Abstract
AffinDB is a database of affinity data for structurally resolved protein–ligand complexes from the Protein Data Bank (PDB). It is freely accessible at http://www.agklebe.de/affinity. Affinity data are collected from the scientific literature, both from primary sources describing the original experimental work of affinity determination and from secondary references which report affinity values determined by others. AffinDB currently contains over 730 affinity entries covering more than 450 different protein–ligand complexes. Besides the affinity value, PDB summary information and additional data are provided, including the experimental conditions of the affinity measurement (if available in the corresponding reference); 2D drawing, SMILES code and molecular weight of the ligand; links to other databases, and bibliographic information. AffinDB can be queried by PDB code or by any combination of affinity range, temperature and pH value of the measurement, ligand molecular weight, and publication data (author, journal and year). Search results can be saved as tabular reports in text files. The database is supposed to be a valuable resource for researchers interested in biomolecular recognition and the development of tools for correlating structural data with affinities, as needed, for example, in structure-based drug design.
INTRODUCTION
Understanding the energetics of biomolecular recognition is of paramount importance for a large variety of biomedical and biotechnological disciplines. One of the most prominent examples is given by structure-based drug design where the 3D structure of a target macromolecule (most frequently a protein) is used to identify, design or optimize small-molecule ligands, which bind tightly to the target. Obviously, such design can only be successful if the structural requirements for energetically favorable interactions and high-affinity binding are known. Much of the current knowledge has been gained from comparative analyses of different complex structures and their affinities (1,2). These analyses, however, were normally restricted to rather small sets of data, and the understanding of protein–ligand recognition is still far from being complete, as illustrated by the recurring surprises during projects of molecular design (3–6). Clearly, more data are instrumental to increase the knowledge about protein–ligand interactions and to improve not only the qualitative understanding but also the quantitative tools for estimating affinities from complex structures, such as empirical, regression-based scoring functions (7–12).
Structural data of protein–ligand complexes are available to a large and rapidly increasing extent through the Protein Data Bank (PDB) (13). This database, however, is a general resource for biomacromolecular structures that had not particularly been designed for protein–ligand complexes. Accordingly, secondary databases, such as Relibase (14,15) and PDBsum (16,17), have been developed which provide more convenient access to specific information about protein–ligand complexes (e.g. search functions for ligand structures and analysis tools for interaction patterns in Relibase). Unfortunately, neither the secondary structural databases nor the PDB contain any information about the binding energetics of the corresponding complex, since this information is not required to be included upon submission of structural coordinates to the PDB. However, some databases exist that collect binding data for enzymes, receptors or protein–ligand complexes in general, such as BindingDB (18) and KiBank (19), but these, in turn, are not limited to complexes with available structure and do not provide a direct link to the available 3D structure of a given complex with measured affinity.
Given the obvious need for databases that establish the missing link between structural information from the PDB and the rather sparse and widely distributed affinity data, we started to develop AffinDB, a database of affinity values collected from the scientific literature for protein–ligand complexes of known structure. Originally intended as a simple tabular collection of affinity values related to PDB codes for in-house use only, the project has grown over time, both with respect to data content and database management, such that it has ultimately been made available to the public as a potentially valuable new resource, despite the recent appearance of other databases of similar scope, most notably PDBbind (20,21) and Binding MOAD (22). In the following, we briefly describe the database architecture and content of AffinDB, as well as the data collection procedure, give a succinct introduction to possibilities for accessing the data through the user interface, and discuss differences and similarities to other databases.
METHODS
Database architecture
AffinDB is based on a MySQL (4.0.24) backend machine. The web interface is written in PHP (4.3.10). The database is designed to provide supplementary information for PDB structures of protein–ligand complexes. Accordingly, AffinDB is structured by PDB code, which is the primary reference for all the data. Basic PDB meta-information about the protein is available for every PDB structure, basic ligand information is provided for ligands with more than five non-hydrogen atoms. Ligand entries of complexes are stored only once in AffinDB, i.e. in case of multiple occurrences of the same ligand in different structures, a pointer to the reference ligand molecule is used. Affinity data and related information are always associated with a specific ligand of a specific PDB structure.
Data collection and database content
The database core is constituted by basic meta-information about all PDB structures. To obtain these data, a helper-application was generated with a Python-based Relibase toolkit and the data were retrieved from Relibase+ (14,15). A further preprocessing step served to store only ligands with more than five non-hydrogen atoms in AffinDB, using a unique and consistently created name for these molecules. The PDB meta-information provided for every entry includes the name of the protein or protein class (as given in the header information of the PDB file), the EC number (for enzymes), the protein source, the resolution of the crystal structure and the names of the authors who determined the structure. In addition, for each PDB code links to the following external databases were added: PDB (13), Relibase (14,15), MSD (23), SCOP (24) and PDBsum (16,17).
The ligand entries consist of the chemical name (as provided in the PDB file), the molecular weight and the SMILES code (25) as basic information. In addition, a 2D molecule drawing of the ligand structure is included. These drawings are generated for every unique ligand with Marvin (3.5.7) (http://www.chemaxon.com/marvin). Babel (1.6) (http://www.eyesopen.com/babel), Corina (3.1) (26), and in-house software was used to harmonize the format of the ligands in order to obtain best results from Marvin. This automated procedure provided correct drawings for most of the ligands. A small proportion which could either not be drawn by Marvin or gave distorted pictures had to be post-processed by hand. Titratable functional groups are always shown in their neutral state, independent of any actual protonation state.
The protein and ligand information described so far is shown by AffinDB regardless whether affinity data are already available for the PDB entry or not. The main purpose of AffinDB, however, is to provide affinity information. Affinity data are exclusively extracted from the scientific literature. Both ‘primary’ and ‘secondary’ references are taken into account. A primary reference is a paper describing the original work of the affinity measurement for the corresponding protein–ligand complex. A secondary reference, instead, is any other paper that reports an affinity value for a PDB complex; this may include publications with compilations of affinity data for the development of scoring functions or similar purposes. In a secondary paper, the affinity value for a PDB complex is often only cited, without specifying further experimental details. So far, more than 740 affinity values covering over 470 PDB complexes could be collected and stored in AffinDB (cf. Discussion).
The input of affinity data into AffinDB is implemented in the form of a wizard. After entering the desired PDB code, AffinDB provides a list of all the ligands of the PDB entry in combination with a 2D molecule drawing and a hint whether affinity information is already stored for the given ligand. After choosing the desired ligand, the user is requested to enter the affinity information. Upon submission of the data, simple checks of the data integrity are performed and the entry is flagged for review by the database curators. Only after a database curator has checked these data, they are released for public access in AffinDB.
The binding affinity is thermodynamically quantified as a free energy of binding ΔGbind or as equilibrium constant (for association: Ka; for dissociation: Kd) for the reversible equilibrium reaction between protein P and ligand L to form the protein–ligand complex PL: P + L ↔ PL. ΔGbind and the equilibrium constants are related by the equation: ΔGbind = −RT ln Ka = RT ln Kd, where T is the temperature (in Kelvin) and R is the ideal gas constant (8.314 Jmol−1 K−1). For enzyme inhibitors, affinities are more frequently quantified in terms of parameters derived from kinetic assays. This may either be the inhibition constant Ki (which to a first approximation may be considered as a Kd for the enzyme–inhibitor complex, thus ΔGbind = RT ln Ki) or the IC50 value, which is the inhibitor concentration leading to 50% inhibition of the enzymatic activity. In AffinDB, the affinity value is stored in the same form as published in the specified reference, i.e. without any conversion of type or unit. If available, experimental uncertainties or error margins are saved as well.
Along with the affinity value itself, also the experimental method and conditions of the affinity measurement are stored in AffinDB, if specified in the corresponding reference. This information is provided to the user through a separate ‘affinity information window’ (Figure 1), which can be opened by activating the ‘Details’ link next to the affinity entry in the main window (Figure 2). The method by which the affinity was determined is characterized by a keyword or a brief statement. Temperature and pH value at which the measurements were carried out are stored separately. In addition, the buffer and any other significant reagents or additives present in the solution are reported. For the literature reference itself, the name of the first author, the title of the journal, as well as volume, year and first page of the publication are stored. The reference is linked to the corresponding PubMed entry, which provides direct access to the abstract. A flag indicates whether the reference is of primary or secondary type. Finally, comments and additional valuable information regarding the method, the reference, the structure or the affinity value itself are saved in a separate data field.
Figure 1.

Affinity information window for one of the affinity entries for PDB complex 1FLR.
Figure 2.
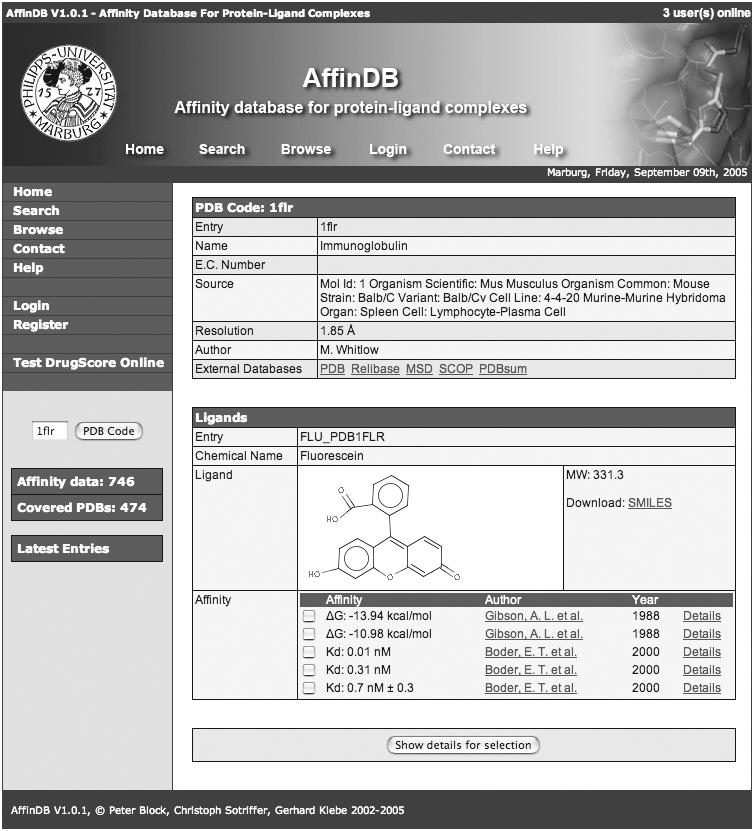
Main window showing a PDB entry with affinity data in AffinDB. PDB complex 1FLR is used as an example. Five different affinity values measured in different studies and under different conditions are available for this complex. The left navigation bar provides fast access to all functionalities of AffinDB.
Database access
The database is freely accessible at http://www.agklebe.de/affinity. Data can be retrieved via the PDB code, by defining specific search queries using the affinity search form, or simply by browsing.
Upon specifying a PDB code in the data entry field on the left navigation bar of the main window (cf. Figure 2), the summary information for the PDB entry is shown. If an affinity value for the ligand is available in the database, it is displayed below the ligand structure, along with the first author and the year of the publication which reports this value. If additional affinity values are available from other references, these are displayed as well, each in a separate line (cf. Figure 2). Further details can be requested for each affinity entry. Searching for a specific PDB entry with the affinity search form (accessible through the ‘Search’ link in the left navigation bar) yields only a result if an affinity value is already associated with the corresponding PDB entry.
In the affinity search form, a variety of queries for affinity data and related information can be defined. It is possible to search for affinities of a certain range of magnitude and for measurements carried out at a specific temperature and/or pH range. Affinities for certain enzyme classes or PDB codes may be retrieved, as well as affinities for ligands of a certain molecular weight range. Also the affinity values published by a certain author or within a specified time frame can be requested, and the retrieved affinity values may be limited to those obtained from primary literature sources.
AffinDB generates tabular reports for displaying affinity search results and for browsing through the database (Figure 3). The format of the tables consists of six columns providing the drawing of the ligand structure; the PDB code (linked to the summary information for the PDB entry), the affinity value in the originally reported form as well as converted to the negative base-10 logarithm [i.e. as pKi, pKd, pIC50 (relative to the standard concentration of 1 mol/l)]; the pH value of the measurement, the first author of the publication (with a link to the PubMed entry); and the year of the publication. Tables reporting search results can be saved as ‘csv’ file, which is an ASCII file with semicolon-separated columns and one affinity entry per line.
Figure 3.
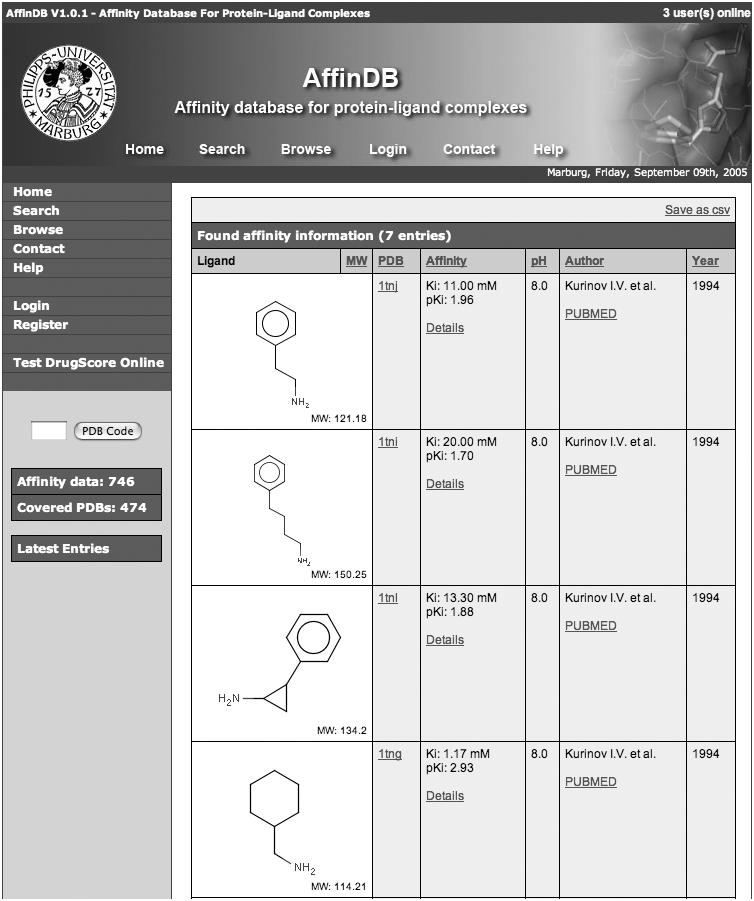
Tabular report, showing part of the search results produced by a query for affinity data published by a specific author (‘Kurinov’) in a primary reference.
DISCUSSION
AffinDB has been designed to provide fast and easy access to affinity data. The popular MySQL backend was chosen as database machine, since MySQL offers a speed-optimized SQL engine. Using the scripting language PHP, special care was taken to generate a clearly structured layout which enables fast and easy navigation. Since all the data can be accessed and retrieved directly via the web browser, the user does not have to install any special software to work with AffinDB.
Using current PC hardware (CPU: Athlon XP 2400+), AffinDB executes search queries in <0.1 s, fairly independent of the complexity of the query. The representation of the tabular report including the 2D molecule drawings needs between 0.1 and 5 s, depending on the number of hits (5 s if all entries are retrieved). The representation of a PDB entry takes up to 0.2 s, depending on the number of affinity data available for that entry. These values reflect only a server-side benchmarking. Obviously, the real speed also depends on the client-side hardware, the Internet connection and the browser.
Data collection for AffinDB is a very time-consuming process which can hardly be automated since scientifically educated readers are required to critically extract the relevant data from the scientific literature. In contrast to other databases (cf. below), we decided to include all affinity data found during literature research for a given PDB complex. Multiple affinity entries may, thus, be available for certain structures. These may reflect measurements with different methods or under different experimental conditions (e.g. PDB 1FLR; cf. Figure 1), or it may be due to additional reports from secondary references, which allows the user to trace back in which context the corresponding complex and its affinity have already been used. Only purely redundant data are not included (e.g. if the value is reported in the same paper as Kd and ΔG derived thereof).
The current coverage of more than 470 PDB structures derives from a priority selection made upon constructing the database. The initial basis was formed by compilations of affinity values from secondary references concerning empirical scoring functions. Owing to discrepancies among some of the values and to obtain more detailed information, primary references were also retrieved for part of this initial set. Subsequently, the database was augmented by seeking affinity data for PDB complexes of different datasets, such as a docking test set of validated structures (27) or datasets for certain target classes (e.g. carbonic anhydrases and trypsin-like serine proteases). Furthermore, published data from our own laboratory were also directly included.
AffinDB is a valuable resource for anyone interested in correlating structural data with binding energetics and complements other databases of similar subject, specifically the Ligand–Protein Database LPDB (28), the Protein–Ligand Database PLD (29), PDBbind (20,21), and—published shortly after first submission of this paper—Binding MOAD (22). LPDB is a compilation of 262 PDB complexes with affinity data. Since it also provides scoring values, docked ligand poses (‘decoys’) and ligand files setup for docking, LPDB is primarily intended to serve as a dataset for testing and developing docking and scoring methods. It does neither provide details nor references for the affinity values. The same is true for PLD, which contains 485 complexes and experimental binding energies for 344 of them. PLD can be searched by using a variety of single search criteria, but no combined search queries are possible. Similar to AffinDB it is freely accessible to anybody over the Internet, whereas PDBbind and Binding MOAD require a registration before granting academic users a free login account. The latter two databases offer by far the largest amount of affinity values, both covering well beyond 1700 complexes in their latest updates. Details about the affinity measurement and experimental conditions, however, are not included, which is an information provided by AffinDB for data retrieved from primary references. In summary, although there is certainly some overlap among the structure-affinity databases recently arosen from independent efforts, there are clear differences in focus, design and content, rendering each database on its own and in mutual combination an indispensable tool for the scientific community as long as affinities are not reported by the PDB and/or no common repository for biomolecular affinity data exists.
AffinDB encourages users to contribute data and submit references to papers with affinity data for PDB complexes. After registering for upload, an input form can be accessed which facilitates the submission of all relevant data in a clear format. Data submitted by users do not directly enter the database, but must first undergo revision by the database curators. This should ensure high fidelity of the affinity data collected from literature and reported by AffinDB.
Acknowledgments
The help of the following project co-workers and students of pharmacy in collecting and processing literature data is gratefully acknowledged: Kathrin Austrup, Petra Cordes, Julia Engel, Silvia Funke, Steffen Hoefft, Stefanie Meseth, Birte Roessing, Sebastian Vollmer. The database has been awarded with the first prize of the web award 2005 of the Molecular Graphics and Modelling Society, German Section (May 2005, Erlangen, Germany). Funding to pay the Open Access publication charges for this article was provided by the Scoring Function Consortium.
Conflict of interest statement. None declared.
REFERENCES
- 1.Klebe G., Bohm H.J. What can we learn from molecular recognition in protein–ligand complexes for the design of new drugs? Angew Chem. Int. Ed. Engl. 1996;35:2588–2614. [Google Scholar]
- 2.Babine R., Bender S. Molecular recognition of protein–ligand complexes: applications to drug design. Chem. Rev. 1997;97:1359–1472. doi: 10.1021/cr960370z. [DOI] [PubMed] [Google Scholar]
- 3.Mueller M.M., Sperl S., Sturzebecher J., Bode W., Moroder L. (R)-3-Amidinophenylalanine-derived inhibitors of factor Xa with a novel active-site binding mode. Biol. Chem. 2002;383:1185–1191. doi: 10.1515/BC.2002.130. [DOI] [PubMed] [Google Scholar]
- 4.Lange U.E.W., Baucke D., Hornberger W., Mack H., Seitz W., Hoffken H.W. D-Phe-Pro-Arg type thrombin inhibitors: unexpected selectivity by modification of the P1 moiety. Bioorg. Med. Chem. Lett. 2003;13:2029–2033. doi: 10.1016/s0960-894x(03)00347-0. [DOI] [PubMed] [Google Scholar]
- 5.Brenk R., Naerum L., Gradler U., Gerber H., Garcia G.A., Reuter K., Stubbs M.T., Klebe G. Virtual screening for submicromolar leads of tRNA-guanine transglycosylase based on a new unexpected binding mode detected by crystal structure analysis. J. Med. Chem. 2003;46:1133–1143. doi: 10.1021/jm0209937. [DOI] [PubMed] [Google Scholar]
- 6.Specker E., Bottcher J., Lilie H., Heine A., Schoop A., Muller G., Griebenow N., Klebe G. An old target revisited: two new privileged skeletons and an unexpected binding mode for HIV-protease inhibitors. Angew Chem. Int. Ed. Engl. 2005;44:3140–3144. doi: 10.1002/anie.200462643. [DOI] [PubMed] [Google Scholar]
- 7.Wang R., Liu L., Lai L., Tang Y. SCORE: a new empirical method for estimating the binding affinity of a protein–ligand complex. J. Mol. Model. 1988;4:379–394. [Google Scholar]
- 8.Wang R., Lai L., Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J. Comput. Aided. Mol. Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- 9.Bohm H.J. The development of a simple empirical scoring function to estimate the binding constant for a protein–ligand complex of known three-dimensional structure. J. Comput. Aided. Mol. Des. 1994;8:243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- 10.Bohm H.J. Prediction of binding constants of protein ligands: a fast method for the prioritization of hits obtained from de novo design or 3D database search programs. J. Comput. Aided. Mol. Des. 1998;12:309–323. doi: 10.1023/a:1007999920146. [DOI] [PubMed] [Google Scholar]
- 11.Head R.D., Smythe M.L., Oprea T.I., Waller C.L., Green S.M., Marshall G.R. VALIDATE: a new method for the receptor-based prediction of binding affinities of novel ligands. J. Am. Chem. Soc. 1996;118:3959–3969. [Google Scholar]
- 12.Eldridge M.D., Murray C.W., Auton T.R., Paolini G.V., Mee R.P. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J. Comput. Aided. Mol. Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 13.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunther J., Bergner A., Hendlich M., Klebe G. Utilising structural knowledge in drug design strategies: applications using Relibase. J. Mol. Biol. 2003;326:621–636. doi: 10.1016/s0022-2836(02)01409-2. [DOI] [PubMed] [Google Scholar]
- 15.Hendlich M., Bergner A., Gunther J., Klebe G. Relibase: design and development of a database for comprehensive analysis of protein–ligand interactions. J. Mol. Biol. 2003;326:607–620. doi: 10.1016/s0022-2836(02)01408-0. [DOI] [PubMed] [Google Scholar]
- 16.Laskowski R.A., Hutchinson E.G., Michie A.D., Wallace A.C., Jones M.L., Thornton J.M. PDBsum: a Web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci. 1997;22:488–490. doi: 10.1016/s0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
- 17.Laskowski R.A., Chistyakov V.V., Thornton J.M. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005;33:D266–D268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X., Liu M., Gilson M.K. BindingDB: a web-accessible molecular recognition database. Comb. Chem. High Throughput Screen. 2001;4:719–725. doi: 10.2174/1386207013330670. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J., Aizawa M., Amari S., Iwasawa Y., Nakano T., Nakata K. Development of KiBank, a database supporting structure-based drug design. Comput. Biol. Chem. 2004;28:401–407. doi: 10.1016/j.compbiolchem.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Wang R., Fang X., Lu Y., Yang C.Y., Wang S. The PDBbind Database: Methodologies and Updates. J. Med. Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 21.Wang R., Fang X., Lu Y., Wang S. The PDBbind database: collection of binding affinities for protein–ligand complexes with known three-dimensional structures. J. Med. Chem. 2004;47:2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 22.Hu L., Benson M.L., Smith R.D., Lerner M.G., Carlson H.A. Binding MOAD (mother of all databases) Proteins. 2005;60:333–340. doi: 10.1002/prot.20512. [DOI] [PubMed] [Google Scholar]
- 23.Boutselakis H., Dimitropoulos D., Fillon J., Golovin A., Henrick K., Hussain A., Ionides J., John M., Keller P.A., Krissinel E., et al. E-MSD: the European Bioinformatics Institute Macromolecular Structure Database. Nucleic Acids Res. 2003;31:458–462. doi: 10.1093/nar/gkg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 25.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31–36. [Google Scholar]
- 26.Gasteiger J., Rudolph C., Sadowski J. Automatic generation of 3D-atomic coordinates for organic molecules. Tetrahedron Comput. Meth. 1990;3:537–547. [Google Scholar]
- 27.Nissink J.W.M., Murray C., Hartshorn M., Verdonk M.L., Cole J.C., Taylor R. A new test set for validating predictions of protein–ligand interaction. Proteins. 2002;49:457–471. doi: 10.1002/prot.10232. [DOI] [PubMed] [Google Scholar]
- 28.Roche O., Kiyama R., Brooks C.L., III Ligand-protein database: linking protein–ligand complex structures to binding data. J. Med. Chem. 2001;44:3592–3598. doi: 10.1021/jm000467k. [DOI] [PubMed] [Google Scholar]
- 29.Puvanendrampillai D., Mitchell J.B.O. L/D protein ligand database (PLD): additional understanding of the nature and specificity of protein–ligand complexes. Bioinformatics. 2003;19:1856–1857. doi: 10.1093/bioinformatics/btg243. [DOI] [PubMed] [Google Scholar]


