of natural trans-splicing are reported in human (11,12), it is generally considered to be rare and will be ignored in this work.
Given the importance of chromosome aberration in cancer detection, prognosis and target identification, it is quite natural to search for chimeric sequences in the GenBank. Alterbi and co-workers (13) developed a screening procedure to identify heterologous, spliced mRNAs with potential origin from chromosomal translocation, mRNA trans-splicing and multi-locus transcription. Hahn et al. (14) extended this approach to include expressed sequence tag (EST) sequences that expanded the search scope significantly. They experimentally verified the predicted IRA1–RGS17 fusion in the breast cancer cell line MCF7. However, they deliberately discarded fusion cases between neighboring genes.
Curated databases are also available from cancer cytogenetics community. NCBI's database of cancer chromosomes (15) integrated the NCI/NCBI SKY/M-FISH and CGH database and the NCI Mitelman Database of Chromosome Aberrations in Cancers. The Cancer Genome Project at the Sanger Institute maintains a list of cancer genes based on published scientific literatures (6). Mutation data and associated information for these cancer genes are stored in the COSMIC database (16). The ‘Atlas of Genetics and Cytogenetics in Oncology and Haematology’ is a peer-reviewed resource that contains concise and updated cards on genes involved in cancer, cytogenetics and clinical entities in oncology, and cancer-prone diseases (17).
In this paper, we describe a new database of fusion genes, ChimerDB. It aims to be a knowledgebase that integrates bioinformatics analysis of transcript sequences (mRNA and EST), literature data from scientific journals (6) and OMIM data on translocation (18). It should be a valuable resource for developing cancer biomarkers and drug targets.
DATABASE CONSTRUCTION
In silico identification of fusion transcripts
All mRNA and EST sequences in the GenBank (Release 148; June 15, 2005) were aligned onto the human genome (NCBI Build 35) using the BLAT program (19). Minimum length and percent identity of valid alignments were 100 bp and 93%, respectively. Transcripts with two non-overlapping, contiguous alignments were selected as fusion candidates. Small overlap (<10 bp) was allowed due to uncertainty in BLAT alignments. Alignments in the same chromosome were restricted to be in opposite orientation to avoid fusion by CoTIS. We found 261 mRNA and 2484 EST sequences as fusion candidates, including artificial chimeras created by accidental ligation of different cDNAs during the cloning procedure. Genuine and artificial chimeras can be distinguished by examining the fusion boundaries. Fusion points in true chimeras usually coincide with a splice site since chromosome breakage tends to take place in long intronic regions rather than in short exons (14). Allowing 10 bp deviation from known splice sites, we obtained 159 mRNA and 258 EST sequences as reliable fusion transcripts. They constitute 355 fusion cases involving 638 genes.
Fusion cases owing to CoTIS can be identified using the ECgene clustering system (20,21). ECgene clusters sequences that share any splice sites in the genomic alignment, taking gene orientation into consideration. Fusion transcripts cause two neighboring genes to join to form a single cluster in the ECgene system. Therefore, we searched for ECgene clusters (Version 1.2) that contained two non-overlapping known genes and identified fusion transcripts. We found 223 mRNA and 396 EST sequences encoding 337 cases of CoTIS. Fusion by CoTIS creates a subtle problem in the genome-based EST clustering procedure that groups sequences sharing any splice sites. They should be identified and removed in advance.
Literature database
Journal publication is the single most important source of scientific knowledge. PubMed search for publications reporting fusion events owing to chromosome translocation gave 2945 manuscripts. Manual inspection of abstract produced 254 fusion cases involving 286 genes. We also imported the list of cancer genes associated with chromosome translocation from the Cancer Genome Project at the Sanger Institute (6). Current cancer gene census contains 257 translocation cases involving 346 genes, most of which coincide with our PubMed search result.
OMIM database is another knowledgebase of human genes and genetic disorders (18). We searched OMIM database for records with chromosome translocations. Manual inspection of ∼850 records gave 320 translocation cases with 597 genes. Literature databases should greatly extend the utility of fusion database by providing literature proof and relevant medical information for each computationally identified event.
Database integration
One of the major problems in dealing with heterogeneous databases, especially the literature data, is the use of aliases for the same gene. This is the source of redundant and fragmented entries. All records use the official HUGO gene to avoid this problem. Alias field in Entrez Gene database (22) was used to deal with different names for the same gene. In silico results from transcript mapping, literature and OMIM data were all integrated according to this official gene symbols. Furthermore, Mitelman's recurrent aberration database and the Atlas Chromosomes in Cancer data were also integrated into ChimerDB.
Table 1 is the summary statistics of ChimerDB. Currently, ChimerDB contains 1258 fusion cases that involve 1777 genes, 381 mRNA and 654 EST sequences. Assuming total number of human genes ∼25 000, this implies that ∼4.4% of human genes are involved in chromosome translocation and another ∼2.7% of human genes show fusion between neighboring genes (CoTIS). It should be noted that overlap between the transcript mapping data and other known databases is not large. This suggests that majority of known chromosome translocations are not supported by transcript data, such as mRNA and EST. Unless transcripts were discarded owing to low alignment quality, they would be from non-sequence-based methods and it would be interesting to obtain clone-based sequence data for those cases.
Table 1.
Summary statistics of ChimerDB
| Data source | Fusion cases | Genesa | mRNA | EST |
|---|---|---|---|---|
| Transcript mappingb | ||||
| Translocation | 355 | 638 | 159 | 258 |
| CoTIS | 337 | 674 | 223 | 396 |
| PubMed literature | 254 | 286 (76) | ||
| Sanger CGP | 257 | 346 (80) | ||
| OMIM records | 320 | 597 (66) | ||
| Mitelman breakpoint | 144 | 158 (54) | ||
| Atlas chromosomes | 307 (61) | |||
| Total (non-redundant) | 1258 | 1777 | 381 | 654 |
| Known genes | 1009 | 1528 | ||
| EST clustersc | 249 | 249 | ||
aNumbers in the parentheses indicate common genes with translocation data.
bTranscript mapping data include EST clusters as well as known genes.
cEST clusters come from 151 translocation and 98 CoTIS cases.
WEB INTERFACE
The contents of ChimerDB can be accessed at http://genome.ewha.ac.kr/ChimerDB. It supports various types of queries such as gene name and cytogenetic band position. Query can be a breakpoint (e.g. AML1) or a fusion event (e.g. BCR–ABL1). We also support searches by site and/or diagnosis as in the NCBI Cancer Chromosomes.
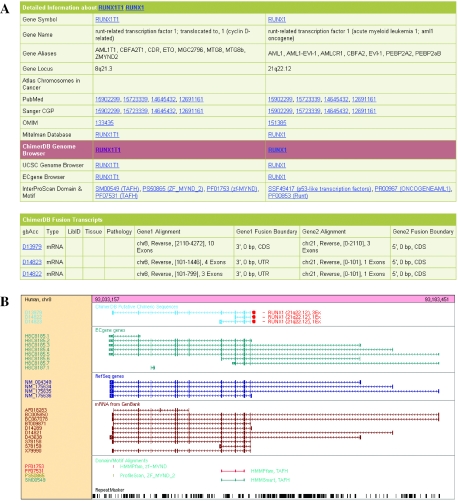
Search result page shows all relevant fusion cases with available types of data. Details page opens an output page for a specific fusion case that consists of a summary table, detailed information table and fusion transcript table as shown in Figure 1A. It includes extensive links to relevant resources, such as the Entrez Gene, OMIM and PubMed databases. Links to NCBI Cancer Chromosomes provide detailed information on SKY/M-FISH and CGH and Mitelman databases—primary databases for cancer cytogenetics. Links to Atlas of Genetics and Cytogenetics in Oncology and Haematology database allow access to community efforts to annotate cancer genes, rich in cytogenetic and clinical information. The transcript table in Figure 1A shows the tissue and pathology information for EST sequences. It also describes properties of the fusion—transcript direction, aligned region, number of exons, deviation of fusion boundary from known splice site and so on. Intact and affected domains before and after translocation are also summarized using the InterPro database (23).
Figure 1.
(A) Part of the output page from ChimerDB. (B) Custom genome browser for RUNX1T1 genomic locus to visualize fusion transcripts. Red dot indicates the fusion point.
Figure 1B is the custom genome browser showing alignment of fusion transcripts in each gene. Breakpoints and fusion partner genes can be immediately recognized in the viewer. It also shows the position of functional domains present in the gene.
Most fusion genes owing to CoTIS do not have detailed information on their functional significance yet. Therefore, we simply provide the minimal information—fused genes, genomic locus, functional domains, alignment browser and exon/intron properties.
FUTURE DIRECTIONS
ChimerDB is an integrated database for fusion sequences that includes bioinformatics analysis, literature data and OMIM data. However, functional significance of fusion events should be examined thoroughly so that these fusion events could serve as drug targets for cancer treatment. Expression analysis of fused transcripts in different histological and pathological conditions should be performed with the bioinformatics analysis such as domain and promoter changes, frame shift and so on. Integrative approach that combines high-throughput techniques, such as SKY, CGH, SNP chip, microarray, proteomics, interactions and pathway analysis, would prove to be powerful in elucidating the functional significance of fusion genes. ChimerDB will continue to integrate relevant data available in public.
Acknowledgments
This work was supported by the Ministry of Science and Technology of Korea through the Bioinformatics Research Program (Grant No. 2005-00201) and the Korea Science and Engineering Foundation through the center for cell signaling research at Ewha Womans University. Funding to pay the Open Access publication charges for this article was provided by MOST Grant No. 2005-00201.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mitelman F., Mertens F., Johansson B. Prevalence estimates of recurrent balanced cytogenetic aberrations and gene fusions in unselected patients with neoplastic disorders. Genes Chromosomes Cancer. 2005;43:350–366. doi: 10.1002/gcc.20212. [DOI] [PubMed] [Google Scholar]
- 2.Albertson D.G., Collins C., McCormick F., Gray J.W. Chromosome aberrations in solid tumors. Nature Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 3.Mitelman F., Mertens F., Johansson B. A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nature Genet. 1997;15:417–474. doi: 10.1038/ng0497supp-417. [DOI] [PubMed] [Google Scholar]
- 4.Kallioniemi A., Kallioniemi O.P., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 5.Schrock E., du Manoir S., Veldman T., Schoell B., Wienberg J., Ferguson-Smith M.A., Ning Y., Ledbetter D.H., Bar-Am I., Soenksen D., et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 6.Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. A census of human cancer genes. Nature Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ar-Rushdi A., Nishikura K., Erikson J., Watt R., Rovera G., Croce C.M. Differential expression of the translocated and the untranslocated c-myc oncogene in Burkitt lymphoma. Science. 1983;222:390–393. doi: 10.1126/science.6414084. [DOI] [PubMed] [Google Scholar]
- 8.Rowley J.D. Chromosome translocations: dangerous liaisons revisited. Nature Rev. Cancer. 2001;1:245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- 9.Mauro M.J., O'Dwyer M., Heinrich M.C., Druker B.J. STI571: a paradigm of new agents for cancer therapeutics. J. Clin. Oncol. 2002;20:325–334. doi: 10.1200/JCO.2002.20.1.325. [DOI] [PubMed] [Google Scholar]
- 10.Communi D., Suarez-Huerta N., Dussossoy D., Savi P., Boeynaems J.M. Cotranscription and intergenic splicing of human P2Y11 and SSF1 genes. J. Biol. Chem. 2001;276:16561–16566. doi: 10.1074/jbc.M009609200. [DOI] [PubMed] [Google Scholar]
- 11.Flouriot G., Brand H., Seraphin B., Gannon F. Natural trans-spliced mRNAs are generated from the human estrogen receptor-alpha (hER alpha) gene. J. Biol. Chem. 2002;277:26244–26251. doi: 10.1074/jbc.M203513200. [DOI] [PubMed] [Google Scholar]
- 12.Finta C., Zaphiropoulos P.G. Intergenic mRNA molecules resulting from trans-splicing. J. Biol. Chem. 2002;277:5882–5890. doi: 10.1074/jbc.M109175200. [DOI] [PubMed] [Google Scholar]
- 13.Romani A., Guerra E., Trerotola M., Alberti S. Detection and analysis of spliced chimeric mRNAs in sequence databanks. Nucleic Acids Res. 2003;31:e17. doi: 10.1093/nar/gng017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn Y., Bera T.K., Gehlhaus K., Kirsch I.R., Pastan I.H., Lee B. Finding fusion genes resulting from chromosome rearrangement by analyzing the expressed sequence databases. Proc. Natl Acad. Sci. USA. 2004;101:13257–13261. doi: 10.1073/pnas.0405490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knutsen T., Gobu V., Knaus R., Padilla-Nash H., Augustus M., Strausberg R.L., Kirsch I.R., Sirotkin K., Ried T. The interactive online SKY/M-FISH and CGH database and the Entrez cancer chromosomes search database: linkage of chromosomal aberrations with the genome sequence. Genes Chromosomes Cancer. 2005;44:52–64. doi: 10.1002/gcc.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bamford S., Dawson E., Forbes S., Clements J., Pettett R., Dogan A., Flanagan A., Teague J., Futreal P.A., Stratton M.R., et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huret J.L., Dessen P., Bernheim A. Atlas of genetics and cytogenetics in oncology and haematology, year 2003. Nucleic Acids Res. 2003;31:272–274. doi: 10.1093/nar/gkg126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim N., Shin S., Lee S. ECgene: genome-based EST clustering and gene modeling for alternative splicing. Genome Res. 2005;15:566–576. doi: 10.1101/gr.3030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim P., Kim N., Lee Y., Kim B., Shin Y., Lee S. ECgene: genome annotation for alternative splicing. Nucleic Acids Res. 2005;33:D75–D79. doi: 10.1093/nar/gki118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]