Abstract
Cap-analysis gene expression (CAGE) Basic and Analysis Databases store an original resource produced by CAGE, which measures expression levels of transcription starting sites by sequencing large amounts of transcript 5′ ends, termed CAGE tags. Millions of human and mouse high-quality CAGE tags derived from different conditions in >20 tissues consisting of >250 RNA samples are essential for identification of novel promoters and promoter characterization in the aspect of expression profile. CAGE Basic Database is a primary database of the CAGE resource, RNA samples, CAGE libraries, CAGE clone and tag sequences and so on. CAGE Analysis Database stores promoter related information, such as counts of related transcripts, CpG islands and conserved genome region. It also provides expression profiles at base pair and promoter levels. Both databases are based on the same framework, CAGE tag starting sites, tag clusters for defining promoters and transcriptional units (TUs). Their associations and TU attributes are available to find promoters of interest. These databases were provided for Functional Annotation Of Mouse 3 (FANTOM3), an international collaboration research project focusing on expanding the transcriptome and subsequent analyses. Now access is free for all users through the World Wide Web at http://fantom3.gsc.riken.jp/.
INTRODUCTION
Cap-analysis gene expression (CAGE) is a high-throughput method to measure expression levels by counting large amounts of sequenced capped 5′ ends of transcripts, termed CAGE tags (1). A similar approach is proposed as 5′ end SAGE (2). The average length of these 5′ end tags of transcripts is 20 bp and the tags are aligned with the genome directly, although original SAGE (3) tags are aligned with 3′ ends of transcripts (4). CAGE tags are an essential resource for profiling transcriptional starting sites and can be used for profiling gene expressions by counting CAGE tags associated with genes. Millions of mouse and human high-quality CAGE tags derived from different conditions in >20 tissues consisting of >250 RNA samples are subjected for analysis in the international collaboration research project, Functional Annotation Of Mouse 3 (FANTOM3). The CAGE tags are used for the analysis of the transcriptional landscape of mammalian genome (5), antisense transcription in the mammalian transcriptome (6), comprehensive promoter analysis (P.Carninci, A.Sandelin, B.Lenhard, S.Katayama, K.Shimokawa, J.Ponjavic, C.A.Semple, M.S.Taylor, P.Engstrom, M.C.Frith, A.R.Forrest, W.B.Alkema, S.L.Tan, C.Plessy, R.Kodzius, T.Ravasi, T.Kasukawa, S.Fukuda, M.Kanamori-Katayama, Y.Kitazume, H.Kawaji, C.Kai, H.Konno, K.Nakano, S.Mottagui-Tabar, P.Arner, A.Chesi, S.Gustincich, F.Persichetti, H.Suzuki, S.M.Grimmond, C.Wells, V.Orlando, C.Wahlestedt, E.T.Liu, M.Harbers, J.Kawai, V.B.Bajic, D.A.Hume and Y.Hayashizaki, manuscript submitted) and subsequent analyses.
We constructed two database systems to utilize the CAGE resource, CAGE Basic and Analysis Databases. Their aims are (i) to manage and trace the CAGE data consistently and (ii) to demonstrate the promoter usage (using CAGE and other data). The former is required to support the novel experimental processes of CAGE and to manage the large amount of RNA samples provided in the FANTOM3 collaboration. The latter is to support subsequent analyses using all of the required data, without influence of our management of the CAGE data. Additionally, we constructed CAGE Expression 3D Viewer for novel type of expression view (K.Shimokawa, Y.Okamura-Oho, C.Kai, P.Carninci and Y.Hayashizaki, manuscript in preparation). The database systems described here were used in FANTOM3 and are now publicly accessible. Here, we present the systems' overview and functions to facilitate the use of the CAGE resource.
DATA BASIS
A consistent and comprehensive dataset is crucial to allow biological analyses in different kinds of viewpoints. Our two database systems are built on the same basis: CAGE tag starting site (CTSS), tag cluster (TC) and transcriptional unit (TU).
CTSS is a nucleotide position on the genome from which an alignment of CAGE tag starts. Counts of CAGE tags sharing the same starting sites represent expression profiles in base pairs level. TC is an operationally defined unit to characterize promoters. It is constructed by clustering 5′ end overlapped region of transcripts (P.Carninci, A.Sandelin, B.Lenhard, S.Katayama, K.Shimokawa, J.Ponjavic, C.A.Semple, M.S.Taylor, P.Engstrom, M.C.Frith, A.R.Forrest, W.B.Alkema, S.L.Tan, C.Plessy, R.Kodzius, T.Ravasi, T.Kasukawa, S.Fukuda, M.Kanamori-Katayama, Y.Kitazume, H.Kawaji, C.Kai, H.Konno, K.Nakano, S.Mottagui-Tabar, P.Arner, A.Chesi, S.Gustincich, F.Persichetti, H.Suzuki, S.M.Grimmond, C.Wells, V.Orlando, C.Wahlestedt, E.T.Liu, M.Harbers, J.Kawai, V.B.Bajic, D.A.Hume and Y.Hayashizaki, manuscript submitted), such as 5′ end 20 bp long of RIKEN full-length cDNA and RIKEN-5′-expressed sequence tag (EST), 5′ end tags of GIS (7) and GSC (4) ditags, DBTSS (8), 5′ end SAGE and CAGE. Of these, overlapping sequences on the genome with at least 1 bp are clustered, and define a TC. Counts of CAGE tags within TCs represent expression profiles on promoter level. TU is also an operationally defined unit proposed in FANTOM2 (9), defined as a region or a set of discontinuous regions in the genome from where all exons of a mature full-length mRNA are derived (10). Counts of CAGE tags within TUs represent expression profiles on gene level. TUs are associated with Entrez Gene (11) and gene ontology term (12) by means of transcripts belonging to them, if possible. CTSS are associated with TCs, and TCs are associated with TUs. Users can access the CAGE resource of interest by searching TUs with their own keywords.
SYSTEM OVERVIEW
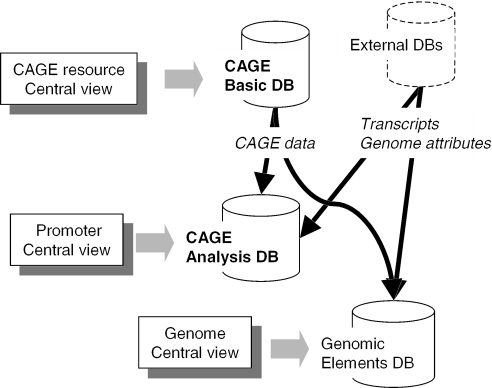
Figure 1 is an overview of the CAGE database systems. CAGE Basic Database is a primary database of the CAGE resource, and provides a central view of CAGE resources. CAGE Analysis Database stores TC related information, and provides a central view of promoters. As a complementary system, Genomic Elements Database is constructed to provide a central view of genome positions. Their main contents are described in Table 1. CAGE Analysis Database would be the most convenient gateway for users, especially new to the CAGE data. Hyperlinks from the database to the others are available depending on their interests, CAGE Basic Database for CAGE sequences themselves and Genomic Elements Database for a conventional genome view.
Figure 1.
An overview of the CAGE supporting systems and data flow among them.
Table 1.
Main contents of the database systems
| Database | Contents |
|---|---|
| CAGE Basic Database | RNA sample information |
| CAGE library information | |
| CAGE clone plate/spot | |
| CAGE clone sequence | |
| CAGE clone sequence quality | |
| CAGE tag sequence | |
| CAGE tag mapping status | |
| Associations of CAGE tags with CTSS | |
| Associations of CTSS with TCs | |
| Associations of TCs with transcript and TUs | |
| CAGE Analysis Database | Base pair level expression profile |
| TC expression profile within TUs | |
| Statistical significance expression fluctuations | |
| Presence of predicted core promoter elementsa in upstream region | |
| Presence of conserved genome region between human and mouse (axtNet) | |
| Presence of CpG islands | |
| Counts of TC related mRNA, 5′-EST, GIS/GSC ditags | |
| Genomic Elements Database | TC |
| Predicted core promoter elementsa | |
| mRNA | |
| GIS/GSC ditag | |
| 5′- and 3′-ESTs | |
| Candidates of imprited transcripts in EICO DB | |
| Transcription factors listed in TFdb | |
| Gene predictionb | |
| CpG islandsb | |
| Repeat detected by repeatmasker and tandem repeats finderb | |
| Assemble gapb | |
| Conserved genome region between human and mouse (axtNet)b |
aTATA box, CCAAT box, GC box and initiator.
bRetrieved from the UCSC Genome Browser Database.
CAGE BASIC DATABASE
In the CAGE protocol, 5′ ends of full-length cDNA synthesized from RNA samples are cleaved with MmeI, a class II restriction enzyme, which cleaves 20/18 bp outside the recognition sequence. The cleaved 5′ end cDNA tags (CAGE tags) are ligated to form concatemers and cloned as CAGE clones in CAGE library. After sequencing the CAGE clone, CAGE tag sequences are extracted and mapped computationally onto the genome.
CAGE clone sequence, CAGE tag location on the clone and its genome mapping information are stored to facilitate their traceability. To manage a broad range of RNA samples provided in the FANTOM3 collaboration, RNA sample ID, tissue name, developmental stage, sample treatment, cell type and collaboration name are stored. The amount of the CAGE data derived from each RNA sample is presented to examine if targeted samples are analyzed with CAGE and to which extent CAGE tags in the samples were sequenced.
CAGE ANALYSIS DATABASE
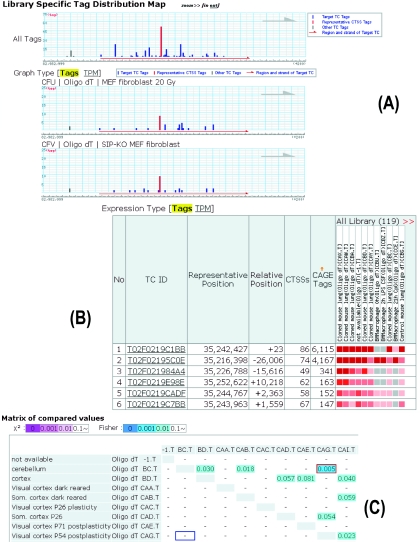
Expression levels are measured by counting associated CAGE tags, and they can be used to measure different levels of expression profiles from base pair to chromosomal band level. Two levels of expression profiles are presented in the CAGE Analysis Database for each RNA sample: base pair scale expressions inside a TC are displayed in histogram (Figure 2A), and TC expressions within a TU are represented by a heat map like representation (Figure 2B). CAGE tag counts and transcripts per million, (tag counts)/(total mapped tag counts in the sample) × 1 000 000, are used as units of expression level. Additionally, statistical significances of expression fluctuations between RNA samples are also accessible in a matrix (Figure 2C). They provide users with graphical views of transcriptional start variation, promoter variation and expression fluctuation of promoters.
Figure 2.
Screenshots of CAGE Analysis Database: (A) a view of base pair scale expression within a TC, where CAGE tag count of each genome position is displayed in histogram, (B) a view of TC expressions within a TU, in which expression levels are represented by a heat map like representation, (C) a view of statistical significances of expression fluctuations between RNA samples, and their E-values are displayed in a matrix.
Rarely expressed promoters contain only a few tags. Although our RACE experiment using an oligo-capping method supported 91% of the tested cases (5), some CAGE tags could be artifacts caused by some errors in library preparation, sequencing and genome mapping. To provide some evidences for promoters, associations of TCs with (genome) conserved regions (13), CpG islands (14), predicted core promoter elements (15–17) and different transcript counts are stored. Users can search TCs with different reliability levels by specifying search conditions.
GENOMIC ELEMENTS DATABASE
Genomic Elements Database is a supplementary database to the two CAGE databases. The aim is to integrate TCs and other data onto the genome and display them in a conventional way. Generic Genome Browser (18) with MySQL DBMS is used to present a genome view. Candidates of imprinted transcripts in EICO DB (19,20), transcription factors in TFdb (21) and other data in the UCSC Genome Browser Database (22) are stored in addition to the utilized data above. This system is also utilized in full-length cDNA annotation in FANTOM3 (5).
CONCLUSION
The CAGE database systems have successfully provided a large amount of mouse and human CAGE tags derived from various RNA samples for the FANTOM3 project, resulting in biological analyses in various viewpoints. The systems have supported these analyses by providing central views of CAGE resource, promoter and genome position depending on the aspects of interests to researchers. They are publicly available now, and are expected to promote subsequent analyses by using the CAGE resource in scientific research community.
AVAILABILITY
The database systems described here are hyperlinked from http://fantom3.gsc.riken.jp/. Their user's guide, glossary and/or database schema are available from their help pages, and their raw data files, table definitions in SQL and tab-delimited data files, are also available for download from http://fantom3.gsc.riken.jp/download.html.
Acknowledgments
We would like to thank A. Hasegawa for data processing; A. Karlsson for English editing; K. Nakano and H. Murakami for building computational systems; K. Yoshida for support; and all members of the FANTOM consortium. This study was supported by Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government to Y.H., a grant of the Genome Network Project from the Ministry of Education, Culture, Sports, Science and Technology, Japan. to Y.H., Grant for the Strategic Programs for R&D of RIKEN to Y.H. Funding to pay the Open Access publication charges for this article was provided by RIKEN.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl Acad. Sci. USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hashimoto S., Suzuki Y., Kasai Y., Morohoshi K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K. 5′-end SAGE for the analysis of transcriptional start sites. Nat. Biotechnol. 2004;22:1146–1149. doi: 10.1038/nbt998. [DOI] [PubMed] [Google Scholar]
- 3.Velculescu V.E., Zhang L., Vogelstein B., Kinzler K.W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 4.Harbers M., Carninci P. Tag-based approaches for transcriptome research and genome annotation. Nature Methods. 2005;2:495–502. doi: 10.1038/nmeth768. [DOI] [PubMed] [Google Scholar]
- 5.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 6.Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 7.Ng P., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., et al. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nature Methods. 2005;2:105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki Y., Yamashita R., Sugano S., Nakai K. DBTSS, DataBase of Transcriptional Start Sites: progress report 2004. Nucleic Acids Res. 2004;32:D78–D81. doi: 10.1093/nar/gkh076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60 770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 10.Kasukawa T., Katayama S., Kawaji H., Suzuki H., Hume D.A., Hayashizaki Y. Construction of representative transcript and protein sets of human, mouse, and rat as a platform for their transcriptome and proteome analysis. Genomics. 2004;84:913–921. doi: 10.1016/j.ygeno.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 15.Smale S.T., Kadonaga J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 16.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 17.Schmid C.D., Praz V., Delorenzi M., Perier R., Bucher P. The Eukaryotic Promoter Database EPD: the impact of in silico primer extension. Nucleic Acids Res. 2004;32:D82–D85. doi: 10.1093/nar/gkh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikaido I., Saito C., Wakamoto A., Tomaru Y., Arakawa T., Hayashizaki Y., Okazaki Y. EICO (Expression-based Imprint Candidate Organizer): finding disease-related imprinted genes. Nucleic Acids Res. 2004;32:D548–D551. doi: 10.1093/nar/gkh093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikaido I., Saito C., Mizuno Y., Meguro M., Bono H., Kadomura M., Kono T., Morris G.A., Lyons P.A., Oshimura M., et al. Discovery of imprinted transcripts in te mouse transcriptome using large-scale expression profiling. Genome Res. 2003;13:1402–1409. doi: 10.1101/gr.1055303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanamori M., Konno H., Osato N., Kawai J., Hayashizaki Y., Suzuki H. A genome-wide and nonredundant mouse transcription factor database. Biochem. Biophys. Res. Commun. 2004;322:787–793. doi: 10.1016/j.bbrc.2004.07.179. [DOI] [PubMed] [Google Scholar]
- 22.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]