Abstract
StellaBase, the Nematostella vectensis Genomics Database, is a web-based resource that will facilitate desktop and bench-top studies of the starlet sea anemone. Nematostella is an emerging model organism that has already proven useful for addressing fundamental questions in developmental evolution and evolutionary genomics. StellaBase allows users to query the assembled Nematostella genome, a confirmed gene library, and a predicted genome using both keyword and homology based search functions. Data provided by these searches will elucidate gene family evolution in early animals. Unique research tools, including a Nematostella genetic stock library, a primer library, a literature repository and a gene expression library will provide support to the burgeoning Nematostella research community. The development of StellaBase accompanies significant upgrades to CnidBase, the Cnidarian Evolutionary Genomics Database. With the completion of the first sequenced cnidarian genome, genome comparison tools have been added to CnidBase. In addition, StellaBase provides a framework for the integration of additional species-specific databases into CnidBase. StellaBase is available at http://www.stellabase.org.
INTRODUCTION
In retrospect, the origin of the Bilateria may have been the most monumental event in the history of animal evolution. The Bilateria comprises more than 99% of all currently identified animal species. Bilaterian animals, including such major phyla as the chordates, arthropods, nematodes, annelids and mollusks have achieved far greater structural and behavioral complexity than non-bilaterian animals. However, in order to understand the genesis of bilaterian diversity and complexity, it is necessary to consult non-bilaterian outgroups taxa such as the phylum Cnidaria (sea anemones, corals, hydras, jellyfishes and their relatives).
Recent EST analyses on cnidarians have revealed surprising complexity in the genomes of these simple animals (1,2). For example, many genes that were previously thought to have originated within vertebrates due to their absence in the genomes of Drosophila and Caenorhabditis elegans have been found in cnidarians. These genes must have been present in the cnidarian-bilaterian ancestor, some 634 million years ago (3).
The starlet sea anemone, Nematostella vectensis, is a small burrowing sea anemone that is found in estuaries along the Atlantic and Pacific coasts of North America and the south of England (4–6). Nematostella is an important new model system for both lab-based and field-based studies of ecology, genomics, development and evolution [reviewed in (7)]. Importantly, Nematostella is the first member of the basal animal phylum Cnidaria, and the first basal animal generally, to have its genome sequenced (Joint Genome Institute; D. Rokshar, PI). Furthermore, among current cnidarian model systems, Nematostella is unsurpassed for the ease with which its entire life cycle may be cultured in the laboratory (8–10). An important advantage of Nematostella and other Cnidaria relative to the major animal models in developmental biology (fruitfly, soil nematode, zebrafish, etc.) is its extensive ability to regenerate. In fact, the adult Nematostella can originate via four distinct developmental pathways, including embryogenesis, regeneration and two forms of asexual fission (7). A systematic comparison of regeneration and embryological development in animals that can regenerate should provide fundamental insights into the genomic basis of regenerative ability. The combination of its informative phylogenetic position, its exceptional experimental tractability, and its impressive developmental flexibility will ensure that Nematostella becomes a widely used genomic model system. As proof of its utility, Nematostella has already provided key insights into the evolution of metazoan body plan traits and developmental gene families (11–16).
NEMATOSTELLA GENE DATABASE
StellaBase houses a gene database comprising a gene library and an assembly of the full genome. The genome assembly was produced using the program Phusion (17); genes were predicted from the assembled genome using GENSCAN (18,19). The predicted genes were classified into putative gene families by comparing them against the complete Pfam library [release 17; (20)] using HMMER, version 2.3.2 [http://hmmer.wustl.edu; (21)]. Exon predictions and gene family predictions are accompanied by estimates of statistical significance (19,22).
Each gene in StellaBase is associated with a unique ID number. Through its ID number, detailed information about the gene may be retrieved including (i) its predicted exon structure, (ii) the statistical significance of the exon predictions and (iii) a listing of all Pfam protein families that match the gene in question with an E-value ≤ 10. Nucleotide and amino acid sequences may be returned to the user in a FASTA format. The definition line of FASTA sequences downloaded through StellaBase includes the ID number, genomic location, best HMM match to a protein family at Pfam (the match with the lowest E-value) and indication of experimental confirmation, if applicable. Genes are considered to be experimentally confirmed if a BLAST search indicates a match against an expressed Nematostella sequence housed at NCBI with an E-value ≤ 10−10.
SEARCHING THE GENE DATABASE USING BLAST
StellaBase uses NCBI's BLAST program (23) to allow users to perform sequence similarity searches against the Nematostella gene database and genome sequence. Results from queries of the gene database return the following information: (i) StellaBase ID number; (ii) the genomic contig on which that gene resides; (iii) the location of the gene on the contig; (iv) whether or not the gene's expression has been confirmed; (v) best Pfam protein family match and associated E-value (as determined by HMMER) and a (vi) blast score with associated E-value. Other information about the gene, including exon structure and other Pfam motif matches may be retrieved through the StellaBase ID number search function.
PROTEIN FAMILY SEARCH
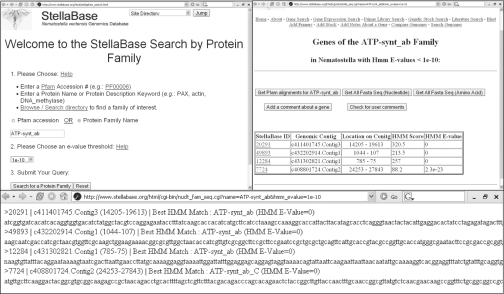
There is great interest in the membership of various protein families in the Cnidaria due to their status as a closely related outgroup to the Bilateria. To identify Nematostella sequences from a particular gene family, users may enter the name or accession number for any of the 7868 protein families included in the Pfam database [http://pfam.wustl.edu/; (20)]. The stringency of the search is determined by selecting a threshold ‘expectation value’ (ranging from 101 to 10−100). The search returns a summary of the predicted genes that match the protein family of interest. For each matching gene, the following information is provided: (i) StellaBase ID number; (ii) the genomic contig on which the gene is found; (iii) HMMER score and associated E-value. Gene sequences in FASTA format may be retrieved for individual genes or for all matches to a particular query. Cross-references to Pfam provide information on the gene family and protein function (Figure 1).
Figure 1.
Screens showing StellaBase interface, clockwise from top left: (i) User interface to search for gene families in the Nematostella genome by keyword and expectation values; (ii) Output for a particular query; (iii) All sequences matching query in FASTA format.
GENETIC STOCKS OF NEMATOSTELLA
Genetic stock data can assist users to identify and obtain isolated DNA or live animals of known geographic origin or genotype. Populations are identified by collection locale, by laboratory and by genetic distance as indicated on a recent intraspecific phylogeny constructed using AFLP data (24). Contact information and population genetics data indicating clonality of the population of interest is returned to the user if available, as are any unique phenotypes represented by that population. The easy availability of genetic stocks is necessary to foster laboratory-based research. To maximize the utilization of this resource, those who collect or culture Nematostella are encouraged to make their own genetic stocks available through StellaBase. This resource will prove particularly useful as unusual phenotypes of particular interest are identified in natural populations or are produced in the laboratory. Currently, StellaBase houses data from 24 available populations of Nematostella.
LITERATURE SEARCH
The rise of Nematostella as a model system is a very recent phenomenon. In the early 1990s, Cadet Hand and Kevin Uhlinger highlighted the merits of this species as a possible model system for developmental biology (8–10). In 1997, the first gene sequences from Nematostella were published (25). The first molecular analyses of Nematostella development were published in 2003 (26). However, while much of the interest in Nematostella is quite recent, there exists a substantial literature on this anemone that would be of great use to the community. From the first published species account in 1935 (4) through 2005, no fewer than 79 publications have directly referenced Nematostella. A wealth of information on the morphology, development and natural history of this species is contained in these articles and book chapters. However, only a small minority of these existing citations (currently 17 of 79) is indexed in electronic literature databases such as PubMed. StellaBase indexes all of them and allows users to perform keyword searches on the complete texts. As future publications on Nematostella are indexed by PubMed, these will automatically be added to the StellaBase literature database. Existing publications that have not been identified, as well as future publications that are not indexed by PubMed, will be added manually.
PRIMER LIBRARY
StellaBase houses a library of 698 oligonucleotide primer sequences. The primer sequences and associated information were gleaned from the literature or obtained directly from researchers. Users may search for primers by gene name; the primer sequence, its melting temperature and usage notes will be returned to the user. Users are encouraged to submit additional primer sequences to this database.
GENE EXPRESSION QUERIES
StellaBase has been integrated into the gene expression search function on CnidBase, The Cnidarian Evolutionary Genomics Database [http://cnidbase.bu.edu; (27)]. Users can search for gene expression data in Nematostella by specifying seven different gene expression parameters: gene name, expression level, life history stage, body region, body layer, assay type and cell type. The gene expression search function of CnidBase facilitates direct comparisons of gene expression between Nematostella and other members of the phylum Cnidaria.
INTEGRATION OF STELLABASE AND CNIDBASE
Genomic data from the Cnidaria are accumulating at a rapid rate. The EST database at NCBI currently lists well over 190 000 cnidarian ESTs from a phylogenetically diverse range of species including corals, jellyfishes, sea anemones and hydras. CnidBase was developed to organize various forms of cnidarian genomic data into a single repository that would facilitate comparative studies among species (27). To further support comparative cnidarian research, we chose not to develop StellaBase as an isolated entity, but rather, we have integrated StellaBase with CnidBase. The synergistic relationship between CnidBase and StellaBase provides a model for the incorporation of future species-specific cnidarian databases into a CnidBase centered network. As more species-specific experimental data is obtained from cnidarians, the need for more species-specific cnidarian databases is likely to arise. Large amounts of data are being amassed for a number of other informative cnidarian species, including Acropora (28), Hydra (29), Hydractinia (30) and Podocoryne (31). To facilitate the inclusion of other species-specific cnidarian databases, we have posted the table structure and an entity-relationship diagram (32) for all genomic data stored within StellaBase.
GENE FAMILY COMPARISONS
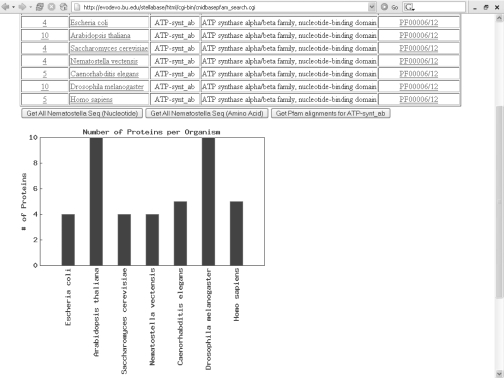
As the phylum Cnidaria enters the genomic age, it will become possible to uncover the full complement of particular gene families present in selective cnidarian species and to compare the complexity of particular gene families in cnidarian and bilaterian models. We have added new functions to CnidBase that facilitate rapid and thorough comparisons of (i) genome content and (ii) gene family content among distantly related organismal lineages. The completed proteomes of Homo sapiens, Caenorhabditis elegans, Saccharomyces cerevisiae, Arabidopsis thaliana, Escheriachia coli and Nematostella vectensis were compared to the Pfam database to identify the number of genes from a particular family present in each lineage. Users can query these data by Pfam protein family name and by genome comparison. The stringency of each search is controlled by specifying a threshold expectation value. The search returns all sequences that match the user defined criteria in each selected species. The full complement of each gene family may quickly be compiled from each taxon, providing a broad overview of the evolution of genomes and particular gene families and a convenient launch point for detailed phylogenetic studies (Figure 2).
Figure 2.
Output from gene family query using the genome comparison tool.
COMMUNITY PARTICIPATION IN STELLABASE AND CNIDBASE
It is a challenge to support researcher initiated databases, regardless of their potential value to the research community—‘consortium-based’ databases are generally more successful in disseminating information than are databases maintained by individual labs or research groups (33). Both StellaBase and CnidBase incorporate explicit opportunities for the cnidarian research community to supply critical content. Users are invited to submit primer sequences and genetic stock. Users are also encouraged to submit comments regarding specific gene sequences or families, such as suggestions for gene annotations. User comments will become available with sequence information as they are added.
In addition to supplying content to these existing databases, we supply relatively simple guidelines for cnidarian researchers to use our model and generate species-specific databases that can be seamlessly integrated into CnidBase. Table structure and an entity-relationship diagram are available on the StellaBase website; code for query interfaces, annotation and database construction are available upon request. As additional cnidarian genomes are sequenced, it is our hope that this model will allow for data to be available to the community quickly and in such a way that interphylum comparisons are facilitated.
FUTURE DIRECTIONS
A number of improvements to both StellaBase and CnidBase will occur in the near future. (i) As StellaBase is used to mine the Nematostella genome, information stored within StellaBase will be updated and new information will be added. We are currently in the process of annotating genes from a number of families; these annotations will be added to StellaBase as completed. (ii) The CnidBase proteomic search function will increase in utility in the future as more species are added. Porifera and Ciona intestinalis will prove to be valuable additions due to their interesting phylogenetic positions and Drosophila melanogaster and Mus musculus will increase the confidence with which estimates are made regarding the proteomes of protostomes and deuterostomes, respectively. (iii) We intend to add a ‘classic literature’ search function to CnidBase. Scanned versions of texts no longer protected by copyright law containing valuable information on cnidarian morphology, development and natural history will be made available.
CONCLUSIONS
StellaBase is the genomics database of Nematostella vectensis. Through it, users may search a wide range of data types, including genomic data, genetic stocks, primers, literature and gene expression patterns. StellaBase provides a launching point for performance of both desktop phylogenetic and genomic analyses and bench-top laboratory research. By developing StellaBase within the framework of CnidBase, we have utilized the inherently comparative nature of CnidBase to develop search functions that facilitate detailed phylogenetic analyses of extremely divergent lineages. We have posted a roadmap for cnidarian researchers to follow in the development of additional species-specific databases that will integrate seamlessly with CnidBase.
Acknowledgments
The authors are grateful to the many researchers who published on Nematostella long before it entered the genomic age (for a complete list, see http://nematostella.org). We are especially grateful to Cadet Hand and Kevin Uhlinger who introduced the PI to this species. This research was funded by the National Science Foundation (grant IBN-0212773 to JRF) and by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. Funding to pay the Open Access publication charges for this article was provided by the National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kortschak R.D., Samuel G., Saint R., Miller D.J. EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr. Biol. 2003;13:2190–2195. doi: 10.1016/j.cub.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y., Cun S., Xie X., Lin J., Wei J., Yang W., Mou C., Yu C., Ye L., Lu Y., Fu Z., Xu A. EST analysis of gene expression in the tentacle of Cyanea capillata. FEBS Lett. 2003;538:183–191. doi: 10.1016/s0014-5793(03)00164-9. [DOI] [PubMed] [Google Scholar]
- 3.Peterson K.J., Butterfield N.J. Origin of the Eumetazoa: testing ecological predictions of molecular clocks against the Proterozoic fossil record. Proc. Natl Acad. Sci. USA. 2005;102:9547–9552. doi: 10.1073/pnas.0503660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephenson T.A. The British Sea Anemones. Volume II. London: The Ray Society; 1935. [Google Scholar]
- 5.Hand C. Another sea anemone from California and the types of certain California species. J. Wash. Acad. Sci. 1957;47:411–414. [Google Scholar]
- 6.Crowell S. A new sea anemone from Woods Hole, Massachusetts. J. Wash. Acad. Sci. 1946;36:57–60. [Google Scholar]
- 7.Darling J.A., Reitzel A.R., Burton P.M., Mazza M.E., Ryan J.F., Sullivan J.C., Finnerty J.R. Rising starlet: the starlet sea anemone, Nematostella vectensis. Bioessays. 2005;27:211–221. doi: 10.1002/bies.20181. [DOI] [PubMed] [Google Scholar]
- 8.Hand C., Uhlinger K.R. The culture, sexual, and asexual reproduction and growth of the sea anemone Nematostella vectensis. Biol. Bull. 1992;182:169–176. doi: 10.2307/1542110. [DOI] [PubMed] [Google Scholar]
- 9.Hand C., Uhlinger K. The unique, widely distributed sea anemone, Nematostella vectensis Stephenson: a review, new facts, and questions. Estuaries. 1994;17:501–508. [Google Scholar]
- 10.Hand C., Uhlinger K.R. Asexual reproduction by transverse fission and some anomalies in the sea anemone Nematostella vectensis. Invertebr. Biol. 1995;114:9–18. [Google Scholar]
- 11.Finnerty J.R., Pang K., Burton P., Paulson D., Martindale M.Q. Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science. 2004;304:1335–1337. doi: 10.1126/science.1091946. [DOI] [PubMed] [Google Scholar]
- 12.Finnerty J.R., Paulson D., Burton P., Pang K., Martindale M.Q. Early evolution of a homeobox gene: the parahox gene Gsx in the Cnidaria and the Bilateria. Evol. Dev. 2003;5:331–345. doi: 10.1046/j.1525-142x.2003.03041.x. [DOI] [PubMed] [Google Scholar]
- 13.Kusserow A., Pang K., Sturm C., Hrouda M., Lentfer J., Schmidt H.A., Technau U., von Haeseler A., Hobmayer B., Martindale M.Q., Holstein T.W. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–160. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- 14.Martindale M.Q., Pang K., Finnerty J.R. Investigating the origins of triploblasty: ‘mesodermal’ gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa) Development. 2004;131:2463–2474. doi: 10.1242/dev.01119. [DOI] [PubMed] [Google Scholar]
- 15.Pang K., Matus D.Q., Martindale M.Q. The ancestral role of COE genes may have been in chemoreception: evidence from the development of the sea anemone, Nematostella vectensis (Phylum Cnidaria; Class Anthozoa) Dev. Genes Evol. 2004;214:134–138. doi: 10.1007/s00427-004-0383-7. [DOI] [PubMed] [Google Scholar]
- 16.Wikramanayake A.H., Hong M., Lee P.N., Pang K., Byrum C.A., Bince J.M., Xu R., Martindale M.Q. An ancient role for nuclear beta-catenin in the evolution of axial polarity and germ layer segregation. Nature. 2003;426:446–450. doi: 10.1038/nature02113. [DOI] [PubMed] [Google Scholar]
- 17.Mullikin J.C., Ning Z. The phusion assembler. Genome Res. 2003;13:81–90. doi: 10.1101/gr.731003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burge C.B., Karlin S. Finding the genes in genomic DNA. Curr. Opin. Struct. Biol. 1998;8:346–354. doi: 10.1016/s0959-440x(98)80069-9. [DOI] [PubMed] [Google Scholar]
- 19.Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 20.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam Protein Families Database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durbin R., Eddy S.R., Krogh A., Mitchison G.J. Biological Sequence Analysis: Probablistic Models of Proteins and Nucleic Acids. Cambridge UK: Cambridge University Press; 1998. [Google Scholar]
- 22.Burset M., Guigo R. Evaluation of gene structure prediction programs. Genomics. 1996;34:353–367. doi: 10.1006/geno.1996.0298. [DOI] [PubMed] [Google Scholar]
- 23.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Darling J.A., Reitzel A.M., Finnerty J.R. Regional population structure of a widely introduced estuarine invertebrate: Nematostella vectensis Stephenson in New England. Mol. Ecol. 2004;13:2969–2981. doi: 10.1111/j.1365-294X.2004.02313.x. [DOI] [PubMed] [Google Scholar]
- 25.Finnerty J.R., Martindale M.Q. Homeoboxes in sea anemones (Cnidaria:Anthozoa): a PCR-based survey of Nematostella vectensis and Metridium senile. Biol. Bull. 1997;193:62–76. doi: 10.2307/1542736. [DOI] [PubMed] [Google Scholar]
- 26.Scholz C.B., Technau U. The ancestral role of Brachyury: expression of NemBra1 in the basal cnidarian Nematostella vectensis (Anthozoa) Dev. Genes Evol. 2003;212:563–570. doi: 10.1007/s00427-002-0272-x. [DOI] [PubMed] [Google Scholar]
- 27.Ryan J.F., Finnerty J.R. CnidBase: The Cnidarian Evolutionary Genomics Database. Nucleic Acids Res. 2003;31:159–163. doi: 10.1093/nar/gkg116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller D.J., Ball E.E. The coral Acropora: what it can contribute to our knowledge of metazoan evolution and the evolution of developmental processes. Bioessays. 2000;22:291–296. doi: 10.1002/(SICI)1521-1878(200003)22:3<291::AID-BIES11>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 29.Steele R.E. Developmental signaling in Hydra: what does it take to build a ‘simple’ animal? Dev. Biol. 2002;248:199–219. doi: 10.1006/dbio.2002.0744. [DOI] [PubMed] [Google Scholar]
- 30.Frank U., Leitz T., Muller W.A. The hydroid Hydractinia: a versatile, informative cnidarian representative. Bioessays. 2001;23:963–971. doi: 10.1002/bies.1137. [DOI] [PubMed] [Google Scholar]
- 31.Galliot B., Schmid V. Cnidarians as a model system for understanding evolution and regeneration. Int. J. Dev. Biol. 2002;46:39–48. [PubMed] [Google Scholar]
- 32.Ramakrishnan R., Gehrke J. Database Management Systems. Third Edition. NY: McGraw-Hill Higher Education; 2003. The Relational Model; pp. 57–94. [Google Scholar]
- 33.Merali Z., Giles J. Databases in peril. Nature. 2005;435:1010–1011. doi: 10.1038/4351010a. [DOI] [PubMed] [Google Scholar]