Abstract
The integrated microbial genomes (IMG) system is a new data management and analysis platform for microbial genomes provided by the Joint Genome Institute (JGI). IMG contains both draft and complete JGI genomes integrated with other publicly available microbial genomes of all three domains of life. IMG provides tools and viewers for analyzing genomes, genes and functions, individually or in a comparative context. IMG allows users to focus their analysis on subsets of genes and genomes of interest and to save the results of their analysis. IMG is available at http://img.jgi.doe.gov.
INTRODUCTION
According to the Genomes Online Database, ∼250 microbial genomes have been sequenced to date, with over 700 other projects ongoing and more in the process of being launched (1). The Department of Energy's (DOE) Joint Genome Institute (JGI) is one of the major contributors of microbial genome sequence data, currently conducting ∼23% of the reported bacterial genome projects worldwide. Individual microbial genomes are sequenced and assembled to draft level at JGI's production facility (PGF), and finished either at PGF, Lawrence Livermore or Los Alamos National Labs. Both draft and finished genomes pass through the automatic Genome Analysis Pipeline (2) at Oak Ridge National Laboratory (ORNL), which generates gene models and associates automatically predicted genes with functional annotations, such as InterPro protein families (3), COG categories (4) and KEGG pathway maps (5). All finished genomes are submitted to GenBank.
Before publication or submission to GenBank, scientific groups interested in a specific genome further review and curate the microbial genome data in collaboration with ORNL's Computational Biology group and JGI's Microbial Genome Analysis Program. The efficiency of microbial genome review, curation and analysis increases substantially when individual microbial genomes are examined in the context of other genomes. Providing such a framework in order to ensure timely analysis of the genomes sequenced at JGI is one of the main goals of the Integrated Microbial Genomes (IMG) system.
IMG provides support for comparative analysis of microbial genomes in an integrated genome data context. This requires a high level of genome diversity from public sources, such as EBI's Genome Reviews (6), NCBI's RefSeq (7) and EMBL's Nucleotide Sequence Database (8). A validation and correction process for gene models ensures data coherence in IMG.
INTEGRATED MICROBIAL GENOMES: DATA
The data model underlying the IMG system provides the structure required for integrating and managing microbial and selected eukaryotic genomic data collected from multiple data sources. The data model incorporates primary genomic sequence information, computationally predicted and curated gene models, pre-computed sequence similarity relationships, functional annotations and pathway information, in a coherent biological context.
Genomes (organisms) are identified via their taxonomic lineage (domain, phylum, class, order, family, genus, species, strain). For each genome, the primary DNA sequence and its organization in scaffolds and/or contigs are recorded. Genomic features, such as predicted coding sequences (CDSs) and some functional RNAs, are recorded with start/end coordinates. Protein-coding genes are further characterized in terms of molecular function and participation in pathways. Proteins are grouped into protein families based on sequence similarity. Pathways, reactions and compounds are included from KEGG. The Gene Ontology (9) is the source for gene functions for the genomes from EBI Genome Reviews, while COGs provide clusters of orthologous groups of genes as further characterization for gene function. Genes are assigned to COGs based on RPS-BLAST (reverse position specific BLAST) and NCBI's Conserved Domain Database (CDD) (10), with an E-value of 10−2. Genes are associated with Pfam (11) in a similar way. Orthologous gene relationships are computed as bidirectional best hits between genomes. Paralogous gene relationships are computed as reciprocal hits within the same genome. Homologs are computed as unidirectional hits with an E-value of 10−2 or better, with IMG providing support for filtering by percent identity, bit score and more stringent E-values.
Before they are loaded into IMG, new JGI finished genomes undergo a gene model validation process for identifying and correcting potential gene model errors. The process involves three steps: editing overlapping CDSs, correcting start codons and identifying missed genes and pseudogenes.
INTEGRATED MICROBIAL GENOMES: ANALYSIS
Microbial genome data analysis in IMG is set in the comparative context of multiple microbial genomes. IMG allows navigating the microbial genome data space along three key dimensions: genomes (organisms), functions (terms and pathways) and genes.
Finding and examining organisms
Organism selections help focus the analysis on a subset of genomes of interest, such as all the strains within a specified genus.
Organisms can be selected using the keyword based ‘Organism Search’ that involves a number of filters, such as Phylum or Sequencing Status. Organisms can also be selected from an alphabetical or phylogenetic list using the ‘Organism Browser’. Selected organisms can be saved in order to provide the context for further analysis. Selected organisms can also be saved to a local file that can subsequently be loaded back into IMG in order to restore organism selections.
Individual organisms can be further explored using the ‘Organism Details’ page, that includes various statistics of interest, such as the number of genes in the organism that are associated with KEGG, COG, Pfam, InterPro or enzyme information. For each organism one can also examine the associated list of scaffolds and contigs. For each coordinate range, the ‘Chromosome Viewer’ allows displaying genes colored according to COG functional categories.
Finding and examining genes
The user can search for genes either by (i) a keyword based ‘Gene Search’, (ii) sequence similarity search or (iii) using the ‘Phylogenetic Profiler’.
‘Gene Search’ finds genes based on partial or exact matches to a string of characters in specified IMG fields such as gene name or locus tag. Similarity searches are implemented via BLASTp (protein versus protein), BLASTx (DNA versus protein), BLASTn (DNA versus DNA) or tBLASTn (protein–DNA versus DNA–protein). Users can define similarity thresholds and select the target database. The ‘Phylogenetic Profiler’ allows the identification of genes in a genome (organism) of interest that have homologs in one group of organisms and lack homologs in another group of organisms. Similarity thresholds can also be defined in order to fine-tune the profile.
All (or some of) the identified genes can then be selected and maintained for further comparative analysis using the ‘Gene Cart’, which is similar to shopping carts of commercial websites. Selected genes can also be saved to a file that can subsequently be loaded back into IMG in order to restore gene selections.
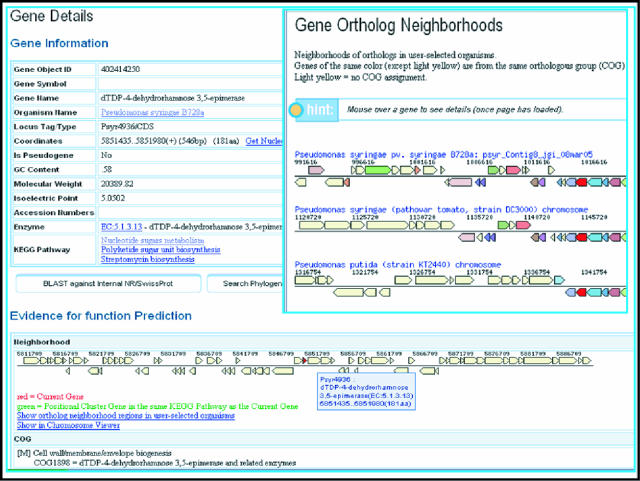
Individual genes can be analyzed using the ‘Gene Details’ page, as illustrated in Figure 1. A Gene Information table includes gene identification, locus information, biochemical properties of the product and associated KEGG pathways. The ‘Gene Details’ page also includes evidence for functional prediction: gene neighborhood (see Figure 1), COG, InterPro, Pfam, and pre-computed lists of orthologs and paralogs. The gene neighborhood displays the gene of interest with its neighboring genes in a 25 kb chromosomal window.
Figure 1.
Gene Details Page and Gene Ortholog Neighborhoods. The gene neighborhood in the ‘Gene Details’ page shows the query gene (centered, in red) and other genes within a 25 kb window. A ‘Gene Ortholog Neighborhoods’ page shows the gene neighborhood of orthologs of the query gene, across several organisms. Each gene's neighborhood appears above and below a single line showing the genes reading in one direction on top and those reading in the opposite direction on the bottom. Genes with the same color indicate association with the same COG group. For each gene, locus tag, scaffold coordinates and COG group number are provided locally (by placing the cursor over the gene), while additional information is available in the Gene Details page that is linked to each gene.
A gene can be examined in the context of its location on the chromosome using the ‘Chromosome Viewer’ link. The neighborhood of the gene can be compared to the corresponding neighborhoods of its orthologs using the ‘Gene Ortholog Neighborhoods’ link (as shown in the right-hand side pane of Figure 1). One can see details for any gene in the displayed (single or multiple) neighborhood by following the link to the associated ‘Gene Details’ page.
A gene can be also examined in the context of its associated pathways, whereby the link embedded in the pathway name listed in the Gene Information table allows the KEGG map associated with the gene to be displayed. On such a map, EC numbers are color-coded and linked to the Gene Details page for the associated genes.
Finding and examining functions
The functional role of genes in IMG is characterized by a variety of annotations, including their COG membership, association with Pfam domains, Gene Ontology assignments and association with enzymes in KEGG pathways. Functional annotations can be searched using keywords and filters, with the selected functions leading to a list of associated genes either directly or via a list of organisms. COG categories and KEGG pathways also can be searched and browsed separately.
Individual COG categories can be further explored using the ‘COG Category Details’ page that lists the COGs of a given category and the number of organisms that have genes belonging to each COG. For a given COG, the ‘organism counts’ are linked to a list of organisms and their associated ‘gene counts’. Gene counts for COGs in a given category can be displayed for multiple organisms using a ‘COG Category Profile’. KEGG pathways can be explored in a similar manner using ‘KEGG Pathway Details’ and ‘KEGG Pathway Profile’.
Comparative analysis
Comparative analysis of genomes is provided in IMG through a number of tools that allow genomes to be compared in terms of organism-specific statistics, genes and sequence conservation.
‘Organism Statistics’ compiles statistics provided through the ‘Organism Details’ page to facilitate comparative analysis of the organisms that have been selected using the ‘Organism Browser’.
Comparative analysis of genes includes gene neighborhood analysis (similar to the gene ortholog neighborhood analysis mentioned above), phylogenetic occurrence profile analysis and multiple sequence alignment, applied on genes selected from the ‘Gene Cart’.
‘Phylogenetic Occurrence Profile’ shows the pattern of occurrence of a specific gene across selected organisms. The occurrence profiles of multiple genes across these organisms can be then visually compared (Figure 2). Occurrence profiles in IMG are based on orthologous relationships. In order to find other genes in the same organism with the same occurrence profile, the ‘Phylogenetic profile similarity search’, available at the ‘Gene Details’ page, can be used.
Figure 2.
Phylogenetic occurrence profile. The occurrence profiles of multiple genes across selected organisms can be visually compared. For each gene, a fixed length ordered vector is displayed in a BLAST-like alignment format. The positions in the vector correspond to the list of selected organisms, whereby the organisms are phylogenetically ordered; presence of a gene or its ortholog in a given organism is indicated by a domain letter (‘B’ for Bacteria, ‘A’ for Archaea and ‘E’ for Eukarya) while the absence of the gene is indicated by a dot (‘.’). One can mouse over the letter or dot to see the organism and phylum names.
Finally, DNA conservation can be explored for a number of organisms in IMG using the VISTA comparative genome analysis tools (12). Selecting an organism from a predefined list invokes the VISTA browser that can then be used for examining conservation.
FUTURE PLANS
The current version of IMG (IMG 1.2, as on September 1, 2005) contains a total of 618 genomes consisting of 318 bacterial, 25 archaeal, 15 eukaryotic genomes and 260 bacterial phages. Among these genomes, 534 are finished and 84 are draft genomes. The finished genomes include 204 bacterial and 21 archaeal genomes from EBI Genome Reviews (version 31, July 18, 2005), 9 eukaryotic genomes from EMBL (as on January 17, 2005), 2 eukaryotic genomes from RefSeq (as on March 21, 2005) and 4 eukaryotic genomes from GenBank (as on July 27, 2005). In addition, IMG 1.2 contains 120 microbial genomes sequenced at JGI, out of which 40 are finished and 80 are draft genomes.
IMG continues to be extended in terms of data content through quarterly updates, whereby it aims at continuously increasing the number of genomes integrated in the system from public and local resources, following the principle that the value of genome analysis increases with the number of genomes available as a context for comparative analysis.
The future versions of IMG will focus on data quality in terms of the coherence of annotations, based on sound validation and correction procedures, as well as corroboration of annotations from other public microbial genome data resources. The comparative analysis context provided by IMG will facilitate the detection and correction of annotation errors.
IMG aims at increasing the coverage (breadth and depth) of functional annotations in the system, the result of providing scientists with tools that implement annotation techniques based on the functional coupling of genes, a hypothesis inspired by observed biological evolutionary phenomena.
Acknowledgments
We thank Annette Greiner, Kristen Taylor, Alla Lapidus and Paul Richardson for their contribution to the development and maintenance of IMG. The work of JGI's production, cloning, sequencing, assembly, finishing and annotation teams is an essential prerequisite for IMG. Eddy Rubin and James Bristow provided support, advice and encouragement throughout this project. This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48, Lawrence Berkeley National Laboratory under Contract No. DE-AC02-05CH11231 and Los Alamos National Laboratory under contract No. W-7405-ENG-36. Funding to pay the Open Access publication charges for this article was provided by the Lawrence Berkeley National Laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bernal A., Ear U., Kyrpides N. Genomes Online Database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Res. 2001;29:126–127. doi: 10.1093/nar/29.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauser L., Larimer F., Land M., Shah M., Uberbacher E. Analysis and annotation of microbial genome sequences. Genet. Eng. 2004;26:225–238. doi: 10.1007/978-0-306-48573-2_11. [DOI] [PubMed] [Google Scholar]
- 3.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tatusov R.L., Koonin E.V., Lipman D.J.A. Genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 5.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kersey P., Bower L., Morris L., Horne A., Petryszak R., Kanz C., Kanapin A., Das U., Michoud K., Phan I., et al. Integr8 and genome reviews: integrated views of complete genomes and proteoms. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts, and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanz C., Aldebert P., Althorpe N., Baker W., Baldwin A., Bates K., Browne P., van den Broek A., Castro A., Cochrane G., et al. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2005;33:D29–D33. doi: 10.1093/nar/gki098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchler-Bauer A., Panchenko A.R., Shoemaker B.A., Thiessen P.A., Geer L.Y., Bryant S.H. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]