Abstract
Alternative splicing is an important regulatory mechanism of mammalian gene expression. The alternative splicing database (ASD) consortium is systematically collecting and annotating data on alternative splicing. We present the continuation and upgrade of the ASD [T. A. Thanaraj, S. Stamm, F. Clark, J. J. Riethoven, V. Le Texier, J. Muilu (2004) Nucleic Acids Res. 32, D64–D69] that consists of computationally and manually generated data. Its largest parts are AltSplice, a value-added database of computationally delineated alternative splicing events. Its data include alternatively spliced introns/exons, events, isoform splicing patterns and isoform peptide sequences. AltSplice data are generated by examining gene-transcript alignments. The data are annotated for various biological features including splicing signals, expression states, (SNP)-mediated splicing and cross-species conservation. AEdb forms the manually curated component of ASD. It is a literature-based data set containing sequence and properties of alternatively spliced exons, functional enumeration of observed splicing events, characterization of observed splicing regulatory elements, and a collection of experimentally clarified minigene constructs. ASD includes a workbench, which is an analysis tool that enables users to carry out splicing related analysis such as characterization of introns for various splicing signals, identification of splicing regulatory elements on a given RNA sequence, prediction of putative exons and prediction of putative translation start codons. The different ASD modules are integrated and can be accessed through user-friendly interfaces and visualization tools. ASD data has been integrated with Ensembl genome annotation project as a Distributed Annotation System (DAS) resource and can be viewed on Ensembl genome browser. The ASD resource is presented at (http://www.ebi.ac.uk/asd).
INTRODUCTION
Alternative pre-mRNA splicing is emerging as one of the most important mechanisms to control eukaryotic gene expression. Recent array data indicate that as much as 76% of genes generate alternatively spliced products (1). Alternative splicing regulates numerous aspects of protein function, such as binding properties, intracellular localization, enzymatic activity, stability and post-translational modifications. Reports in literature indicate that protein isoforms generated by alternative splicing show in most cases only subtle differences. However, in some cases alternative splicing can lead to large functional differences, e.g. by generating dominant negative isoforms (2). Finally, 10–15% of genes could be switched off due to the coupling between nonsense-mediated decay and alternative splicing (3). This indicates that alternative splicing controls both transcript composition and abundance.
Despite intense research, the mechanisms leading to splice site selection are not fully understood. Currently, it is not possible to accurately predict alternative exons from genomic sequences. It is not at all possible to predict the tissue or developmental expression profile of an alternative exon. The major obstacle for an accurate prediction is the lack of conservation in the regulatory sequences of the pre-mRNA that can only be described by consensus sequences or expectation matrices. However, in vivo, alternative exons are recognized and regulated with high fidelity, because numerous proteins bind to pre-mRNA and help in exon recognition. Due to the combination of multiple weak protein–protein and protein–RNA interactions, alternative exons can be faithfully recognized (4,5). The importance of proper splicing site recognition is underlined by an increasing number of diseases that are caused or associated with the selection of wrong splicing sites (6,7).
Alternative splicing events have been compiled previously in different databases [reviewed in (8)]. Here, we present the continuation and upgrade of the alternative splicing database (ASD) [the previous version was reported earlier in (9)] as a bioinformatics resource that integrates data on alternative splicing, derived from computational as well as literature based approaches, and bioinformatics analysis tools.
ASD
The different component data sets of ASD
Data generated from computational approaches and data reported in the literature are the two major sources for databases on alternative splicing. The major advantage of data from computational approaches, such as EST comparison, is the large size of the data sets. However, these data sets lack biological information about the alternative splicing events. In contrast, data sets derived from the literature contain biologically relevant information, but these data sets are smaller. In order to combine these two approaches, we carried out two activities namely, (i) we developed a computational pipeline (AltSplice) that generates genome-wide value-added data on alternative exons, and (ii) we developed a procedure (AEdb) that manually collects data on alternative exons from literature. In addition, data on motifs, functions and minigenes described in the literature were collected in databases. We then built an integrated database (Figure 1) from these heterogeneous data resources. The integrated database is named ASD for which we developed query interfaces that are flexible enough to handle the heterogeneity. A single-query bar provides a quick access to all of ASD data, allowing retrieval of data using keyword search or sequence comparison searches. In addition, each data set can be queried using a data set-specific interface. Finally, all data sets can be downloaded as flat file distributions.
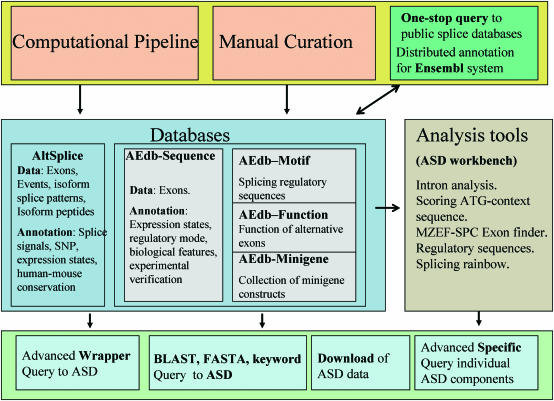
Figure 1.
Structure of ASD. We used computational pipelines and manual curation (top, pink) to create the modular databases of ASD (middle, blue). The individual databases are integrated, cross-linked and are available through a variety of interface tools (bottom, sky blue). Currently the databases are the computer generated Altsplice and the manually-curated AEdb-Sequence, AEdb-Motif, AEdb-Function and AEdb-Minigene databases. The ASD data are integrated with Ensembl genome annotation system and is visible from Ensembl genome browser; Publicly-available databases on alternative splicing are accessible from ASD interfaces (top right, dark green) The databases are connected to analysis tools that are collected in the ASD workbench (middle right, grey).
The statistics on different data sets of the ASD are listed in Table 1.
Table 1.
Statistics on ASD data
| AltSplice data statistics (Release 2 of AltSplice-Human and AltSplice-Mouse) | |
| Genes with alternative splicing | 9929 (61%) of 16 293 human genes |
| 8211 (50%) of 16 391 (mouse) genes | |
| Alternative splicing events per gene | 2.6 (human); 2.2 (mouse) |
| Alternative splicing patterns per gene | 3.9 (human); 3.7 (mouse) |
| Distribution of the observed event types | Cassette exon (52%) |
| Alternate acceptor or donor (27%) | |
| Intron retention (17%) | |
| Mutually exclusive (4%) | |
| Flanking exons often undergo extension or truncation | 26% of Cassette exon events |
| 23% of Retained introns | |
| 19% of Mutually exclusive exons | |
| Human-mouse conservation | Orthologous gene pairs = 5176 |
| Number of conserved events = 1177 | |
| Number of conserved exons = 28 276 | |
| Entries with peptide sequence annotation for at least 1 splicing pattern | 6848 (human); 4366 (mouse) |
| Entries with peptide sequence annotation for ≥2 splicing patterns | 2896 (human); 977 (mouse) |
| Entries for which variant peptide data are available in UniProt | 2083 (human); 896 (mouse) |
| Entries presenting data for ≥2 isoform peptide sequences (by either of the above two sources) | 3994 (human); 1573 (mouse) |
| Entries associated with AEdb-Sequence database | 367 (human); 118 (mouse) |
| AEdb-Sequence data statistics-2255 entries | |
| Distribution | Number of entries |
| Organism distribution | Human (1283); mouse (413); rat (232); drosophila (100); others (227) |
| Event type distribution | Cassette exon (1281) |
| Alternative acceptor or donor (395) | |
| Intron retention (154) | |
| Mutually exclusive exons (130) | |
| Alternative 3′ exon by polyA variant (71) | |
| Regulation associated with disease | 295 |
| Regulation associated with development | 282 |
| Regulation associated with tissue type | 312 |
| Regulation causing frameshift | 151 |
| Regulation introducing stop codons | 260 |
| Alternative exon being noncoding exon | 222 |
| Entries associated with AltSplice | 1198 (human and mouse entries) |
| AEdb-Function data statistics-354 entries | |
| Functional role | Number of entries |
| Modulation of protein interaction | 136 |
| Internal structural change | 119 |
| Novel carboxyl terminus | 87 |
| Novel amino terminus | 38 |
| Association with disease | 81 |
| Intracellular location | 76 |
| Enzymatic activity | 64 |
| Channel activity | 54 |
| Others | 37 |
| AEdb-Motif data statistics-255 entries | |
| Type of regulator sequence | Number of entries |
| Exon enhancer | 97 |
| Exon silencer | 44 |
| Intron enhancer | 56 |
| Intron silencer | 37 |
| AEdb-Minigene data statistics—82 entries | |
| Distribution | Number of entries |
| Organism distribution | Human (46); mouse (17); rat (15); others (9) |
| Splicing mechanism distribution | Cassette exon [single exon, 45; multiple casette exons (3); incremental combinatorial exons (2)]; Alternative acceptor or donor sites (17); Mutually exclusive exons (13); Intron retention (2) |
| Reported tissue specificity | 55 |
| Known regulatory factors | 32 |
| Deduced enhancer and silencer sequences | 97 |
| Hyperlinks to AEdb-Sequence database | 78 (to 105 AEdb-Sequence entries) |
AltSplice
AltSplice data are generated by an automated computational pipeline. The basal data includes transcript-confirmed introns/exons, alternative splicing events and isoform splicing patterns. The basal data are generated through computational comparison of EST/mRNA alignments with genomic sequences [see (9,10) for details on the computational methods]. The data are annotated for biological features (such as splicing site characteristics, expression state of the isoforms, allele usage at SNP positions, conservation of intron/exon-events across species and peptide isoforms) by various computational modules that are part of AltSplice pipeline. AltSplice data (Table 1) indicates that up to 61% of human genes (and 50% of mouse genes) undergo alternative splicing. The available transcriptome data indicates that in an average 3.9 isoform splicing patterns can be expressed from a single human gene. Cassette exon events outnumber the other event types and one in every four cassette exon events is accompanied by extension/truncation of the flanking exons. The number of human-mouse orthologous gene pairs (with data on alternative splicing) present in AltSplice is around 5200, and such a large data set is valuable for studies on evolution of alternative splicing. Finally, AltSplice presents data on isoform peptide sequences for around 4000 human genes, and such a large data set of variant peptides is valuable for studies aimed at deciphering splicing mediated functional and structural changes in proteins.
AEdb-Sequence
AEdb-Sequence is a literature based, manually curated database of alternative exons. We used ‘alternative splicing’ as a keyword to search PubMed bibliography data and collected information on the following features from the resultant research articles: organism, splicing mechanism, tissue-specificity, regulation during development stages, disease association, regulatory features of the exon and the sequence of the alternatively spliced exon as well as its flanking constitutive exons. It is seen that more than half the number of AEdb-Sequence entries are from human (Table 1). As is in the case of AltSplice data, cassette exon events outnumber other event types. The data set reports splicing events that are specific to cellular states, such as tissue type, development stage and disease state. Roughly 10% of the entries report events that introduce premature stop codons and this data set can serve the studies on nonsense mediated decay of transcripts. Finally, 10% of the reported exons are from non-coding regions of the genes.
AEdb-Function
The function database is a literature based, manually curated database of known functions of the alternative exons. Functional differences between the protein isoforms generated by alternative splicing are enumerated from the literature and are organized into 11 well-defined categories, such as ‘Modulation of protein interaction’ or ‘Internal structural change’ (Table 1). An analysis of the function of alternative exons based on this data set has been published previously (2).
AEdb-Motif
Alternative splicing site selection is partially regulated by weak binding of proteins to highly degenerate regulatory sequences. As a first attempt to understand the combinatorial control behind this regulation, we collected splicing regulatory motifs described in literature and expanded upon the previous collections of intronic regulatory sequences (11), exonic regulatory sequences (12,13) and disease-causing mutations (6). The collection reports 153 enhancer sequences and 81 silencer sequences (Table 1). The entries are annotated with value-added information, such as the experimental technique used, the nucleotide sequence of the motif, mutations that are studied and the protein that binds at the motif.
AEdb-Minigenes
A minigene is a genomic fragment that includes the alternative exon and the surrounding introns as well as the flanking constitutively spliced exons. Constructs derived by cloning the insert in an eukaryotic expression vector are increasingly used to study alternative splicing (14,15). We compiled all minigenes described in the literature. The splicing patterns and deduced regulatory sequences are represented in a graphic format. The minigene collection includes 82 entries for which a total of 97 regulatory sequences are ascribed. The reported minigene constructs representing cassette exon events outnumber those for other event types (Table 1). The minigene entries are linked to appropriate entries in AEdb-Sequence data set, which allows the user to quickly identify experimentally useful minigenes by searching the database.
DATA INTEGRATION
Integration of data across the different data sets of ASD
Extensive integration has been made between AltSplice and AEdb-Sequence. Alternative exons and events that are common in AltSplice and AEdb-Sequence are identified and are annotated in both the databases. This allows associating the manually-collected annotations to the computationally generated data in AltSplice. Related entries among AEdb-Sequence, -Function and -Minigenes are identified and are annotated. Table 1 shows that from the 1700 AEdb-Sequence entries, as much as 1200 are associated with AltSplice entries; and as much as 78 of 82 entries from AEdb-Minigenes data set are associated with AEdb-Sequence data set.
Integration with other resources
Ensembl (16) and UniProt (17) are among the most important resources on sequence data. Both include significant data relating to alternative splicing, e.g. Ensembl reports alternative transcripts while UniProt reports curated data on isoform peptide sequences. AltSplice uses Ensembl genes as the starting gene set for deriving splicing patterns. Therefore, the AltSplice data are intrinsically associated with Ensembl annotation of alternate transcripts and related information. Data on peptide variants collected in UniProt are integrated with AltSplice and they complement the set of AltSplice-derived peptide isoform data.
ACCESS TO DATABASES
ASD interfaces
ASD contains heterogeneous data sets—AltSplice is created through computational analysis of gene-transcript alignments, and AEdb is created by manual curation of literature data. Furthermore, there are differences in the extent of annotation and in the adopted vocabularies. We therefore generated interfaces that are flexible enough to handle this heterogeneity and that allow an easy retrieval of information. Different layers of interfaces are available and they provide either a single-point access to all the data sets or advanced searches of individual data sets.
Single-query bar and wrapper interfaces
Both these interfaces provide a quick access to all of ASD data. The single-query bar accepts commonly-used search terms, such as keywords, gene symbols (or their synonyms) and database cross-references. The wrapper interface queries all of ASD data against a given search term; these terms include the above-mentioned commonly used terms and splicing event type. Queries can be selectively restricted to specific sets of gene entries, such as set of human-mouse orthologous gene pairs or set of gene entries for which data on isoform peptide sequences is available or integrated set of AltSplice-AEdb entries.
Advanced search query interfaces
The individual data sets differ in terms of the type of data and annotation—e.g. AltSplice reports splicing events, splicing patterns and introns/exons while AEdb-Motif reports splicing regulatory sequences. Thus specialized query interfaces have been built for individual data sets.
Interface for AltSplice
Genes can be queried by chromosomal location, gene names and synonyms, protein keywords and database cross-references [such as EMBL (18) and UniProt accession nos (17), HUGO gene symbols (19), Gene Ontology identifiers (20) and protein identifiers], types of splicing events and allele specificity at SNP positions. Browsers allowing selection of eVOC standard vocabularies for EST library annotation (21) and GO classifiers facilitate querying through expression states and through protein function/process/location. Queries can be selectively restricted to specific sets of gene entries, such as to the set of human-mouse orthologous gene pairs, or to the set of gene entries for which data on isoform peptide sequences is available, or to the integrated set of AltSplice-AEdb entries. A particularly useful query for experimentalists is ‘Library Subtraction Tool’, which let users retrieve gene entries with splicing patterns that are differentially expressed in different cell states.
Interface for AEdb-Sequence
The data can be queried by gene names and synonyms, database cross-references, type of splicing events and type of regulatory roles, such as introducing premature termination codons or frameshifts. Further, the data can be queried for disease association and developmental specificity.
Interface for AEdb-Function
The data can be queried by gene names, protein keywords and database cross-references. Further, queries based on the functional enumeration of the isoform peptide sequence can be raised by selecting from a predefined list of functional categories (for the list of functional categories see Table 1).
Interface for AEdb-Motif
The interface allows free-text search. The search items include gene names, sequence of the regulatory motifs and type of regulatory sequence (enhancer or silencer).
BLAST and FASTA searches to ASD
The nucleotide and peptide sequences from ASD can be searched through both BLAST (WU-BLAST2) (http://blast.wustl.edu) and FASTA utilities (22). The objective of these search programs is to identify sequence similarities between novel sequences and alternatively spliced sequences collected in ASD. BLAST reports regions of high similarity. FASTA can be very useful to identify long regions of low similarity between highly diverged sequences. Further, FASTA is helpful when the query sequence is short, since BLAST usually fails to report results for short query sequences.
One-stop query system to access publicly available databases on alternative splicing
Several databases on alternative splicing are publicly available. To facilitate extraction of all known information about splicing of a gene, we generated a single interface that queries various databases simultaneously. Presently, seven alternative splicing databases namely, ASD, ASG, PALS, SpliceInfo, MAASE and HASDB (23–27) are made available from this interface. The interface accepts typical search terms (such as keywords, gene names and cross-references) and queries all these databases. The results that are obtained from the individual database servers are presented with hyperlinks to the individual databases.
Example of data search and of data content
Figures 2 and 3 illustrate the results of searching the database for all entries of tra2-alpha and tra2-beta, two important splicing regulators. Querying the ASD using wrapper query interface for ‘tra2a* | tra2b*’ as keyword produces an output page (Figure 2) that lists entries from the different component data sets. As can be seen from the figure, related data entries across the different data sets are hyperlinked to one another. Figure 3a and b illustrate the presentation of some of the data items from AltSplice for tra2-beta; Figure 3a shows sections on gene information, on evidences for alternative splicing of tra2-beta, and on the observed splicing events in AltSplice. Figure 3b shows splicing patterns presented in textual form (as Splice Pattern Table) and in graphical form (as Splice Pattern view). Individual data items in the display page are hyperlinked to pages that list detailed information. For example, the splicing pattern entry number in the table is linked to a page that lets the user to perform multiple alignments on the sequences of all the observed isoform splicing patterns or peptides. Figure 3c shows the display page of tra2-beta entry from AEdb-Function data set; it illustrates the wealth of information that is captured from published literature.
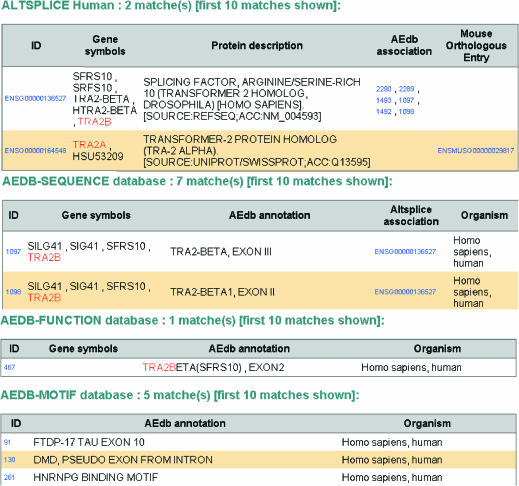
Figure 2.
Result page of query to all of ASD data. The ASD was queried using the wrapper interface with the term ‘tra2a* | tra2b*’ and this resulted in the retrieval of data entries from AltSplice-Human (two entries), AEdb-Sequence (seven entries), AEdb-Function (one entry) and AEdb-Motif (five entries). This figure illustrates the integration among the different data sets of ASD—(i) the AltSplice-Human entries are seen associated with entries from AEdb-Sequence, and from AltSplice-Mouse; (ii) the AEdb-Sequence entries are seen associated with entries from AltSplice-Human. These associations are hyperlinked.
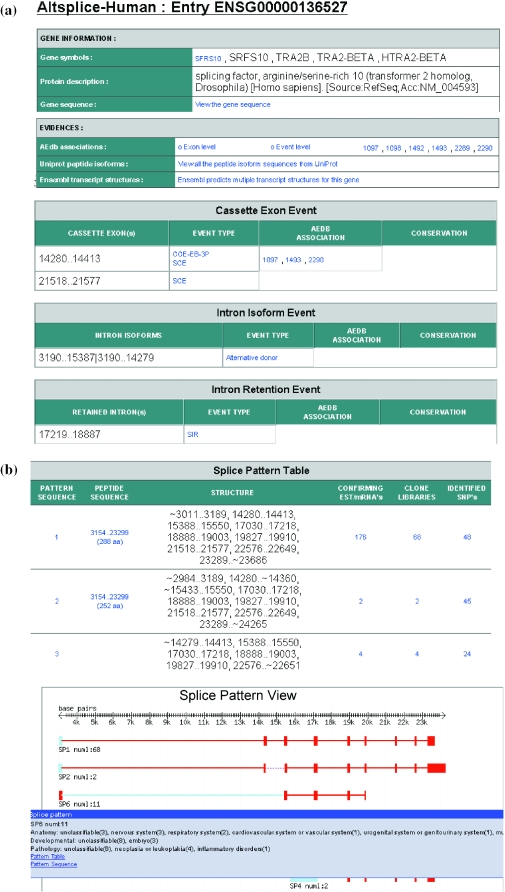
Figure 3.
Display of data on alternative splicing of human tra2-beta gene as seen in AltSplice and AEdb-Function data sets. (a) This figure presents a display of data sections on Gene Information, Evidences and Splicing events as seen in AltSplice. Gene information section provides hyperlinks to a page listing the gene entry from HUGO Gene Nomenclature database and to a page which lists the sequence of the gene. The evidences section provides hyperlinks to the associated entries from AEdb-Sequence, to pages that list variant peptide sequences for the gene from UniProt or to pages that list the Ensembl transcript sequences for the gene. The events section lists all the splicing events that AltSplice has identified for the gene. Column 1 lists the gene coordinates of alternatively spliced exons/introns. Column 2 indicates whether the event involves modifications in the flanking exons as well; entries are hyperlinked to pages listing detailed information on the event. Column 3 indicates the identifier of the associated entry from AEdb-Sequence (if any) and the entry is hyperlinked. Column 4 indicates the identifier of the orthologous gene (if any) and the coordinates of the exon orthologous to the one presented in column 1; the entry is hyperlinked to the orthologous gene entry. (b) This figure presents the textual and graphical display of observed splicing patterns for tra2-beta gene as seen in AltSplice data. Splice Pattern Table: Entry in column 1 is hyperlinked to a page listing the sequence of the splicing pattern. Entry in column 2 gives the coding start and end positions on the gene and the length of the translated peptide sequence and is hyperlinked to a page listing the peptide sequence. Entry in column 3 lists the structure of the splicing pattern as a string of exons (exon boundaries are presented in gene coordinates). Entry in column 4 is hyperlinked to pages listing detailed information on the confirming transcript sequences. Entry in column 5 is hyperlinked to pages listing expression states. Entry in column 6 is hyperlinked to pages listing allele specificity of the splicing pattern. Splice Pattern View: Exons are indicated by boxes and introns by lines. Exons/introns that are variants are indicated in blue color. Browsing the cursor over various elements of the pattern displays pop-up's giving detailed information on the elements. The displayed pop-up in this example shows information on the expression state of Splicing Pattern 6. (c) This figure presents data on the functional changes due to alternative splicing in tra2-beta as seen in AEdb-Function data set. The data are organised into three sections namely, gene information, bibliography information and functional information. This figure illustrates the wealth of knowledge captured from literature.
ASD WORKBENCH
The workbench provides a set of online tools that enable users to carry out analysis of pre-mRNA sequences. It includes tools for intron analysis, scoring ATG-context sequence, finding exons and identifying splicing regulatory sequences. These tools are accessed either through a single wrapper interface or through interfaces that are specialised for individual tools.
Intron analysis
The tool examines intron sequences (as provided by the user) for putative branch point (BP) sites and polypyrimidine tracts (PPT). It further calculates the strength of the donor and acceptor sites. The methods are described elsewhere (10). The user has a choice of weight matrices for donor and acceptor sites tailored for different intron types, such as U2-type GT-AG and GC-AG and U12-type GT-AG and AT-AC.
Scoring ATG-context sequence
This tool examines each occurrence of ATG in a given transcript sequence for its ability to act as translation start codon. Each ATG is scored for Kozak's ATG-context sequence (28) using a weight matrix that we built from experimentally confirmed translation initiation sites. The sequence of translated peptide from each occurrence of ATG is presented along with the ATG-context score. FASTA/BLAST searches against UniProt sequence data can be launched for each of the translated peptide sequence.
MZEF-SPC exon finder
This tool identifies potential exons in a given nucleotide sequence. It integrates Michael Zhang's Exon Finder (29) and Thanaraj's SpliceProximalCheck (30). MZEF identifies putative exons using quadratic discriminant analysis. SPC is a decision tree implementation of splicing signals that differentiate genuine human splicing sites from the proximal false sites and thus specialises in validating the predicted exon boundary for exactness.
Detection of short regulatory sequences
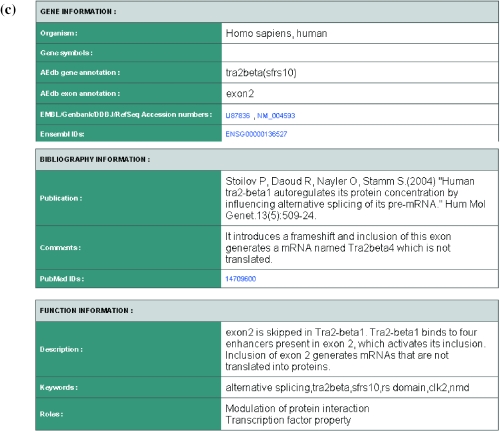
Exons are regulated by short, degenerative sequences that bind to interacting splicing factors and proteins. These sequences are collected in the AEdb-Motif database. The Regulatory Sequence tool uses these motifs to examine a given nucleotide sequence for their presence. Users have a choice to specify the extent of allowed mismatches. The identified motifs are hyperlinked to the corresponding entries in AEdb-Motif database (See Figure 4 for illustration of identified motifs in tra2-beta gene). The splicing rainbow is a visualizations tool that colour-codes presence of different regulatory motifs in a user-supplied sequence.
Figure 4.
Example output from workbench tool that detects splicing regulatory sequences. The figure displays a portion of the page that reports splicing regulatory sequences in tra2-beta gene. The names of identified motifs are hyperlinked to appropriate entries in AEdb-Motif database. Matches against tra2-beta regulatory sequences are also seen. It is known that tra2-beta1 auto regulates its protein concentration by influencing alternative splicing of its pre-mRNA (31).
SUMMARY OF UPDATES AND ENHANCEMENTS
The current release of ASD includes a large number of improvements over that reported earlier (9). AltSplice (the production pipeline) supersedes AltExtron (the research and development pipeline) in functionalities, data content, integrations and data presentations. As a result, AltExtron has become redundant and is not maintained any further; however for archival purposes, the earlier versions of AltExtron data are still presented in ASD web pages. Examples of enhancements in AltSplice data content include data on evidences for alternative splicing and isoform peptide sequences; those in integrations include related data from UniProt and Ensembl; those in query tools include differential library expression profiler; and those in presentation of data include a complete redesign of both the query and data (textual and graphical) results pages and multiple alignment view of isoform splice pattern and peptide sequences. In addition to the AEdb-Sequence data set presented in (9), further AEdb data sets (namely AEdb-Function, AEdb-Motif and AEdb-Minigenes) are presented. The ASD workbench is a new addition that complements the data content. The ASD sequence data are now available for search through BLAST and FASA tools. The current version of ASD provides three levels of search facilities, namely single-query bar, wrapper and data set specific advanced search query page. Further, a one-stop query system that enables users to query many publicly-available databases (including all the data sets of ASD) on alternative splicing is now provided. Integrations with splice variant data from UniProt and Ensembl enhance the value of ASD resources; AltSplice data are presented on Ensembl genome annotation browsers as DAS (Distributed Annotation Server) tracks. Since the last report, the ASD pipeline has matured to high production standards. The pipeline and the database are now handled by EBI database-production team which is committed to making regular data releases, to expanding the repertoire of organisms for which the data are made available, and to make seamless integration and hyperlinks both among the different data sets of ASD and to various other external databases.
CONCLUSIONS
We present here ASD, a bioinformatics resource for alternative splicing. The individual components of the resource are (i) AltSplice—a value-added data set generated by our AltSplice, (ii) AEdb—a manually curated data set of alternatively spliced exons and their properties, and (iii) ASD Workbench—a collection of tools that carry our various analyses on pre-mRNA sequences. These individual components are integrated to one another and to related data from UniProt and Ensembl. The resource also provides a one-stop query system that accesses various other publicly available databases on alternative splicing. The integrated resource is available to the community (from http://www.ebi.ac.uk/asd) through user-friendly interfaces. The future releases will contain data that are being generated by other members of the splicing community, e.g. array expression data on alternative splicing of genes.
General enquiries on the ASD database can be mailed at asd-ebi@ebi.ac.uk.
Acknowledgments
T.A.T. and S.S. thank European Commission for the ASD grant (QLRT-CT-2001-02062). Involvement of Francis Clark and Juha Muilu in the earlier stages of the project is acknowledged. NLB-M acknowledges Portugal Foundation for Science and Technology for financial support (Fellowship SFRH/BD/2914/2000). Juan Valcarcel is acknowledged for mentoring NLB-M in developing the Splicing Rainbow tool. Funding to pay the Open Access publication charges for this article was provided by the ASD grant from European Commission.
Conflict of interest statement. None declared.
REFERENCES
- 1.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mrna splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 2.Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T.A., Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 3.Lewis B.P., Green R.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith C.W., Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 5.Caceres J.F., Kornblihtt A.R. Alternative splicing regulation: multiple control mechanisms and involvement in human diseases. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 6.Stoilov P., Meshorer E., Gencheva M., Glick D., Soreq H., Stamm S. Defects in pre-mRNA processing as causes and predisposition to diseases. DNA Cell Biol. 2002;21:803–818. doi: 10.1089/104454902320908450. [DOI] [PubMed] [Google Scholar]
- 7.Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 8.Thanaraj T.A., Stamm S. Prediction and statistical analysis of alternatively spliced exons. Prog. Mol. Subcell. Biol. 2003;31:1–31. doi: 10.1007/978-3-662-09728-1_1. [DOI] [PubMed] [Google Scholar]
- 9.Thanaraj T.A., Stamm S., Clark F., Riethoven J.J., Le Texier V., Muilu J. ASD: the Alternative Splicing Database. Nucleic Acids Res. 2004;32:D64–D69. doi: 10.1093/nar/gkh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark F., Thanaraj T.A. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum. Mol. Genet. 2002;11:1–14. 451–464. doi: 10.1093/hmg/11.4.451. [DOI] [PubMed] [Google Scholar]
- 11.Ladd A.N., Cooper T.A. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 2002;3:8.1–8.16. doi: 10.1186/gb-2002-3-11-reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Z.M. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 2004;11:278–294. doi: 10.1159/000077096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourgeois C.F., Lejeune F., Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:37–88. doi: 10.1016/S0079-6603(04)78002-2. [DOI] [PubMed] [Google Scholar]
- 14.Tang Y., Novoyatleva T., Benderska N., Kishore S., Thanaraj T.A., Stamm S. Analysis of alternative splicing in vivo using minigenes. In: Westhof E., Bindereif A., Schön A., Hartmann K., editors. Handbook of RNA Biochemistry. Vol. 2. Verlag, Weinheim: Wiley-VCH; 2005. pp. 755–782. [Google Scholar]
- 15.Stoss O., Stoilov P., Hartmann A.M., Nayler O., Stamm S. The in-vivo minigene approach to analyse tissue-specific splicing. Brain Res. Protoc. 1999;4:383–394. doi: 10.1016/s1385-299x(99)00043-4. [DOI] [PubMed] [Google Scholar]
- 16.Hubbard T., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The universal protein resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanz C., Aldebert P., Althorpe N., Baker W., Baldwin A., Bates K., Browne P., van den Broek A., Castro M., Cochrane G., et al. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2005;33:D29–D33. doi: 10.1093/nar/gki098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wain H.M., Lush M., Ducluzeau F., Povey S. Genew: the human gene nomenclature database. Nucleic Acids Res. 2002;30:169–171. doi: 10.1093/nar/30.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburner M., Ball C.A., Blake J.A., Butler H., Cherry J.M., Corradi J., Dolinski K., Janan T., Eppig J.T., Harris M., et al. Creating the Gene Ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelso J., Visagie J., Theiler G., Christoffels A., Bardien-Kruger S., Smedley D., Otgaar D., Greyling G., Jongeneel V., McCarthy M.I., et al. eVOC: a controlled vocabulary for gene expression data. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leipzig J., Pevzner P., Heber S. The alternative splicing gallery (ASG): bridging the gap between genome and transcriptome. Nucleic Acids Res. 2004;32:3977–3983. doi: 10.1093/nar/gkh731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y.H., Chen Y.T., Lai J.J., Yang S.T., Yang U.C. PALS db: putative alternative splicing database. Nucleic Acids Res. 2002;30:186–190. doi: 10.1093/nar/30.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang H.D., Horng J.T., Lin F.M., Chang Y.C., Huang C.C. SpliceInfo: an information repository for mRNA alternative splicing in human genome. Nucleic Acids Res. 2005;33:D80–85. doi: 10.1093/nar/gki129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng C.L., Nair T.M., Gribskov M., Kwon Y.S., Li H.R., Fu X.D. A database designed to computationally aid an experimental approach to alternative splicing. Pac. Symp. Biocomput. 2004:78–88. doi: 10.1142/9789812704856_0008. [DOI] [PubMed] [Google Scholar]
- 27.Modrek B., Resch A., Grasso C., Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M.Q. Using MZEF to find internal coding exons. In: Baxevanis A.D., Davison D.B., editors. Current Protocols in Bioinformatics. Vol. 1. New York: John Wiley & Sons, Inc.; 2003. pp. 4.2.1–18. [DOI] [PubMed] [Google Scholar]
- 30.Thanaraj T.A., Robinson A. Prediction of exact boundaries of exons. Brief Bioinform. 2000;1:343–356. doi: 10.1093/bib/1.4.343. [DOI] [PubMed] [Google Scholar]
- 31.Stoilov P., Daoud R., Nayler O., Stamm S. Human tra2-beta1 autoregulates its protein concentration by influencing alternative splicing of its pre-mRNA. Hum. Mol. Genet. 2004;13:509–524. doi: 10.1093/hmg/ddh051. [DOI] [PubMed] [Google Scholar]