Abstract
pSTIING (http://pstiing.licr.org) is a new publicly accessible web-based application and knowledgebase featuring 65 228 distinct molecular associations (comprising protein–protein, protein–lipid, protein–small molecule interactions and transcriptional regulatory associations), ligand–receptor–cell type information and signal transduction modules. It has a particular major focus on regulatory networks relevant to chronic inflammation, cell migration and cancer. The web application and interface provide graphical representations of networks allowing users to combine and extend transcriptional regulatory and signalling modules, infer molecular interactions across species and explore networks via protein domains/motifs, gene ontology annotations and human diseases. pSTIING also supports the direct cross-correlation of experimental results with interaction information in the knowledgebase via the CLADIST tool associated with pSTIING, which currently analyses and clusters gene expression, proteomic and phenotypic datasets. This allows the contextual projection of co-expression patterns onto prior network information, facilitating the identification of functional modules in physiologically relevant systems.
INTRODUCTION
The advent of high-throughput genomic and proteomic approaches coupled with annotation efforts has generated a comprehensive ‘parts list’ of biomolecules encoded by the genome, resulting in the establishment of a number of publicly available repositories and databases, each focussing on a certain specialized aspect of biological data collection, ranging from gene information [e.g. UniGene (1)], protein entries [e.g. UniProt (2)] and structural domain information [e.g. Pfam (3), InterPro (4) and CATH (5)] to molecular interactions [e.g. MINT (6) and BIND (7)] and pathways [e.g. BioCarta (www.biocarta.com) and KEGG (8)].
The extraction of meaningful information in the context of physiological systems requires characterization of not only the function of individual genes or proteins but also the physical interactions and functional associations in which they participate. To this end, recent genome-wide interactome projects involving two-hybrid (9), co-immunoprecipitation, protein chip analyses (10), affinity purification (11,12) and mass spectrometry (13) have produced a wealth of protein–protein interaction data.
Cellular and physiological processes are complex systems (14) modulated by signals from the extracellular environment and coordinated by the interplay of intracellular regulatory networks assembled into functional modules (15) and pathways. Regulatory networks comprise molecular (protein–protein, protein–lipid, protein–small molecule) interactions and transcriptional regulatory associations between transcription factors and the protein products of genes they trans-activate.
In order to understand these processes as interconnected and interdependent systems, there is a need to derive new associations from not only molecular interaction data but also integrating this with transcriptional regulatory associations, signal transduction modules and other information with corroborating evidence from multiple diverse sources, which often entails bringing together heterogeneous data. This is necessary to facilitate effective cross-correlation with expression and functional analyses in physiologically relevant investigations.
In light of this, we developed Protein, Signalling, Transcriptional Interactions and Inflammation Networks Gateway (pSTIING), a publicly accessible knowledgebase and web application to facilitate an integrative ‘systems’ approach towards understanding inflammation, cell migration and cancer. The knowledgebase integrates protein–protein, protein–lipid, protein–small molecule, ligand–receptor interactions, receptor–cell type associations, disease information and transcriptional associations with signal transduction modules. Experimental data from gene expression or proteomic analyses can also be directly cross-correlated with network information in pSTIING via CLADIST, a clustering and data analysis tool associated with pSTIING. pSTIING-CLADIST is accessible at http://pstiing.licr.org.
DATA SOURCES AND DATABASE ACCESS
pSTIING contains data manually curated from the literature and publicly available data derived from multiple sources as listed under six broad categories in Table 1. As pSTIING is growing rapidly, we expect the numbers and data sources listed in Table 1 to change very quickly. Because our current strategy for building pSTIING is to adopt a selective approach rather than taking the ‘all-inclusive’ route towards data integration, we highlight in Table 1 the various subsets of data that were included or not incorporated into pSTIING as it stands currently.
Table 1.
Data sources
| Categories | Data sources | Data incorporated into pSTIING |
|---|---|---|
| General protein and gene-related information | UniProt (2) | 180 823 entries from key organismsa and S.pombe. |
| UniRef90 (2) | 111 850 entries from key organismsa and S.pombe. | |
| UniRef50 (2) | 74 650 entries from key organismsa and S.pombe. | |
| InterPro (4) | 7309 entries. | |
| Entrez Gene (30) | 1 280 648 entries. | |
| Genew/HGND (31) | 23 914 entries. | |
| UniGene (1) | 53 256 clusters. | |
| GOA (18) | 19 141 entries. | |
| Orthologous groups | KOG/COG (19) | 4852 orthologous groups involving 26 477 proteins from H.sapiens, M.musculus, D.melanogaster, C.elegans. Entries lacking SwissProt/TrEMBL identifiers are excluded. |
| Human disease | OMIM (20) | 3466 human disease entries. |
| Molecular interactions | MINT (6) | 47 403 interactions involving 17 774 interacting entities from key organismsa. |
| BIND (7) | 11 568 interactions involving 6161 interacting proteins from key organismsa. Self-interactors and entries lacking SwissProt/TrEMBL identifiers are excluded. Currently, non-protein–protein interactions are not included. Protein-small molecule interactions from BIND will be included soon. | |
| Reactome (32) | 11 452 interactions (‘reaction’ type) involving 520 interacting entities from H.sapiens. Self-interactors and indirect interactions excluded. Macromolecular complexes from Reactome will be included soon. | |
| HPRD (33) | 7011 interactions involving 5516 interacting entities from H.sapiens. Self-interactors or entries lacking SwissProt/TrEMBL identifiers and interactors ambiguously classified as ‘mammalian’ rather than from a clearly defined species are excluded. Macromolecular complexes from HPRD will be included soon. | |
| In-house curated set | 709 protein–protein [including ligand–receptor (34,35)], protein–small molecule, protein–lipid interactions manually curated from the literature. | |
| MIPS (36) | 461 interactions from H.sapiens, R.norvegicus, M.musculus. Self-interactors lacking SwissProt/TrEMBL identifiers and interactions with low-confidence MIPS score are excluded. | |
| Transcriptional and Signalling modules and pathways | In-house curated set | Currently 11 signalling modules (H.sapiens) involving 561 interactions manually curated from the literature. Other reference sources: BioCarta, KEGG, STKE, AfCS. |
| TRED (16) | 2200 experimentally verified transcriptional associations (H.sapiens) were incorporated, of which 568 have been placed into transcriptional modules in pSTIING. Associations determined by indirect evidence or prediction methods are excluded. | |
| KEGG (8) | 21 signalling pathways involving 1441 proteins (H.sapiens). | |
| BioCarta | 241 signalling pathways involving 1165 proteins (H.sapiens) from BioCarta (www.biocarta.com). | |
| Receptor, CD marker and Cell type information | In-house curated set HCDM/HLDA8 | 797 CD marker/receptor-cell type associations manually curated from the literature and PathologyOutlines (www.pathologyoutlines.com). 358 CD marker entries from HCDM (www.hlda8.org). |
aKey organisms: H.sapiens, R.norvegicus, M.musculus, D.melanogaster, C.elegans and S.cerevisiae.
For example, we incorporated 2200 experimentally verified transcriptional associations from TRED that have been ranked highly for promoter and binding quality (16). Lower ranked entries such as from computational predictions or those supported by indirect or unreliable evidence are not incorporated.
To facilitate efficient storage and retrieval of interaction data, we constructed a relational database in an SQL (Oracle) server. The model underlying the database structure at its core consists of modules, interactions and interacting entities. A module (e.g. IL6 signalling module or STAT3 transcriptional module) is a biologically or functionally relevant grouping of interactions or associations. For example, we are building a curated collection of signalling modules by not only providing curated information at the pathway level [as with BioCarta, KEGG (8), STKE (www.stke.org) and AfCS (www.signaling-gateway.org)] but also focussing on individual interactions and associations within signalling modules. The assembly of interactions into functionally relevant signalling modules is an on-going effort involving manual literature curation and also using BioCarta, KEGG, STKE and AfCS as general reference resources.
Interactions between interacting entities can be direct (e.g. physical, stimulatory or inhibitory) or indirect (e.g. transcriptional associations). Each interaction has a source (e.g. MINT, HPRD or our in-house curated set) and is supported by experiments which have Pubmed ID links. Currently, most interactions in pSTIING are binary but can also involve more than two interacting entities, which can be any small molecule (e.g. Ca2+) or biomolecules (including proteins and lipids). Small molecules and lipids are linked to KEGG compound identifiers. Proteins are linked to protein-related information [via UniProt (2) ID], protein domain information [via InterPro (4) accession], protein structure [via PDB (17) ID], gene ontology (GO) [via GOA (18) ID], orthology [via KOG (19) ID], human disease [via OMIM (20) ID] and taxonomy classification [via Taxonomy (1) ID]. Organisms represented in pSTIING include Homo sapiens (Taxonomy ID: 9606), Rattus norvegicus (10116), Mus musculus (10090), Drosophila melanogaster (7227), Caenorhabditis elegans (6239) and Saccharomyces cerevisiae (4932).
The pSTIING web interface provides a number of starting points from which to navigate the database (Supplementary Data 1). Users may perform single or multiple (batch) searches using protein or gene name, synonyms or identifiers (such as SwissProt or TrEMBL accessions) which retrieve information about interacting entities, interactions, transcriptional associations and pathway modules, with supporting experimental evidence and extensive cross-references to external databases (Supplementary Data 2). Users can also query across species, via GO annotations, InterPro domains, motifs, human diseases or using datasets from gene expression or proteomic experiments.
VISUALIZING INTERACTIONS, TRANSCRIPTIONAL ASSOCIATIONS AND EXTENDING PATHWAYS
pSTIING dynamically generates graphical representations of interaction networks and displays maps in both compressed and scalable vector graphics (SVG) formats. With a suitable SVG viewer plug-in for browsers such as Corel SVG or Adobe SVG viewers, users will be able to zoom in and out, or move (pan) a network map around within the viewing screen. Nodes represent interacting components (e.g. proteins, macromolecular assemblies, lipids or small molecules) and edges represent interaction or transcriptional regulatory relationships between components.
In signalling modules, these edges may be directed, denoting stimulatory or inhibitory interactions, or undirected, representing molecular interactions that are neither stimulatory nor inhibitory, or where their stimulatory/inhibitory status is not known. In pSTIING, the representation of a transcriptional association is abstracted to indicate the relationship between two proteins, one of which is a transcription factor that trans-activates the expression of the gene encoding the other protein. Such associations are represented by dashed edges. Depending on the respective display mode selected, edges are colour-coded to highlight a specific module or a cellular compartment, within which an interaction occurs, or to indicate the number of documented experimental types supporting an interaction.
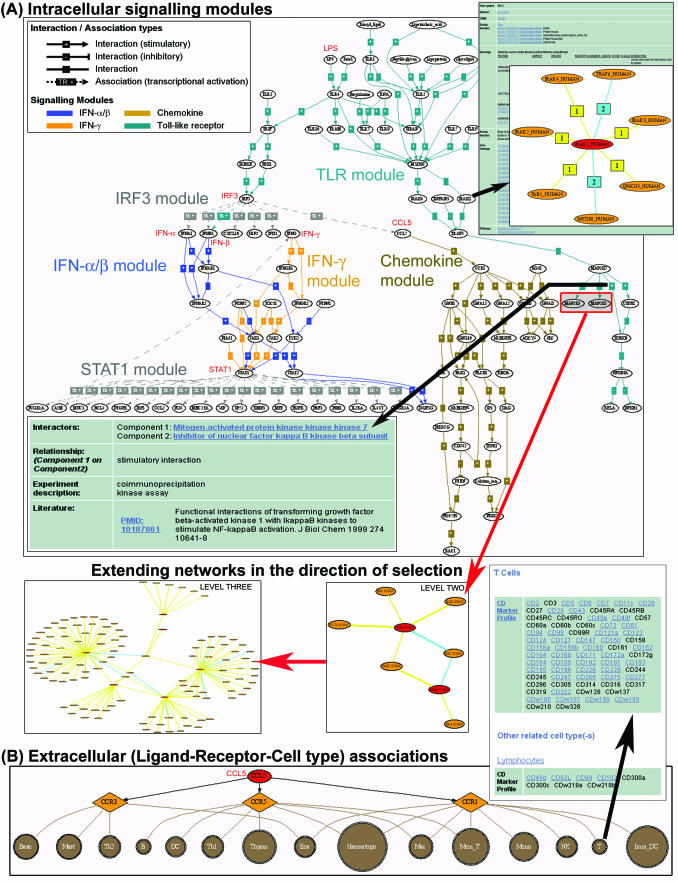
Interaction maps can also be progressively expanded outwards to display the next interaction neighbourhood or subsequent levels beyond, thus, making it possible to ‘grow’ networks or extend signalling pathway modules in a desired direction. For example, by selecting mitogen-activated protein kinase kinase 3 (MAP2K3) and MAP2K6 from a list of interactors in the Toll-like receptor (TLR) signalling module, the network can be extended to display subsequent interaction levels with MAP2K3 and MAP2K6 at the centre of the interaction network (Figure 1A). By clicking the ‘Display Network (‘Backbone’)’ button, it is possible to display only the query nodes and their immediate interaction partners that link query nodes together. This feature is useful for simplifying complex network maps.
Figure 1.
Graphical maps generated in pSTIING showing (A) cross-connectivity between four intracellular signalling modules (TLR, IFN-α/β, IFN-γ and Chemokine) and two transcriptional regulatory modules (STAT1 and IRF3). Interactions are represented by solid edges while transcriptional associations by dashed lines. Each component node (e.g. IRAK1) is linked to more extensive information about the component (including interaction partners, InterPro domains, GO, homologous proteins and cross-links to external databases). Clicking on an interaction node provides interaction details and experimental evidence supporting that interaction. Pathways can be extended in a desired direction by displaying subsequent interaction neighbourhood levels centered on interactors selected by users (e.g. mitogen-activated kinase kinases, MAP2K3 and MAP2K6). (B) Representation of ligand–receptor–cell type associations. The ligand CC-chemokine CCL5 activates a subset of leukocytes and lymphocytes by binding to chemokine receptors CCR1, CCR3 and CCR5 expressed on these cell types. Each ligand and receptor node is linked to detailed information about the respective protein and their interaction partners. Information about each cell type, its related or parental cell types and the cell surface markers (CD antigens) that phenotypically identifies the cell type can be accessed by clicking on the cell type node.
LINKING TRANSCRIPTIONAL ASSOCIATIONS TO SIGNALLING MODULES
Regulatory networks comprise both transcriptional regulatory modules and molecular interaction cascades organized within signal transduction modules. Therefore, the incorporation of transcriptional regulatory associations with molecular interaction information is necessary to connect seemingly distinct signalling modules into functional regulatory systems. In pSTIING, multiple signalling and transcriptional regulatory modules can be displayed in combination, facilitating the identification of important ‘cross-talk’ points (or ‘hubs’) between modules.
As an example, Figure 1A illustrates this cross-module connectivity between TLR, interferon (IFN) α/β, IFN-γ and chemokine signalling modules participating in an inflammatory response against gram-negative bacteria infection. Bacterial lipopolysaccharide interacts with TLR4 and triggers a cascade of signalling events in the TLR pathway, leading to activation of the transcription factor, IRF3. IRF3 binds to its cis element in the promoter of genes encoding type I IFNs (IFN-α and IFN-β) and C–C chemokine, CCL5, inducing their expression. The type I IFN signalling cascade induces the production and secretion of type II IFNs (IFN-γ) via the transcription factor STAT1. IFN-γ triggers a second wave of STAT1 activation which reinforces the first in a positive feed-back loop. Also, CCL5 promiscuously activates three G-protein-coupled receptors CCR1, CCR3 and CCR5 which are expressed on a variety of immunologically relevant cell types, thus transmitting extracellular cues to appropriate effector cells as represented in pSTIING (Figure 1B and Supplementary Data 3). Clicking on a ‘cell type’ node retrieves information about cell surface (CD or Cluster of Differentiation) markers that phenotypically identify this cell type.
PROJECTING AND MAPPING INTERACTIONS ACROSS SPECIES
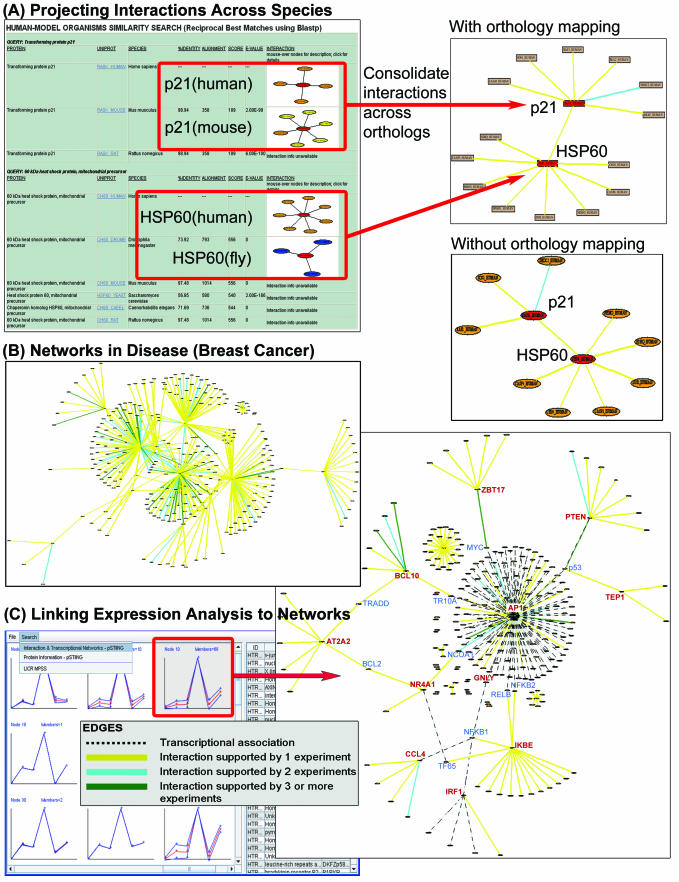
The integration of homology information with molecular interaction data allows us to project and map interactions known for a protein onto homologous (and where possible, orthologous) proteins across species in which the interaction information is not as extensive or not available (Figure 2A). Our aim is to obtain as much additional insights as it is possible about the human interactome by inferring from model organisms.
Figure 2.
(A) The identification of potential orthologues using human-model organism reciprocal similarity (BLASTP) searches allows additional interaction information to be projected from model organism proteins onto orthologous proteins in human. The sequence similarity between orthologous proteins (percent identity, BLAST score and E-value) for each query protein is displayed alongside their interaction information and when consolidated across species, these interactions are mapped onto human proteins. As a comparison, the corresponding interaction map without orthology mapping is shown. (B) pSTIING supports querying by human disease, facilitating the exploration of interaction and transcriptional regulatory associations between proteins implicated in disease (e.g. breast cancer). (C) Linking of expression analysis to molecular interaction networks (represented by solid edges) and transcriptional regulatory modules (dashed lines) using pSTIING-CLADIST. The CLADIST tool associated with pSTIING provides various clustering algorithms including SOM, which was used to identify a cluster of co-ordinately expressed genes (represented by nodes with red labels) from MPSS expression analyses of tumour and non-tumour samples. These were then cross-correlated directly with interaction and transcriptional information in pSTIING to detect any underlying physical or functional associations.
pSTIING provides three approaches whereby users can choose to query interaction information across species. The first uses orthology groupings from Clusters of Orthologous Groups (COG/KOG) (19). The second approach involves UniRef 50/90 (2) clusters of proteins with at least 50 or 90% identity, respectively. The third employs a reciprocal best hit approach to map protein interaction information (where available) in model organisms (rat, mouse, fly, worm and yeast) onto human orthologues. Sequence similarity (BLASTP) searches against human proteins were performed for each model organism protein, and reciprocal searches carried out for each top BLAST hit. Each reciprocal best match is selected as a potential orthologue and its interactions displayed graphically. Also shown is the extent of sequence similarity (percent identity, BLAST score and E-value) between orthologous proteins, which provides an indication of the level of confidence associated with the inferred or projected interactions (Figure 2A and Supplementary Data 4).
INTERACTION NETWORKS VIA ONTOLOGY, MOTIFS AND INTERPRO DOMAINS
GO annotation (18) provides a well-structured set of controlled common vocabularies to describe the molecular function of gene products and proteins, the biological processes with which they are associated and the cellular compartments in which they are located. The inclusion of GO annotation terms and category identifiers into pSTIING facilitates the construction of interaction networks associated with a chosen GO annotation.
For example, a search using a phrase for a biological process like ‘regulation of apoptosis’, returns several possible matching terms, including ‘positive regulation of apoptosis’ (GO:0043065) and ‘negative regulation of apoptosis’ (GO:0043066) from which users could refine and select the appropriate term (Supplementary Data 5). There are several display options available. The ‘protein’ option generates graphical maps of interactions between proteins associated with the selected GO term. As for ‘UniRef 50/90’ and ‘COG/KOG’ options, the GO search is combined with cross-species homology information, producing graphical representations of interactions between UniRef or COG/KOG clusters, respectively.
It is also possible to explore interaction networks of proteins with similar InterPro domains or motifs. pSTIING supports searches by InterPro accession, InterPro name or regular expression specifying conserved motifs (Supplementary Data 6).
HUMAN DISEASE AND INTERACTION NETWORKS
Many human diseases including inflammatory and autoimmune disorders (21–23), atherosclerosis (24) and cancers (25,26) have complex aetiologies associated with a variety of environmental cues feeding into a number of interaction networks or modules leading to aberrant perturbations at the ‘systems’ level.
A list of diseases was selected from the OMIM (20) morbid map initiative, and the information about proteins whose de-regulation or mutation have association or causal implication in disease was integrated with interaction information in pSTIING. For example, searching ‘colorectal cancer’ or ‘breast cancer’ will return information and OMIM links about the disorder (such as clinical synopses) and associated chromosomal loci. pSTIING will graphically represent interaction networks of proteins implicated in the disease (Figure 2B).
LINKING GENE EXPRESSION AND PROTEOMIC EXPERIMENTS TO INTERACTION AND REGULATORY NETWORKS
Filtered and normalized datasets from gene expression [including microarrays and massively parallel signature sequencing (MPSS)] and proteomic experiments can be imported into pSTIING via CLADIST, a clustering tool associated with pSTIING. pSTIING-CLADIST features various clustering algorithms including hierarchical clustering, k-means and self-organizing maps (SOM) from which users could select the clusters of co-regulated genes or proteins and directly cross-correlate these with molecular interaction and transcriptional information in pSTIING.
This feature allows the contextual projection of co-expression patterns onto prior network information, facilitating the assembly of protein interaction and transcriptional regulatory networks into functional modules. For example, Figure 2C shows the identification of a set of genes that are co-ordinately expressed across tumour and non-tumour cells. When linked to information in pSTIING, preliminary data reveals an underlying network of protein–protein interactions and transcriptional regulatory associations, suggesting their involvement in the same functional module or programme within the context of the physiological state examined.
FUTURE DEVELOPMENTS
Building on molecular interaction data provided by publicly available databases [including MINT, MIPS, BIND, IntAct (27), STRING (28) and DIP (29)], pSTIING offers a unique resource in that it integrates transcriptional regulatory associations with molecular interaction data and signalling modules, allowing researchers and students to explore and extend regulatory networks using experimental data or by searching on protein domains/motifs, GO annotations, human diseases and orthologous proteins across species.
We will continue to assemble molecular interaction and transcriptional regulatory networks into functional pathways, particularly those that underpin inflammatory processes, cell migration and tumourigenesis. Hence, we welcome and encourage contributions from investigators towards this open resource via the pSTIING website.
We will also expand the knowledgebase to include quantitative data from directed expression and functional studies to examine the temporo-spatial dynamics of regulatory networks as determined by cell type, the state of a cell, its microenvironment and extracellular cues, in the context of a specific physiological process or pathological condition. This is critical for the development of realistic and predictive quantitative models of biological systems, which we envisage pSTIING to facilitate in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors wish to thank Anne Ridley and Buzz Baum for useful discussions and critical reading of the manuscript. pSTIING is a project within the MAIN-NOE (Network of Excellence for Cell migration in Chronic Inflammation) consortium supported and funded by the EU 6th Framework Programme (EU project no. FP6-502935). Funding to pay the Open Access publication charges for this article was provided by the Ludwig Institute for Cancer Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Church D.M., DiCuccio M., Edgar R., Federhen S., Helmberg W., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearl F., Todd A., Sillitoe I., Dibley M., Redfern O., Lewis T., Bennett C., Marsden R., Grant A., Lee D., et al. The CATH Domain Structure Database and related resources Gene3D and DHS provide comprehensive domain family information for genome analysis. Nucleic Acids Res. 2005;33:D247–D251. doi: 10.1093/nar/gki024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 7.Alfarano C., Andrade C.E., Anthony K., Bahroos N., Bajec M., Bantoft K., Betel D., Bobechko B., Boutilier K., Burgess E., et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S., Armstrong C.M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., et al. A map of the interactome network of the metazoan C.elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 11.Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 12.Bouwmeester T., Bauch A., Ruffner H., Angrand P.O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nature Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 13.Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 14.Kitano H. Computational systems biology. Nature. 2002;420:206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 15.Hartwell L.H., Hopfield J.J., Leibler S., Murray A.W. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 16.Zhao F., Xuan Z., Liu L., Zhang M.Q. TRED: a Transcriptional Regulatory Element Database and a platform for in silico gene regulation studies. Nucleic Acids Res. 2005;33:D103–D107. doi: 10.1093/nar/gki004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camon E., Magrane M., Barrell D., Lee V., Dimmer E., Maslen J., Binns D., Harte N., Lopez R., Apweiler R. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with gene ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ollier W.E. Cytokine genes and disease susceptibility. Cytokine. 2004;28:174–178. doi: 10.1016/j.cyto.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 22.Aderem A., Smith K.D. A systems approach to dissecting immunity and inflammation. Semin. Immunol. 2004;16:55–67. doi: 10.1016/j.smim.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Lehner B., Semple J.I., Brown S.E., Counsell D., Campbell R.D., Sanderson C.M. Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics. 2004;83:153–167. doi: 10.1016/s0888-7543(03)00235-0. [DOI] [PubMed] [Google Scholar]
- 24.King J.Y., Ferrara R., Tabibiazar R., Spin J.M., Chen M.M., Kuchinsky A., Vailaya A., Kincaid R., Tsalenko A., Deng D.X., et al. Pathway analysis of coronary atherosclerosis. Physiol Genomics. 2005;23:103–118. doi: 10.1152/physiolgenomics.00101.2005. [DOI] [PubMed] [Google Scholar]
- 25.Lin B., White J.T., Lu W., Xie T., Utleg A.G., Yan X., Yi E.C., Shannon P., Khrebtukova I., Lange P.H., et al. Evidence for the presence of disease-perturbed networks in prostate cancer cells by genomic and proteomic analyses: a systems approach to disease. Cancer Res. 2005;65:3081–3091. doi: 10.1158/0008-5472.CAN-04-3218. [DOI] [PubMed] [Google Scholar]
- 26.Khalil I.G., Hill C. Systems biology for cancer. Curr. Opin. Oncol. 2005;17:44–48. doi: 10.1097/01.cco.0000150951.38222.16. [DOI] [PubMed] [Google Scholar]
- 27.Hermjakob H., Montecchi-Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S., Vingron M., Roechert B., Roepstorff P., Valencia A., et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Mering C., Jensen L.J., Snel B., Hooper S.D., Krupp M., Foglierini M., Jouffre N., Huynen M.A., Bork P. STRING: known and predicted protein–protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wain H.M., Lush M.J., Ducluzeau F., Khodiyar V.K., Povey S. Genew: the Human Gene Nomenclature Database, 2004 updates. Nucleic Acids Res. 2004;32:D255–D257. doi: 10.1093/nar/gkh072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peri S., Navarro J.D., Kristiansen T.Z., Amanchy R., Surendranath V., Muthusamy B., Gandhi T.K., Chandrika K.N., Deshpande N., Suresh S., et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:D497–D501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fitzgerald K.A., O'Neill L.A.J., Gearing A.J.H., Callard R.E. The Cytokine Factsbook, 2nd edn. San Diego: Academic Press; 2001. [Google Scholar]
- 35.Isacke C.M., Horton M.A. The Adhesion Molecule Factsbook, 2nd edn. San Diego: Academic Press; 2000. [Google Scholar]
- 36.Pagel P., Kovac S., Oesterheld M., Brauner B., Dunger-Kaltenbach I., Frishman G., Montrone C., Mark P., Stumpflen V., Mewes H.W., et al. The MIPS mammalian protein–protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.